- *Corresponding Author:
- Wei Zhang
Department of Oncology,
Traditional Chinese Medicine Hospital of China Three Gorges University,
Yichang Hospital of Traditional Chinese Medicine,
Yichang,
Hubei Province 443000,
China
E-mail: a13377664449@126.com
| This article was originally published in a special issue, “Novel Therapeutic Approaches in Biomedicine and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2021:83(6) Spl Issue “247-254” |
Abstract
Bromodomain and extra-terminal domain inhibitors target bromodomain-containing proteins and are currently being evaluated as anti-cancer agents. In this study, we found that bromodomain and extraterminal domain inhibitor OTX015 can effectively inhibit the proliferation of hepatocellular carcinoma cells and promote the apoptosis of hepatocellular carcinoma cells. At the same time, the anticancer effect of OTX015 is better than that of JQ1, a similar bromodomain and extra-terminal domain inhibitor. In addition, we also found that OTX015, like JQ1 can inhibit the expression of programmed death-ligand 1 in hepatocellular carcinoma cells induced by interferon alpha. We also demonstrated that this inhibition was achieved by inhibiting the binding of bromodomain and extra-terminal domain protein to the promoter of programmed death-ligand 1 and inhibiting the transcriptional activity of programmed death-ligand 1. When treated hepatocellular carcinoma cells with bromodomain and extra-terminal domain inhibitor, peripheral blood mononuclear cells can kill hepatocellular carcinoma cells. Finally, we also detected the expression levels of bromodomain and extra-terminal domain protein, bromodomain-containing protein 2, bromodomain-containing protein 4 and programmed death-ligand 1 in patient samples. The results showed that the expression levels of bromodomain-containing protein 2 and bromodomain-containing protein 4 were correlated with that of programmed death-ligand 1 in some patients. Therefore, bromodomain and extraterminal domain protein inhibitor OTX015 may be a small molecule drug with dual anti-tumor effects.
Keywords
Bromodomain-containing protein 2, bromodomain-containing protein 4, OTX015, programmed death-ligand 1, hepatocellular carcinoma
Many small molecule inhibitors of chromatin-related enzymes or binding proteins cannot only play an antitumor role by promoting apoptosis, but also promote the anti-tumor activity of host immune cells by improving the activity of host immune cells[1-3]. In autoimmune function, immune checkpoints play an important role[4]. There are active and inhibitory immuno-checkpoint molecules (CD40L, CD137/4-1BB or CTLA4, CD279/ Programmed Cell Death-1 (PD-1)) on the surface of immune cells. These immuno-checkpoints can bind with ligand/receptor molecules on the surface of presenting cells or tumor cells to regulate the immune activity of immune cells[4-8]. In the microenvironment of tumors, immune cells may be depleted, which is mainly due to the high expression of Programmed Death-Ligand 1 (PD-L1) in tumour cells, leading to PD with immune cells[9]. Therefore, by blocking the interaction between PD-1 and PD-L1, monoclonal antibody drugs that can promote immune cells to kill cancer cells have become a research and development hotspot in recent years[10,11].
The Bromodomain and Extra-Terminal Domain (BET) family of epigenetic “reader” proteins bind acetylated histone lysine residues to facilitate the recruitment of transcriptional elongation complexes such as Positive Transcription Elongation Factor b (P-TEFb) [12]. Bromodomain-Containing Protein 2 (BRD2) and Bromodomain-Containing Protein 4 (BRD4) are associated with active promoters and enhancers and loading of BRD4 onto “super-enhancers” drives oncogenic transcription programs in lymphoma, particularly where immunoglobulin gene switch translocations are juxtaposed to Myc[13]. Therefore, researchers have developed small molecular inhibitors specifically targeting BET proteins, such as JQ1 and OTX015. And researchers have proved that JQ1 and OTX015 can not only inhibit the proliferation of lymphoma cells and breast cancer cells, but also promote the apoptosis of cancer cells[14,15]. More importantly, in recent years, researchers have also found that JQ1 can promote the immune system to kill tumor cells by inhibiting the expression of PD-L1 in tumor cells[16].
Here, we demonstrated that OTX015, a BET inhibitor can inhibit the proliferation of hepatocellular carcinoma cells and promote the apoptosis of hepatocellular carcinoma cells through pharmacological effection, which is time-dependent and dose-dependent. At the same time, we also proved that the effect of OTX015 is superior to that of JQ1, an inhibitor of the same type. In addition, we also demonstrated for the first time that OTX015 can inhibit the expression of PD-L1 induced by Interferon Alpha (IFN-α) at the transcriptional level in hepatocellular carcinoma cells. Because OTX015 is a BET inhibitor, we demonstrated that BRD2 and BRD4 can bind to the promoter of PD-L1 through Chromatin Immunoprecipitation (ChIP) experiments, thus promoting the transcription of PD-L1. On the other hand, we also found that OTX015 can not only kill hepatocellular carcinoma cells through pharmacological action, but also promote the killing effect of Peripheral Blood Mononuclear Cells (PBMCs) and both of them have synergistic effect. Finally, we detected the expression levels of BRD2, BRD4 and PD-L1 in patient samples. The results showed that there was a positive correlation between the expression of BRD2 and BRD4 and the expression of PD-L1 in some patients.
Materials and Methods
Cell culture:
The Hepatoma G2 (HepG2) cells were cultured using Dulbecco's Modified Eagle Medium (DMEM) supplemented with 20 % of fetal bovine serum, 1 % sodium pyruvate (100 mM), 1 % non-essential amino acids (10 mM) and 1 % penicillin/streptomycin solution (Invitrogen, Carlsbad, CA). Cells were maintained at 37° at 5 % CO2 atmosphere. The cells were passaged every 5 d.
Western blot:
Cells were harvested, lased and subject to sodium dodecyl sulphate–polyacrylamide gel electrophoresis gels, which were prepared depending on the protein size. Electrophoresis and transmembrane were carried out on a protein electrophoresis and blotting system (Liuyi, Beijing, China), followed by the incubation with primary antibody. Membranes were visualized using the Immun-Star WesternC Chemiluminescence Kit (Liuyi, Beijing, China) and images were captured using a ChemiDoc XRS+ System and processed using Image Lab software (Liuyi, Beijing, China).
Real-time Polymerase Chain Reaction (PCR):
Complementary Deoxyribonucleic Acid (cDNA) templates were prepared after Ribonucleic Acid (RNA) extraction and reverse transcription. Amplification was performed on a real-time PCR system (Applied Biosystems 7500, USA). The whole procedure was according to the manual of SYBR® Premix Ex Taq™ kit (Takara RR420A, Japan). Relative expression was calculated using the formula of 1/2△Ct.
Chromatin immunoprecipitation and sequencing analysis:
HepG2 cells were cultured in the presence or absence of 1 μM OTX015 for 2 h. Cells were cross-linked with 1 % formaldehyde for 20 min at room temperature and quenched with the addition of 1.25 M glycine. Cells were washed three times (ice-cold Phosphate Buffered Saline (PBS), 1400 rpm, 5 min) and lysed in ChIP lysis buffer (20 mM Tris hydrochloride (Tris- HCl) [pH 8], 150 mM Sodium Chloride (NaCl), 2 mM Ethylenediaminetetraacetic Acid (EDTA) [pH 8], 1 % Nonidet P40 (NP40), 0.3 % sodium dodecyl sulfate in water (H2O)). Lysates were sonicated in a Covaris ultrasonicator to achieve a mean DNA fragment size of 300-500 bp. Immunoprecipitation was performed in ChIP dilution buffer (20 mM Tris-HCl [pH 8], 150 mM NaCl, 2 mM EDTA, 1 % Triton-X and protease inhibitors in H2O) for at least 12 h at 4°. The following antibodies were using for ChIP assays: Anti-Flag (ab128874, Abcam). An equal volume of protein A and G magnetic beads (Life Technologies) were used to bind the antibody and associated chromatin. Reverse crosslinking of DNA was performed by DNA purification using QIAquick PCR purification kits (QIAGEN).
Cell apoptosis detection:
The percentage of cells actively undergoing apoptosis was quantified by means of a combined staining with annexin and propidium iodide by using the Annexin V-FITC Apoptosis Detection Kit (Keygene, Nanjing, China) according to the manufacturer’s instructions. Briefly, according to results of cell counting kit- 8, following treatment with 15 μmol/l gefitinib and gefitinib derivative LPY-9, cells were harvested and washed with PBS twice. Then we added 5 μl Annexin V-FITC and 5 μl propidium iodide, after mixing for 5 min was incubated at room temperature and analyzed by using Magnetic Resonance Flow (MR FLOW) software.
Cell counting kit-8:
HepG2 cells (2×104 cells/well) were seeded in 96- well plates and incubated for 24 h at 37° followed by treating with different concentrations of gefitinib and LPY-9. Then, 10 μL of CCK-8 solution (Dojindo) was added to each well and incubated for 4 h. The samples were then detected at 450 nm by a microplate reader (Bio-Rad, Hercules, CA, USA) and analyzed using the mathematical formula [(Optical Density (OD) value of test-OD value of blank)/(OD value of control-OD value of blank)] to quantify the cell viability.
Statistical analysis:
Statistical analysis was performed using Statistical Package for the Social Sciences (SPSS) 17.0 software. The differences were evaluated by using two-ways analysis of variance or t-test. Error bars represent standard errors and three independent experiments were performed for each assay. Statistical significance was indicated at *p<0.05,**p<0.01 or ***p<0.001. All statistical analyses and comparisons were made between with control group. Histograms were drawn using GraphPad Prism 5.01 and every figure was combined using Photoshop C S6.
Results and Discussion
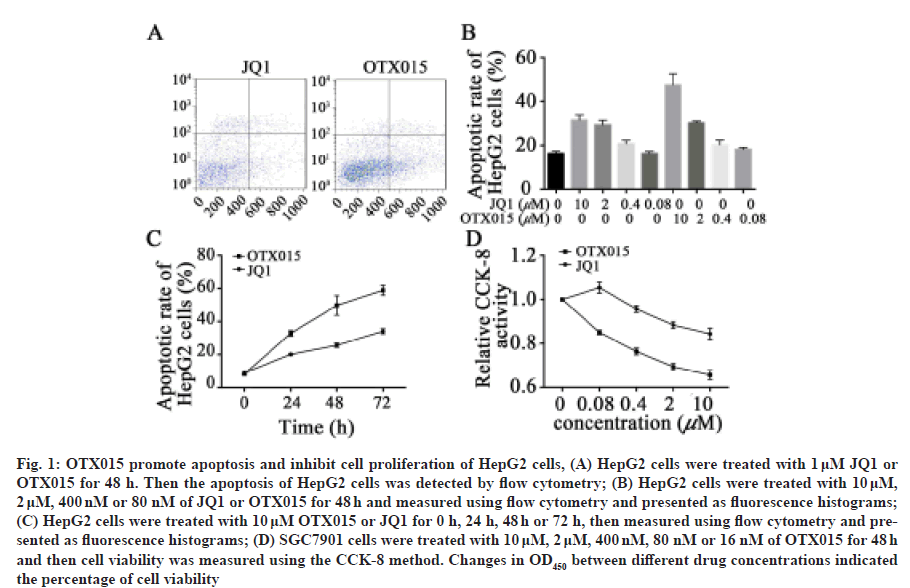
In order to detect whether OTX015 can induce apoptosis of hepatocellular carcinoma cells, we first carried out experiments on HepG2 cell line. After treatment with 1 μM OTX015 and JQ1 for 48 h, we found that 30.1 % and 29.9 % of HPG2 cells showed apoptotic phenotype (fig. 1A). Then, in order to further explore the timedose relationship between OTX015 and JQ1 on HepG2 apoptosis, we conducted a time- and dose-dependent experiment. The results showed that with the increase of OTX015 and JQ1 concentration, the apoptotic rate of HepG2 cells also increased synchronously (fig. 1B). In addition, over time the apoptotic effect of OTX015 and JQ1 on HepG2 cells increased gradually (fig. 1C). In addition to promoting apoptosis, previous studies have shown that BET inhibitors can inhibit the proliferation of cancer cells. Therefore, we further explored the effects of OTX015 and JQ1 on the proliferation of HepG2 cells through CCK8 assay. The results showed that both OTX015 and JQ1 could inhibit the proliferation of HepG2 cells and the effect of OTX015 was better than that of JQ1 (fig. 1D). These results indicate that both JQ1 and OTX015 have potential antineoplastic drugs through pharmacological effect.
Fig. 1: OTX015 promote apoptosis and inhibit cell proliferation of HepG2 cells, (A) HepG2 cells were treated with 1 μM JQ1 or OTX015 for 48 h. Then the apoptosis of HepG2 cells was detected by flow cytometry; (B) HepG2 cells were treated with 10 μM, 2 μM, 400 nM or 80 nM of JQ1 or OTX015 for 48 h and measured using flow cytometry and presented as fluorescence histograms; (C) HepG2 cells were treated with 10 μM OTX015 or JQ1 for 0 h, 24 h, 48 h or 72 h, then measured using flow cytometry and presented as fluorescence histograms; (D) SGC7901 cells were treated with 10 μM, 2 μM, 400 nM, 80 nM or 16 nM of OTX015 for 48 h and then cell viability was measured using the CCK-8 method. Changes in OD450 between different drug concentrations indicated the percentage of cell viability
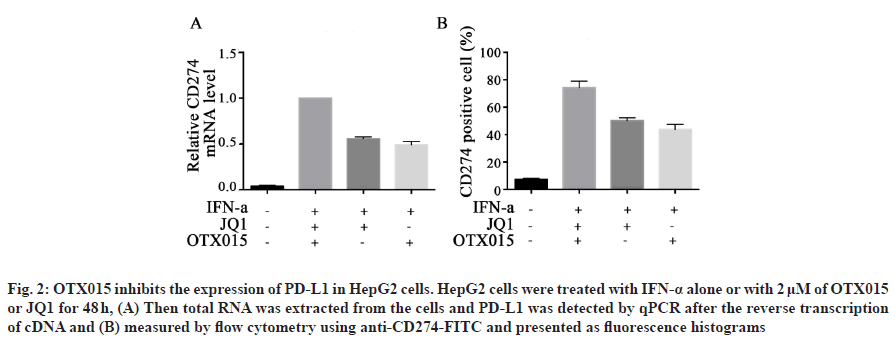
Under the effect of IFN-α, tumor cells escape the immune response by overexpressing PD-L1 in tumor microenvironment[17]. In previous studies, we have known that BET inhibitor JQ1 can inhibit the expression of PD-L1 in lymphoma[18]. Therefore, we want to further explore whether BET inhibitor OTX015 can inhibit the expression of PD-L1 in hepatocellular carcinoma cells. Firstly, HepG2 cells were treated with IFN-α and then different concentrations of JQ1 and OTX015 were added to detect the transcription level of PD-L1 in HepG2 cells. The results showed that the transcription level of PD-L1 in HepG2 cells increased significantly after IFN-α treatment, but decreased in different degrees after JQ1 and OTX015 treatment (fig. 2A). Then, we detected the expression of PD-L1 protein on HepG2 cells by flow cytometry. The results showed that the expression of PD-L1 on HepG2 cells increased significantly after treatment with IFN-α, but decreased significantly after treatment with OTX015 and JQ1 (fig. 2B). These results suggest that BET inhibitor OTX015 cannot only inhibit the transcription of PD-L1 in HepG2 cells induced by IFN-α at the transcriptional level, but also ultimately reach the protein level, thus inhibiting the immune escape of tumor cells.
Fig. 2: OTX015 inhibits the expression of PD-L1 in HepG2 cells. HepG2 cells were treated with IFN-α alone or with 2 μM of OTX015 or JQ1 for 48 h, (A) Then total RNA was extracted from the cells and PD-L1 was detected by qPCR after the reverse transcription of cDNA and (B) measured by flow cytometry using anti-CD274-FITC and presented as fluorescence histograms
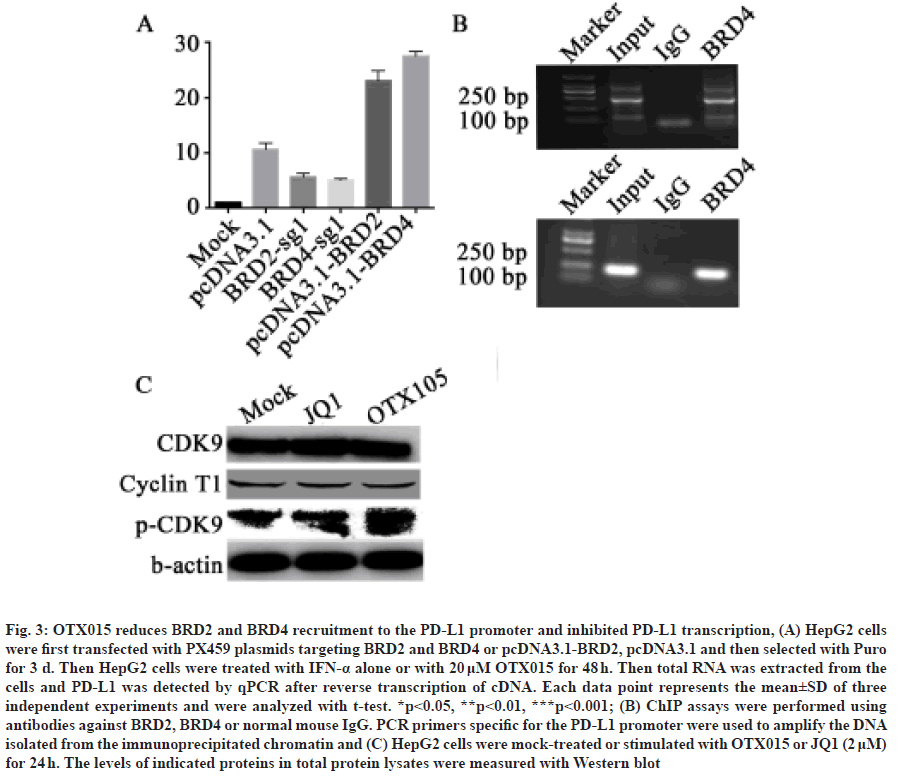
Because BET inhibitors compete with BET proteins such as BRD2 and BRD4 to bind to the promoter region of target genes, we speculate that OTX015 also acts on PD-L1 through BRD2 and BRD4. Therefore, in order to explore whether the transcriptional expression of PD-L1 is related to BRD2 and BRD4, we first constructed BRD2 and BRD4 overexpression plasmids and Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR)-based knockout plasmids. In HepG2 cells, we transfected BRD2 and BRD4 knockout and overexpression plasmids instantaneously and then treated the cells with IFN-α, respectively. The results showed that the expression of PD-L1 could be significantly increased by adding IFN-α, but knocking out BRD2 or BRD4 could inhibit the trend, while overexpression of BRD2 and BRD4 could increase the trend (fig. 3A).
Fig. 3: OTX015 reduces BRD2 and BRD4 recruitment to the PD-L1 promoter and inhibited PD-L1 transcription, (A) HepG2 cells were first transfected with PX459 plasmids targeting BRD2 and BRD4 or pcDNA3.1-BRD2, pcDNA3.1 and then selected with Puro for 3 d. Then HepG2 cells were treated with IFN-α alone or with 20 μM OTX015 for 48 h. Then total RNA was extracted from the cells and PD-L1 was detected by qPCR after reverse transcription of cDNA. Each data point represents the mean±SD of three independent experiments and were analyzed with t-test. *p<0.05, **p<0.01, ***p<0.001; (B) ChIP assays were performed using antibodies against BRD2, BRD4 or normal mouse IgG. PCR primers specific for the PD-L1 promoter were used to amplify the DNA isolated from the immunoprecipitated chromatin and (C) HepG2 cells were mock-treated or stimulated with OTX015 or JQ1 (2 μM) for 24 h. The levels of indicated proteins in total protein lysates were measured with Western blot
BET protein family is a protein that can bind to the promoter position of the gene. Therefore, after clarifying the expression of BRD2 and BRD4 on PD-L1, we want to further explore whether BRD2 and BRD4 can bind to the promoter of PD-L1. We transfected BRD2 and BRD4 plasmids into HepG2 cells instantaneously and then ChIP experiments were carried out using anti- Flag antibodies. After that, we used primers specific to the promoter of PD-L1 for PCR. The results showed that both BRD2 and BRD4 could bind to the promoter region of PD-L1 (fig. 3B). In addition, in order to further explore whether OTX015 acts by inhibiting the binding of BRD2/4 to the PD-L1 promoter, we used ChIP experiments to detect the changes in the binding level of BRD2/4 and PD-L1 promoter in cells before and after OTX015 treatment. It was found that after treating HepG2 cells with OTX015, quantitative PCR (qPCR) detection revealed that the level of BRD2/4 binding to the PD-L1 promoter was significantly reduced. This shows that OTX015 inhibits the transcription of PD-L1 by inhibiting BRD2/4 (fig. 3C and fig. 3D). Because BET protein promotes gene transcription by recruiting P-TEFb, we also want to know whether BRD2 and BRD4 also promote transcriptional extension through P-TEFb. The results showed that the expression of Cyclin T1 in HepG2 cells treated with OTX015 and JQ1 did not change significantly after IFN-α induced PD-L1 expression. However, the phosphorylation level of Cyclin T1 in HepG2 cells increased significantly in IFN-α alone induced group and decreased after adding inhibitors (fig. 3C). This suggests that OTX015 inhibits the transcriptional extension of PD-L1 by inhibiting P-TEFb after the action of BRD2 and BRD4 proteins.
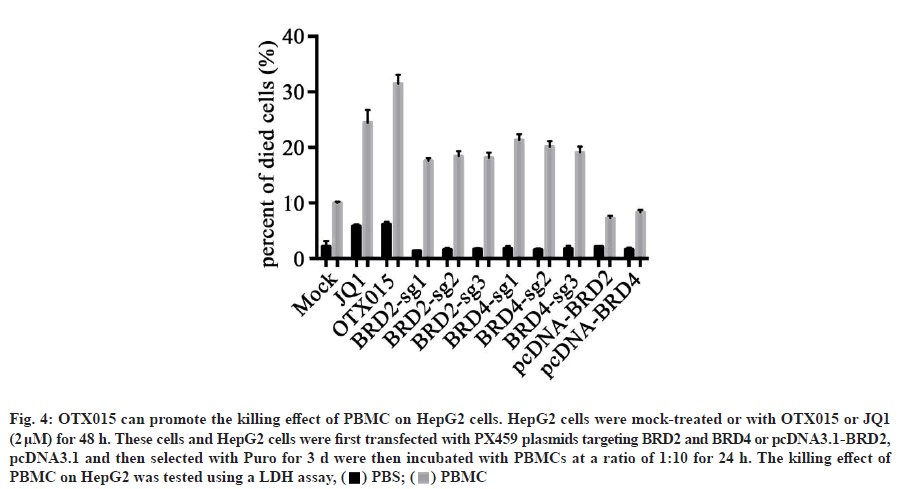
In tumor microenvironment, PD-L1 is one of the most important mechanisms for tumor cells to escape immune response. Drugs targeting the interaction between PD-1 and PD-L1 also show good therapeutic effects on tumors. In our previous studies, OTX015 has been shown to reduce IFN-α-induced PD-L1 expression in HepG2 cells. Therefore, we speculate that OTX015 can promote immune cells to kill cancer cells? To test our hypothesis, we isolated PBMC from healthy people. Afterwards, HepG2 cells induced by IFN-α treated with OTX015 and JQ1 were co-incubated with PBMC or not. The results showed that OTX015, JQ1 or PBMC alone could kill HepG2 cells partially, but the killing effect of PBMC on HepG2 cells treated with OTX015 was significantly better than that of any single group and the killing effect was greater than that of the two groups alone. This indicates that OTX015 cannot only kill HepG2 cells, but also promote the killing of PBMC (fig. 4). The above results suggest that the killing effect of OTX015 on tumors may not only be limited to pharmacological action, but also promote the killing effect of immune cells on tumors by improving the immune response.
Fig. 4: OTX015 can promote the killing effect of PBMC on HepG2 cells. HepG2 cells were mock-treated or with OTX015 or JQ1
(2 μM) for 48 h. These cells and HepG2 cells were first transfected with PX459 plasmids targeting BRD2 and BRD4 or pcDNA3.1-BRD2,
pcDNA3.1 and then selected with Puro for 3 d were then incubated with PBMCs at a ratio of 1:10 for 24 h. The killing effect of
PBMC on HepG2 was tested using a LDH assay, (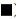 ) PBS; (
) PBS; ( ) PBMC
) PBMC
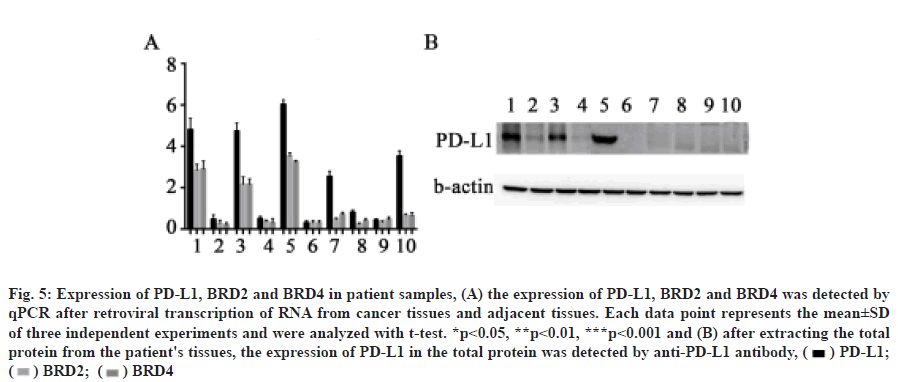
We confirmed that OTX015 inhibits PD-L1 expression by BET protein in HepG2 cells, but whether BET protein is related to PD-L1 expression in patient samples needs further verification. Through qPCR and Western blot, we found that the expression of PD-L1 in cancer tissues of patients 1, 3, 5, 7 and 10 was significantly higher than that in normal tissues. In addition, the expression of PD-L1 in patients 1, 3 and 5 increased significantly, as did the expression of BRD2 and BRD4 (fig. 5A and fig. 5B). This indicated that the expression of BRD2 and BRD4 was correlated with the expression of PD-L1 in some patients.
Hepatocellular carcinoma is one of the most common cancers in China and it is a complex disease with a high mortality rate[19-21]. Currently, the main treatment of hepatocellular cancer is surgical resection, but patients after surgery also need to take medicine to control the metastasis and spread of cancer cells[22-24]. In the past, controlling the spread and recurrence of hepatocellular cancer was mainly achieved by chemotherapeutic drugs[25]. However, in recent years, more and more attention has been paid to targeted therapy and the BET protein has become one of the most popular targets for cancer treatment. Previous studies have found that BET protein inhibitors can control cancer cell proliferation and promote cancer cell apoptosis by inhibiting the expression of oncogenes such as c-Myc[26,27]. BET protein inhibitors have now entered the clinical research stage due to their good therapeutic effects in vitro.
Fig. 5: Expression of PD-L1, BRD2 and BRD4 in patient samples, (A) the expression of PD-L1, BRD2 and BRD4 was detected by
qPCR after retroviral transcription of RNA from cancer tissues and adjacent tissues. Each data point represents the mean±SD
of three independent experiments and were analyzed with t-test. *p<0.05, **p<0.01, ***p<0.001 and (B) after extracting the total
protein from the patient's tissues, the expression of PD-L1 in the total protein was detected by anti-PD-L1 antibody, ( ) PD-L1;
(
) PD-L1;
( ) BRD2; (
) BRD2; ( ) BRD4
) BRD4
In addition to targeted therapy, immune checkpoints such as PD-1 and CTAL4 are also important targets in immunotherapy research. In previous studies, researchers have found that the PD-1 receptor molecule is expressed in immune cells and tumor cells can achieve immune escape by up-regulating the expression of PD-L1[28]. Interestingly, in 2017 researchers found that JQ1, one of the BET inhibitors, can down-regulate the expression of PD-L1 in lung cancer cells, which will likely increase the use of BET inhibitors in the treatment of cancer[29].
However, is JQ1 only a BET inhibitor, or does it have another mechanism of action that impacts PD-L1? Is it a specific treatment for lung cancer alone, or does it have the same effect on multiple types of cancer? This information has not been elucidated. Therefore, in this study, we intended to preliminarily explore the answers to the above questions using OTX015, another BET inhibitor, in hepatoma carcinoma cells.
In this study, we first showed that OTX015, a BET inhibitor, could promote the apoptosis of HepG2 cells and inhibit the proliferation of HepG2 cells at high concentrations. Furthermore, we discovered that the apoptotic and inhibition effects of the drug are achieved by inhibiting the expression of proliferating genes and promoting the expression of apoptotic genes. In addition, we found that OTX015 could also inhibit the expression of PD-L1 in HepG2 cells and this inhibition was achieved on the RNA level. We further noted that BRD2 and BRD4 of the BET protein family can bind directly to the promoter region of PD-L1. After knocking out these two proteins, the transcription of PD-L1 is relatively reduced, which indicates that BRD2 and BRD4 can promote the expression of PDL1, which is why BET inhibitors reduce the expression of PD-L1. We then found that when HepG2 cells were treated with OTX015, the expression of PD-L1 was decreased and the killing effect of PBMCs on HepG2 cells was increased, which indicated that OTX015 had a dual anti-tumor effect on hepatocellular cancer.
Although we have preliminarily demonstrated the dual anti-tumor effect of OTX015, there are still some shortcomings of this study. First, the effect of OTX015 on hepatoma carcinoma cells is mainly focused on the HepG2 cell model and we did not analyze any other models in this study. Secondly, the conclusions we have drawn were all obtained using in vitro experiments and the results may differ if performed in vivo. However, despite these shortcomings, our research lays the groundwork for further studies and provides many useful references for subsequent researchers.
In conclusion, we confirmed for the first time that OTX015, a BET inhibitor, has dual antineoplastic effects on hepatoma carcinoma cells, suggesting that OTX015 is not only an epigenetic targeting drug, but also a potential promoter of cellular immunotherapy.
Conflict of interests:
The authors declared no conflicts of interest.
References
- Bode AM, Dong Z, Wang H. Cancer prevention and control: Alarming challenges in China. Natl Sci Rev 2016;3(1):117-27.
- Nie Y, Wu K, Yu J, Liang Q, Cai X, Shang Y, et al. A global burden of gastric cancer: The major impact of China. Expert Rev Gastroenterol Hepatol 2017;11(7):651-61.
- Zhang XY, Zhang PY. Gastric cancer: Somatic genetics as a guide to therapy. J Med Genet 2017;54(5):305-12.
- Roy PS, Saikia BJ. Cancer and cure: A critical analysis. Indian J Cancer 2016;53(3):441-2.
- Goldberg AD, Allis CD, Bernstein E. Epigenetics: a landscape takes shape. Cell 2007;128(4):635-8.
- Egger G, Liang G, Aparicio A, Jones PA. Epigenetics in human disease and prospects for epigenetic therapy. Nature 2004;429(6990):457-63.
- Nebbioso A, Tambaro FP, Dell’Aversana C, Altucci L. Cancer epigenetics: Moving forward. PLoS Genet 2018;14(6):e1007362.
- You JS, Jones PA. Cancer genetics and epigenetics: Two sides of the same coin? Cancer cell 2012;22(1):9-20.
- Sokratous G, Polyzoidis S, Ashkan K. Immune infiltration of tumor microenvironment following immunotherapy for glioblastoma multiforme. Hum Vaccin Immunother 2017;13(11):2575-82.
- Wu Y, Sarkissyan M, Vadgama JV. Epigenetics in breast and prostate cancer. Methods Mol Biol 2015;1238:425-66.
- Pan Y, Liu G, Zhou F, Su B, Li Y. DNA methylation profiles in cancer diagnosis and therapeutics. Clin Exp Med 2018;18(1):1-4.
- West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest 2014;124(1):30-9.
- Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov 2014;13(9):673-91.
- Filippakopoulos P, Knapp S. Targeting bromodomains: Epigenetic readers of lysine acetylation. Nat Rev Drug Discov 2014;13(5):337-56.
- Lovén J, Hoke HA, Lin CY, Lau A, Orlando DA, Vakoc CR, et al. Selective inhibition of tumor oncogenes by disruption of super-enhancers. Cell 2013;153(2):320-34.
- da Motta LL, Ledaki I, Purshouse K, Haider S, De Bastiani MA, Baban D, et al. The BET inhibitor JQ1 selectively impairs tumour response to hypoxia and downregulates CA9 and angiogenesis in triple negative breast cancer. Oncogene 2017;36(1):122-32.
- Coudé MM, Braun T, Berrou J, Dupont M, Bertrand S, Masse A, et al. BET inhibitor OTX015 targets BRD2 and BRD4 and decreases c-MYC in acute leukemia cells. Oncotarget 2015;6(19):17698-712.
- Peedicayil J. The role of DNA methylation in the pathogenesis and treatment of cancer. Curr Clin Pharmacol 2012;7(4):333-40.
- West AC, Johnstone RW. New and emerging HDAC inhibitors for cancer treatment. J Clin Invest 2014;124(1):30-9.
- Falkenberg KJ, Johnstone RW. Histone deacetylases and their inhibitors in cancer, neurological diseases and immune disorders. Nat Rev Drug Discov 2014;13(9):673-91.
- Pardoll DM. The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 2012;12(4):252-64.
- Topalian SL, Drake CG, Pardoll DM. Immune checkpoint blockade: A common denominator approach to cancer therapy. Cancer cell 2015;27(4):450-61.
- Stromnes IM, Hulbert A, Pierce RH, Greenberg PD, Hingorani SR. T-cell localization, activation and clonal expansion in human pancreatic ductal adenocarcinoma. Cancer Immunol Res 2017;5(11):978-91.
- Gomes-Silva D, Mukherjee M, Srinivasan M, Krenciute G, Dakhova O, Zheng Y, et al. Tonic 4-1BB costimulation in chimeric antigen receptors impedes T cell survival and is vector-dependent. Cell Rep 2017;21(1):17-26.
- Öckinger J, Hagemann-Jensen M, Kullberg S, Engvall B, Eklund A, Grunewald J, et al. T-cell activation and HLA-regulated response to smoking in the deep airways of patients with multiple sclerosis. Clin Immunol 2016;169:114-20.
- Lu YC, Robbins PF. Cancer immunotherapy targeting neoantigens. Semin Immunol 2016;28(1):22-7.
- Nicodemus CF. Antibody-based immunotherapy of solid cancers: progress and possibilities. Immunotherapy 2015;7(8):923-39.
- Prasad V, Kaestner V. Nivolumab and pembrolizumab: Monoclonal antibodies against Programmed Cell Death-1 (PD-1) that are interchangeable. Semin Oncol 2017;44(2):132-5.
- Hogg SJ, Vervoort SJ, Deswal S, Ott CJ, Li J, Cluse LA, et al. BET-Bromodomain Inhibitors Engage the Host Immune System and Regulate Expression of the Immune Checkpoint Ligand PD-L1. Cell Rep 2017;18(9):2162-74.