- *Corresponding Author:
- Hui Zeng
National & Local Joint Engineering Research Center of Orthopaedic Biomaterials, Peking University Shenzhen Hospital, Shenzhen, 518036, PR China.
E-mail: zenghui_36@163.com
| This article was originally published in a special issue, “Clinical Research in Pharmaceutical and Biomedical Sciences” |
| Indian J Pharm Sci 2021:83(1)spl issue “132-140” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
Abstract
Beta catenin gene can inhibit the chondrogenic differentiation of human adipose tissue derived stem cells, while sex determining region Y-box9 gene can promote the chondrogenic differentiation of human adipose tissue derived stem cells. However, it is still necessary to study the role of the combination of the two genes in the early chondrogenic differentiation of human adipose tissue derived stem cells. The purpose of this study was to investigate whether the knockdown of Beta catenin gene and overexpression of sex determining region Y-box9 gene combined with modification of human adipose tissue derived stem cells could promote the early chondrogenic differentiation of stem cells. Therefore, human adipose tissue derived stem cells was divided into four groups with knockdown or overexpression lentivirus vectors, including negative control group (empty vector), Beta-catenin knockdown group (lentivirus-RNA interference-Betacatenin), sex determining region Y-box9 overexpression group (lentivirus-FLAG-sex determining region Y-box9) and union group (lentivirus-RNA interference-Beta-catenin+lentivirus-FLAG-sex determining region Y-box9). After 24 h of transfection, quantitative reverse transcription Polymerase Chain Reaction and Western blot were used to detect the knockdown efficiency of Beta catenin gene and the overexpression efficiency of sex determining region Y-box9 gene. Furthermore the pellets were cultured in chondrogenic differentiation medium for 21 d and the tissue was stained with safranin-O-green and toluidine blue. The messenger RNA expression of cartilage related genes was detected by quantitative reverse transcription polymerase chain reaction. Quantitative reverse transcription polymerase chain reaction and Western blot results showed that both Beta catenin knockdown gene and sex determining region Y-box9 overexpression gene were successfully transfected and expressed (p< 0.05). quantitative reverse transcription polymerase chain reaction results showed that type-II collagen alpha 1 chain and aggrecan messenger RNA expression levels in the union group were higher than those in the first three groups (p<0.05), while type-X collagen alpha 1 chain and type-I collagen alpha 1 chain in the union group were lower than in the negative control group (p<0.05). The results of staining showed that the absorbance of union group was higher than that of the former three groups (p<0.05). Immunohistochemical results showed that the protein expression of type-II collagen alpha 1 chain and aggrecan in the union group was higher than that in the first three groups (p<0.05). Therefore, the combination of Beta-catenin gene knockdown and sex determining region Y-box9 overexpression can promote the early chondrogenic differentiation of human adipose tissue derived stem cells.
Keywords
hADSCs, Sox9, western blot, chondrogenic differentiation medium
Osteoarthritis (OA) is a disease characterized by articular cartilage degeneration and changes in ligaments, synovial bursa and synovial membrane, which is also the most common degenerative joint disease at present. Due to the aging of the population, its occurrence frequency increases year by year [1].
Since there is no blood supply in cartilage tissue, the joint cavity can only rely on synovial fluid to provide nutrition. Therefore, once cartilage tissue is damaged, it is difficult to induce progenitor cells in blood or bone marrow to enter the defect for repair and the damage is difficult to heal itself [2].
In addition to the traditional methods of articular cartilage repair, the development of regenerative medicine has opened up a new path for cartilage regeneration and repair. The basic method is to inoculate autologous or allogeneic tissue cells into a kind of biomaterial with good histocompatibility and degradable absorption after being cultured and expanded in vitro. The complex is implanted into the tissue defect site in vivo and the biomaterial is gradually degraded, absorbed and the implanted cells continue to propagate to form new functional cartilage tissue, which achieves the purpose of repairing cartilage defect [3]. The three basic elements of tissue engineering are seed cells, biomaterials and tissue construction. However, the problems from laboratory to clinical are still difficult, such as aging of seed cells, phenotypic changes of differentiated chondrocytes, poor mechanical properties of formed cartilage tissue, poor fusion with host tissue and late degradation and separation [4]. Therefore, the selection of seed cells and the acquisition, culture and differentiation of seed cells are the premise and basis for repairing articular cartilage defects with tissue engineered cartilage.
It has been shown that human adipose tissue derived stem cells (hADSCs) are pluripotent adult stem cells rather than simple mixed colonies of mononuclear progenitor cells. Compared with bone marrow mesenchymal stem cells, hADSCs have a wide range of sources, easy to obtain and have stronger proliferation ability. Previous studies have shown that after implantation of hADSCs in the cartilage defect site, the morphology and function of the cartilage forming tissue are not consistent with that of hyaline cartilage tissue and the repair tissue is difficult to completely and stably fuse with the surrounding intact cartilage [5-7]. Therefore, an effective way to overcome these problems is to modify hADSCs gene before implantation into the injured site of cartilage, which may be through enhancing the cartilage repair resurrection of cells.
A variety of genes have been identified as potential chondrogenic markers, among which sex determining region Y-box9 (Sox9) gene has been confirmed to be a key gene in cartilage formation [8,9]. The role of Sox9 gene in cartilage differentiation is achieved by inhibiting Beta catenin (β-catenin) gene and there is a complex relationship between them. Many studies have shown that β-catenin gene and Sox9 gene play an antagonistic role in chondrogenic differentiation of mesenchymal stem cells [10]. Therefore, lentiviral transfection was used to create a double gene modification model with overexpression of Sox9 and knock down of β-catenin in this study. It is known that Sox9 gene can inhibit Wnt/β-catenin signaling pathway during cartilage differentiation and inhibition of Wnt/β-catenin signaling pathway can increase Sox9 expression during cartilage differentiation. In this study, we verified that the cumulative effect was better than single gene modification through double gene synergistic modification.
According to the comparison with independent treatment, the activation of multiple therapeutic genes is more valuable for stimulating the chondrogenic activity of stem cells [11,12]. Therefore, in this study, lentivirus encoding Sox9 gene and β-catenin knockdown lentivirus were union to act on human hADSCs to detect the chondrogenic activity of hADSCs under the condition of dual gene co coding.
Materials and Methods
Experimental design
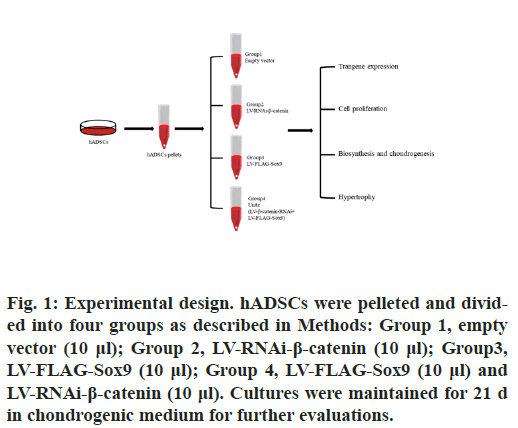
hADSCs were pelleted (2×105 cells/pellet) and kept in chondrogenic medium [13,14] for 24 h prior to transduction. The hADSCs pellets were next treated with the various Lentivirus vectors or vector combinations according to the following 4 conditions for maintenance in chondrogenic medium over a period of 21 d (fig. 1): Group 1, pellets transduced with 10μl empty vector; Group 2, pellets immediately transduced with 10 μl lentivirus (LV)-RNA interference (RNAi)-β-catenin.; Group3, pellets immediately transduced with 10 μl LVFLAG- Sox9; Group 4, pellets immediately transduced with 10 μl LV-FLAG-Sox9 and 10μl LV-RNAi-β- catenin.
Figure 1: Experimental design. hADSCs were pelleted and divided into four groups as described in Methods: Group 1, empty vector (10 μl); Group 2, LV-RNAi-β-catenin (10 μl); Group3, LV-FLAG-Sox9 (10 μl); Group 4, LV-FLAG-Sox9 (10 μl) and LV-RNAi-β-catenin (10 μl). Cultures were maintained for 21 d in chondrogenic medium for further evaluations.
Chemicals and Reagents
All reagents including alginate (low viscosity alginic acid sodium salt from brown algae) were from Sigma. The anti-Aggrecan antibody (ACAN) and anti-β- catenin antibodies were from Abcam. The anti-type- II collagen (COL2A1), anti-SOX9 antibody was from ABclonal. Biotinylated secondary antibodies were purchased from Thermo. Human Adipose derived Mesenchymal Stem Cell (HUXMD-01001), Human Adipose-derived Stem Cell Chondrogenic Differentiation Basal Medium(HUXMD-9004) and Human Adipose-derived Mesenchymal Stem Cell Basal Medium(HUXMD-90011) were from Cyagen.
Construction of Small interfering RNA (siRNA) lentiviral vector
AgI and EcoRI digested and linearized gv112 vector 1 μL, annealed double stranded DNA (dsDNA) 1 μL, mixed with T4 phage DNA ligase buffer 2μL, T4 phage DNA ligase 1 μL, double-distilled water (ddH2O) 15 μL to form a 20 μL reaction system, which was connected overnight at 16°. The ligation product was transformed into Escherichia coli DH5 α and the recombinant clone was selected for Polymerase chain reaction (PCR) identification. The upstream primer was 5’-CCATGATTCCTTCATATTTGC-3’. The downstream primer was 5’-GTAATACGGTTATCCACGCG-3’ (designed and synthesized by Shanghai Jikai Company). The total PCR reaction system was 20 μL, including 10×PCR buffer 2 μL, 2.5 mmol/L dNTPs 0.8 μL, 10 μmol/L upstream and downstream primers 0.4 μL, 5 U/μL Taq polymerase 0.2 μL, DNA template 1 μL, ddH2O 15.2 μL. PCR cycle parameters were denaturation at 94° for 30 s, 94° for 30 s, 60° for 30 s, 72° for 30 s and 72° for 6 min. PCR amplified products were identified by 10 g/L agarose gel electrophoresis. Positive clones were selected and sent to Shanghai Jikai Company for sequencing analysis.
Construction of overexpression lentivirus vector
In the plasmid containing Sox, the target gene was amplified by PCR, and the primers were synthesized by Shanghai Jikai Company. The upstream sequence (f) was 5’- GAGGATCCCCGGGT ACCGGTCGCCACCATG AATCTCCTGG ACCCCTTC-3’. The downstream sequence (R) was 5’-TCCTTGTAGT CCATACCAGGTCGAGTGAG CTGTGTGTAG-3’. The PCR reaction system was 5×Taq buffer 4 μL, 2.5 mmol/L dNTPs 1.6 μL, 10μmol/L upstream and downstream primers 0.4 μL, 10 ng/μL ldna templates 1 μL, Taq polymerase 0.2 μL, ddH2O supplement to 20 μL. The PCR reaction conditions were denaturation at 94° for 5 min, 94° for 30 s, 58° for 30 s, 72° for 25 s, with a total of 30 cycles and extension at 72° for 10 min. The PCR products were linked to the linearized gv358 vector after digestion with Agel. The purified products were transformed into Escherichia coli DH5 α and the positive colonies were detected by PCR. The upstream sequence (f) is 5’-GGAGG AAGTCG GTGAA GAAC-3’. The downstream sequence (R) was 5’-CCTTAT AGTCCT TATCA TCGTC-3’. The PCR reaction system was the same as above. The PCR reaction conditions were as follows: 94° 2 min; 94° 30 s, 55° 30 s, 72° 40 s, a total of 30 cycles; 72° extension for 6 min. PCR positive clones were sent to Shanghai Jikai company for sequencing and comparative analysis. The target plasmid was successfully constructed for screening effective interference targets.
In vitro pellet culture
For the induction of chondrogenesis, the in vitro pellet culture was carried out using 2.5×105 hADSCs at passage 3 transduced with LV (10 or 20 μl each pellet, 4×105 or 8×105 functional recombinant viral particles, respectively, multiplicity of infection=2 or 4) in chondrogenic medium (CM; Dulbecco’s Modified Eagle Medium (DMEM)/F-12 supplemented with 1 % insulintransferrin- selenium, 10-7 μM L-dexamethasone, 50 μM L-ascorbate-2-phosphate, 50 μM L-proline, and 1 mM sodium pyruvate). The hADSCs pellets were cultured with 10 ng/mL of transforming growth factor (TGF)-β1 (R&D Systems Inc.)
For a pellet culture, 1 mL of the cell suspension was aliquoted into 15 mL polypropylene centrifuge tubes and spun in a benchtop centrifuge (Hanil) at 500 ×g for 5 min. The tubes were placed in an incubator at 37° in a humidified atmosphere containing 95 % air and 5 % CO2 for 21 d. The caps of the tubes were loosened to allow air exchange. The medium was changed every 3rd d. Pellets were taken for analysis at 21 d after culture.
Real time RT-PCR analysis.
RNA was isolated using the standard guanidine isothiocyanate Tri-Reagent® (Sigma Chemical) according to the manufacturer’s recommendations. The isolated RNA samples were converted to complementary DNA (cDNA) using reverse transcriptase (SuperScript III®; Invitrogen) and oligo (dT) primers. All of the polymerase chain reaction (PCR) were performed on the LightCycler 480 system® (Roche Diagnostics) in standard 20 μL reactions. The reactions were performed for SOX9, β-catenin, ACAN, COL1A1, COL2A1, (COL10A1 and GAPDH as a housekeeping gene. The primers and reaction conditions used for amplification are listed in Table 1. After polymerase activation (95° for 10 min), 45 cycles were run with 10 s denaturation at 95°, 10 s annealing at 65°, and 10 s extension at 72°. Meltcurve analysis was performed immediately after the amplification protocol under the following conditions: 5 s at 95° (hold time on reaching temperature), 1 min at 65° and 1 s at 97°. The temperature change rates were 20° per sec (except for the final step, which had a temperature change rate of 0.1° per sec). The peak melting temperature obtained represented the specific amplified product. To guarantee the reliability of the results, all samples were processed in triplicate. The test was considered positive if the signal from the amplified product was clearly visible in both samples. Each assay was performed using the positive and negative controls. The data were standardized to the housekeeping H-actin values for all samples using the delta Ct method (User bulletin #2 Applied Biosystems; Roche Molecular System). Seven pellets were used for 7 genes per donor.
| Target Gene | Forward Sequence (5’-3’) | Reverse Sequence (5’-3’) |
|---|---|---|
| β-catenin | GGAGGAAGGTCTGAGGAGCA | AGTAGCCATTGTCCACGCTG |
| Sox9 | GAGGAAGTCGGTGAAGAACGG | CCTCTCGCTTCAGGTCAGC |
| COL2A1 | AGGCTGGCAGCTGTGTGCAG | GAGGTCAGTTGGGCAGATGG |
| ACAN | ACTGGCGAGCACTGTAACAT | GTAGCTGGGAAGGCATAAGCA |
| COL1A1 | TTTGGATGGTGCCAAGGGAG | CACCATCATTTCCACGAGCA |
| COL10A1 | GATACCAAATGCCCACAGGC | AGGACTTCCGTAGCCTGGTT |
Table 1: Primers for Real-Time RT-PCR Of HADSCS
Toluidine blue
After 21 d of chondrogenic differentiation of hADSCs pellet, toluidine blue staining was performed. Toluidine blue staining was diluted to 0.1 % with 20 % ethanol solution. 4 % paraformaldehyde was added into the six well plate to fix the cells and the diluted toluidine blue staining solution was dropped for staining. After dyeing for 5 min, wash with Phosphate-buffered saline (PBS) three times, 5 min each time. After that, the average absorbance values were measured and compared by Image Pro Plus 6.0 software.
Saffron fast green staining
After 21 d of chondrogenic differentiation of hADSCs pellet, fresh Weigert staining solution was added for staining for 35 min, washing with acid differentiation solution for 15 s, PBS washing for 10 min, soaking in solid green staining solution for 5 min and washing sections with weak acid solution for 10-15 s to remove the residual solid green, add safranine to dye for 5min, then wash it with PBS for three times, observe under microscope and then compare the average absorbance value with Image Pro Plus 6.0 software.
Immunohistochemistry
The relative protein expression of COL2A1 and ACAN was detected by immunohistochemistry. First, PBS was added to wash the differentiated hADSCs pellet three times, 1 min each time. After that, 4 % paraformaldehyde was fixed, 5 % Bovine serum albumin (BSA) was blocked and primary antibody was incubated at 4° overnight. The next day after cleaning, the second antibody working solution was incubated at 37° and then incubated in SABC wet box and then the color was developed by diaminobenzidine (DAB). Under the light protection, it was observed to be brown under the microscope. After that, the average absorbance value was measured and compared by Image Pro Plus 6.0 software.
Statistical Analysis
Graphpad Prism 8.0 software was used for statistical analysis of experimental data. The mean±standard deviation was used to represent all the data. After the homogeneity test of variance, one-way Analysis of variance (ANOVA) statistical method was used to analyze the measurement data between groups. The independent test was used to analyze the measurement data between the two groups. When p<0.05 the difference was statistically significant.
Results and Discussion
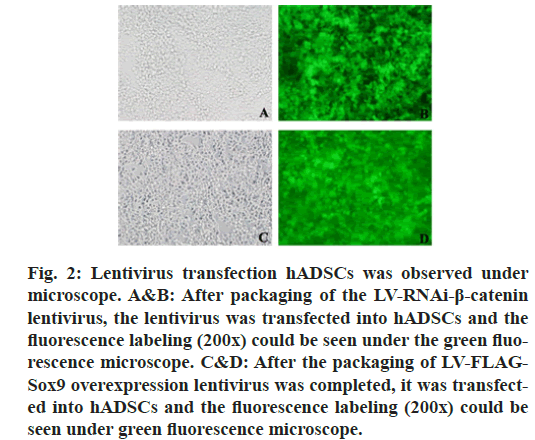
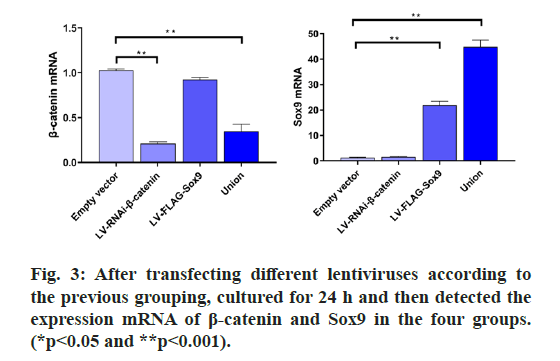
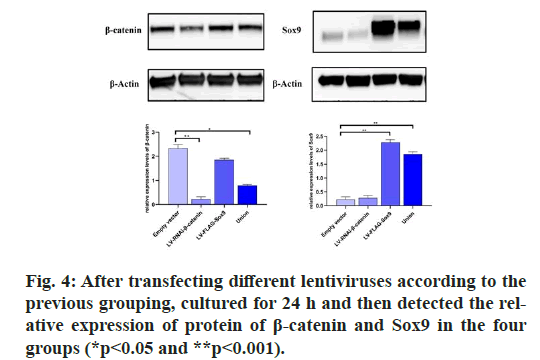
The results showed that there was no expression of GFP in the untransfected group, but the expression of GFP was found in the transfected group, which indicated that the Green fluorescent protein (GFP) gene carried by lentivirus could be normally expressed in cells. RT-PCR was used to detect the expression of β-catenin and Sox9. At the same time, β-catenin was distinctly depressed and Sox9 was prominently expressed and WB results also confirmed the same conclusion, which proved that lentivirus transfection was successful, which can be used for subsequent experiments. (fig. 2, fig. 3 & fig. 4).
Figure 2: Lentivirus transfection hADSCs was observed under microscope. A&B: After packaging of the LV-RNAi-β-catenin lentivirus, the lentivirus was transfected into hADSCs and the fluorescence labeling (200×) could be seen under the green fluorescence microscope. C&D: After the packaging of LV-FLAGSox9 overexpression lentivirus was completed, it was transfected into hADSCs and the fluorescence labeling (200×) could be seen under green fluorescence microscope.
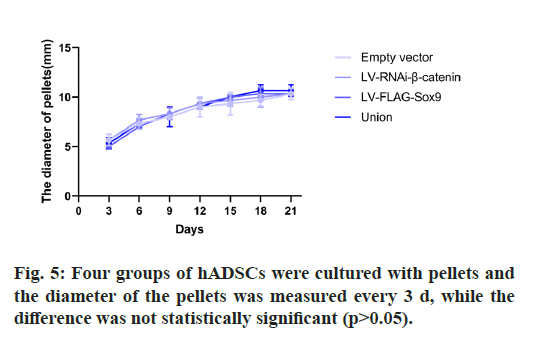
The results showed that there was no significant difference in the diameter of microspheres in each group. There was no change in the size of microspheres at the later stage of chondrogenic culture. (Table 2, fig. 5).
| Days | Empty vector | LV-RNAi-β-catenin | LV-FLAG-Sox9 | Union |
|---|---|---|---|---|
| 3 | 5.66±0.58 | 5.66±0.58 | 5.0 | 5.33±0.58 |
| 6 | 7.33±0.58 | 7.33±0.58 | 7.0 | 7.33±0.58 |
| 9 | 8.0 | 8.33±0.58 | 8.33±0.58 | 8±1 |
| 12 | 9±1 | 9.33±0.58 | 9.33±0.58 | 9.0 |
| 15 | 9.33±1.15 | 9.67±0.58 | 10.0 | 10.0 |
| 18 | 9.67±0.58 | 10±1 | 10.33±0.58 | 10.67±0.58 |
| 21 | 10±1 | 10.33±0.58 | 10.33±0.58 | 10.67±0.58 |
Table 2: The Diameter of Pellets (mm)
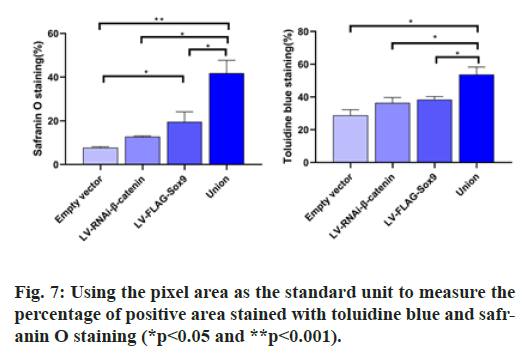
After chondrogenic differentiation, the average absorbance was 7.764±1.31 in the negative control group, 12.78±1.14 in the β-catenin knockdown group, 19.58±3.92 in the Sox9 overexpression group and 41.83±4.73 in the union group of β-Catenin gene knockdown and Sox9 overexpression group. The average absorbance of the union group was significantly higher than that of the other three groups (p<0.05), so the content of proteoglycan in the union group was higher than that in the other three groups. After toluidine blue staining, the average absorbance of negative control group was 28.84±2.33 that of β-catenin knockdown group was 36.34±2.17, that of Sox9 overexpression group was 38.41±1.75 and that of combination group was 53.69±3.14. The average absorbance of the union group was significantly higher than that of the other three groups (p<0.05). After adding COL2A1 and aggrecan first antibody into the six well plate, immunohistochemistry was performed.
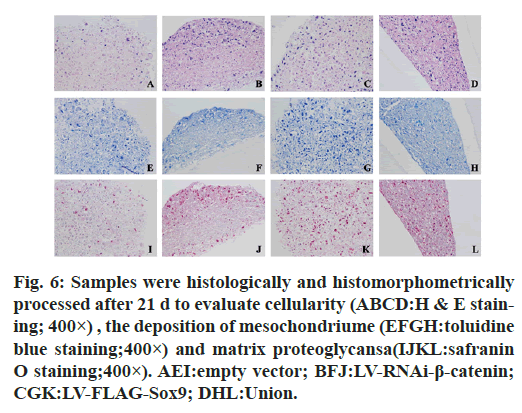
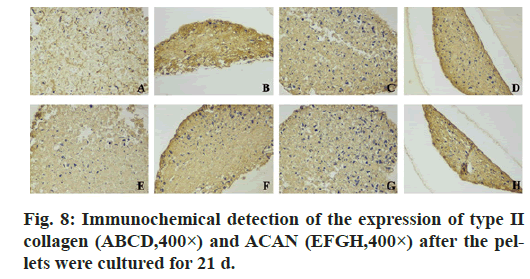
In the immunohistochemistry of COL2A1, the absorbance of negative control group was 15.93±1.11 that of β-catenin knockdown group was 25.0±2.0, that of Sox9 overexpression group was 25.61±1.79 and that of union group was 28.47±1.53. In aggrecan immunohistochemistry, the absorbance of the above groups was 16.40±2.40, 26.80±2.26, 29.64±3.0, 35.37±2.98 respectively. In the immunohistochemistry of COL2A1 and ACAN, the average absorbance value of the union group was higher than that of the other three groups (p<0.05) (fig. 6, fig. 7 & fig. 8).
Figure 6: Samples were histologically and histomorphometrically processed after 21 d to evaluate cellularity (ABCD:H & E staining; 400×) , the deposition of mesochondriume (EFGH:toluidine blue staining;400×) and matrix proteoglycansa(IJKL:safranin O staining;400×). AEI:empty vector; BFJ:LV-RNAi-β-catenin; CGK:LV-FLAG-Sox9; DHL:Union.
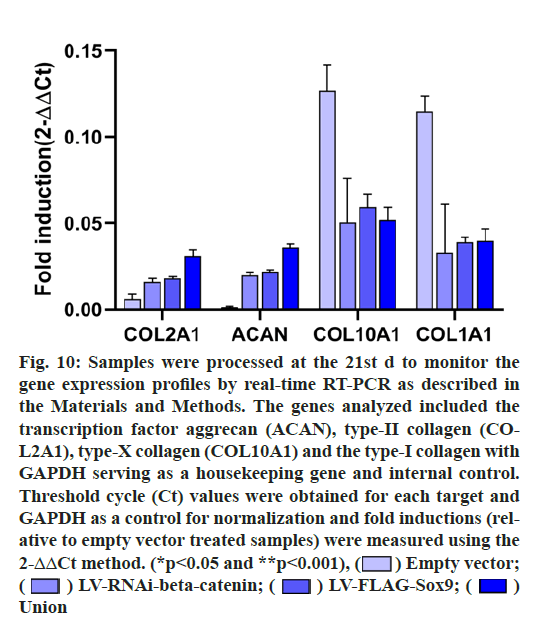
The mRNA expressions of COL2A1, ACAN, COL10A1 and COL1A1 were detected after 21 d of chondrogenic differentiation of hADSCs. The results showed that COL2A1 and ACAN of the union group were higher than those of the other three groups, COL2A1 was 5.21 higher than that of the control group, ACAN was 21.18 higher than that of the control group and the difference was statistically significant (p<0.05). It should be noted that after co-transfection of the two genes, the expression of COL10A1 and COL1A1 in the union group decreased by 0.41 and 0.34 times compared with the control group on the 21st d respectively, indicating that the osteogenic differentiation and chondrocyte hypertrophy levels were reduced after gene modification. (fig. 9 & fig. 10).
Figure 10: Samples were processed at the 21st d to monitor the gene expression profiles by real-time RT-PCR as described in the Materials and Methods. The genes analyzed included the transcription factor aggrecan (ACAN), type-II collagen (COL2A1), type-X collagen (COL10A1) and the type-I collagen with GAPDH serving as a housekeeping gene and internal control. Threshold cycle (Ct) values were obtained for each target and GAPDH as a control for normalization and fold inductions (relative to empty vector treated samples) were measured using the 2-ΔΔCt method. (*p<0.05 and **p<0.001),  Empty vector;
Empty vector; LV-RNAi-beta-catenin;
LV-RNAi-beta-catenin;  LV-FLAG-Sox9;
LV-FLAG-Sox9;  Union
Union
In recent years, with the discovery that articular cartilage plays a key role in OA and cartilage can be repaired, transplantation and culture of autologous chondrocytes for repairing deep cartilage defects of knee joint is an important and potential therapeutic method. Good chondrogenic differentiation ability of genetically modified mesenchymal stem cells and good biocompatibility of various scaffold materials make them show important value and good prospects in cartilage injury repair [15-18]. In this experiment, we cultured hADSCs in microspheres, which can better simulate the chondrogenesis environment of mesenchymal stem cells in vivo than adherent culture [19,20]. According to the experimental results, we can know that the size of hADSCs microspheres in different groups does not show obvious difference in cartilage differentiation medium, which may be due to the fact that the volume change has no obvious effect on the chondrogenesis of mesenchymal stem cells.
Many studies have shown that Sox9 inhibits the activation of β-catenin/T-cell factor/lymphoid enhancer factor (TCF/LEF) by competing with TCF/LEF n to bind to the β-catenini site, promotes the degradation of β-catenin through the formation of Sox9-β-catenin complex through ubiquitination/26S proteasome pathway and co-acts with cyclin D1, a target gene downstream of wnt signaling pathway, to inhibit chondrocyte proliferation and delay hypertrophic chondrocyte [21]. Previously, Topol L et al. [22] found that during cartilage formation, N-terminal of Sox9 could promote β-catenin degradation, while C-terminal could inhibit β-catenin transcription activity without affecting its stability. Therefore, Sox9 affects chondrocyte differentiation process by inhibiting Wnt/β-catenin signaling pathway transduction [10]. In addition, Zhang Y et al. [23] found that miR-410 increased the expression of Sox9 during chondrogenic differentiation of mesenchymal stem cells by targeting Wnt3A to inhibit Wnt/β-catenin signaling pathway, and also increased the mRNA and protein levels of type II collagen (COL2A1), aggrecan (ACAN) and hyaluronic acid synthase 2 (Has2). Akiyama et al. [24] found that the phenotype of chondroplasia abnormality in mutant mice with Sox9 overexpression in chondrocytes was similar to that in mice with β-catenin silenced in chondrocytes. On the contrary, the phenotypic changes caused by the stable expression of β-catenin in chondrocytes were similar to those of mice whose Sox9 allele was silenced in chondrocytes. Venkatesan et al. [10] and Frisch et al. [25] both found that Sox9 transfection would inhibit the expression of β-catenin in hMSCs. It was also found that Sox9 overexpression could inhibit the level of β-catenin during cartilage repair in vivo [26]. Therefore, the mutual inhibition between Sox9 expression and classical Wnt/β-catenin signaling pathway indicates that chondrocyte differentiation may be regulated by a positive feedback loop mechanism: inhibition of classical Wnt signaling pathway causes Sox9 expression; Sox9 further inhibited Wnt/β-catenin signal and osteogenic differentiation, thus promoting the complete differentiation of chondrocytes [27].
In this study, we transfected hADSCs with lentiviral vector harboring the genes related to knockdown of β-catenin and over-expression of Sox9 gene separately or jointly. After culture for 21 d in cartilage microsphere differentiation medium, the absorbance value of Annona Red O/Fast Green staining showed that the joint group was 41.83±4.73, which was higher than that of the other three groups and the difference was statistically significant, indicating that the cells modified with double genes synthesized more type 2 collagen than the cells modified with single gene. Similarly, the results of toluidine blue staining showed that the staining absorption value in the joint group was 35.37±2.98 and the color was darker than that in the other three groups, so it can be concluded that the cells modified with double genes group expressed more cartilage matrix. Immunohistochemistry of COL2A1 and ACAN also showed that the expression of COL2A1 and ACAN in the joint group was higher than that in the other three groups. The relative expression of-β-catenin, SOX9, COL2A1, ACAN, COL1A1 and COL10A1 mRNA was detected by RT-qPCR and it was found that the expression of COL2A1 and ACAN in the joint group (β-catenin RNAi+Sox9) was significantly higher than that in the control group, indicating more chondrogenic differentiation in the joint group. COL1A1 and COL10A1 were lower than those in the control group, suggesting that the combined modification with the two genes advantageously delayed the premature hypertrophy and differentiation of hADSCs, which might be caused by the anti-hypertrophy effect of exogenous SOX9 expression [28]. Meanwhile, knockdown of β-catenin restricted osteogenesis [29], thus further slowing down the hypertrophy phenomenon after cartilage formation. In this study, it could be confirmed that the interaction between β-catenin and Sox9 was very important in the chondrogenic differentiation of stem cells, which also was verified by our experiments using lentiviral transfection technology to modify the genes related to hADSCs differentiated into cartilage. The dynamic balance between classical Wnt signaling pathway and Sox9 may play a decisive role in osteogenic or chondrogenic differentiation of stem cells. Inhibition of Wnt/β-catenin signaling may be a key factor to promote chondroblasts differentiated by stem cell and resist osteoblast differentiation, thus participating in the management of chondrocyte proliferation and differentiation.
Therefore, it is found that the chondrorepair activity of progenitor cells can be stimulated by the modification of various genes in stem cells by the independent gene transduction of slow virus vector, which provides the basis for controlling the cartilage repair process after hADSCs are implanted into cartilage defects.
The benefit of co-transduction of therapeutic lentiviral vector gene repairer hADSCs provides evidence that enhancing the expression of Sox9 and knockdown β-catenin can significantly improve its cartilage repair activity in vitro.
Acknowledgements
This study was supported by grants from “San- Ming” Project of Medicine in Shenzhen (No. SZSM201612092), Natural Science Foundation of Guangdong Province (No. 2019A1515011290) and Research and Development Projects of Shenzhen (No. JCYJ20170307111755218).
Conflict of interests
The authors declared no conflicts of interest.
References
- Ibarra A, Zhuang J, Zhao Y, Salathia NS, Huang V, Acosta AD, et al. Non-invasive characterization of human bone marrow stimulation and reconstitution by cell-free messenger RNA sequencing. Nat Commun 2020;11(1):1-1.
- Lee J, Jeon O, Kong M, Abdeen AA, Shin JY, Lee HN, et al. Combinatorial screening of biochemical and physical signals for phenotypic regulation of stem cell-based cartilage tissue engineering. Sci Adv 2020;6(21):eaaz5913.
- Sun Y, You Y, Jiang W, Wang B, Wu Q, Dai K. 3D bioprinting dual-factor releasing and gradient-structured constructs ready to implant for anisotropic cartilage regeneration. Sci Adv 2020;6(37):eaay1422.
- Griffith LG, Naughton G. Tissue engineering-current challenges and expanding opportunities. Science 2002;295(5557):1009-14.
- Hwang NS, Im SG, Wu PB, Bichara DA, Zhao X, Randolph MA, et al. Chondrogenic priming adipose-mesenchymal stem cells for cartilage tissue regeneration. Pharm Res 2011;28(6):1395-405.
- Choi SW, Hong KY, Minn KW, Chang H. Chondrogenesis of adipose-derived stem cells on irradiated cartilage. Plast Reconstr Surg 2020;145(2):409-18 .
- Kim JS, Kim TH, Kang DL, Baek SY, Lee Y, Koh YG, et al. Chondrogenic differentiation of human ASCs by stiffness control in 3D fibrin hydrogel. Biochem Biophys Res Commun 2020;522(1):213-9.
- Weißenberger M, Weißenberger MH, Wagenbrenner M, Heinz T, Reboredo J, Holzapfel BM, et al. Different types of cartilage neotissue fabricated from collagen hydrogels and mesenchymal stromal cells via SOX9, TGFB1 or BMP2 gene transfer. Plos one 2020;15(8):e0237479.
- Haengseok S, Keun-Hong P. Regulation and function of SOX9 during cartilage development and regeneration. Semin Cancer Biol 2020;67:12-23.
- Venkatesan JK, Ekici M, Madry H, Schmitt G, Kohn D, Cucchiarini M. SOX9 gene transfer via safe, stable, replication-defective recombinant adeno-associated virus vectors as a novel, powerful tool to enhance the chondrogenic potential of human mesenchymal stem cells. Stem Cell Res Ther 2012;3(3):1-5.
- Jing H, Zhang X, Gao M, Luo K, Fu W, Yin M, et al. Kartogenin preconditioning commits mesenchymal stem cells to a precartilaginous stage with enhanced chondrogenic potential by modulating JNK and ß-catenin–related pathways. FASEB J 2019;33(4):5641-53.
- Li J, Dong S. The signaling pathways involved in chondrocyte differentiation and hypertrophic differentiation. Stem Cells Int 2016;2016.
- Steinert AF, Palmer GD, Pilapil C, Nöth U, Evans CH, Ghivizzani SC. Enhanced in vitro chondrogenesis of primary mesenchymal stem cells by combined gene transfer. Tissue Eng Part A 2009;15(5):1127-39.
- Liu P, Sun L, Chen H, Sun S, Zhou D, Pang B, et al. Lentiviral-mediated multiple gene transfer to chondrocytes promotes chondrocyte differentiation and bone formation in rabbit bone marrow-derived mesenchymal stem cells. Oncol Rep 2015;34(5):2618-26.
- Ikeda T, Kamekura S, Mabuchi A, Kou I, Seki S, Takato T, et al. The combination of SOX5, SOX6, and SOX9 (the SOX trio) provides signals sufficient for induction of permanent cartilage. Arthritis Rheum 2004;50(11):3561-73.
- Pagnotto MR, Wang Z, Karpie JC, Ferretti M, Xiao X, Chu CR. Adeno-associated viral gene transfer of transforming growth factor-ß 1 to human mesenchymal stem cells improves cartilage repair. Gene Ther 2007;14(10):804-13.
- Cucchiarini M, Ekici M, Schetting S, Kohn D, Madry H. Metabolic activities and chondrogenic differentiation of human mesenchymal stem cells following recombinant adeno-associated virus–mediated gene transfer and overexpression of fibroblast growth factor 2. Tissue Eng Part A 2011;17(15-16):1921-33.
- Kim HJ, Im GI. Electroporation-mediated transfer of SOX trio genes (SOX-5, SOX-6, and SOX-9) to enhance the chondrogenesis of mesenchymal stem cells. Stem Cells Dev 2011;20(12):2103-14.
- Buckwalter JA, Hunziker EB. Healing of bones, cartilages, tendons, and ligaments: a new era. The Lancet 1996;348:S18.
- Johnstone B, Hering TM, Caplan AI, Goldberg VM, Yoo JU. In vitro chondrogenesis of bone marrow-derived mesenchymal progenitor cells. Exp Cell Res 1998;238(1):265-72.
- Held A, Glas A, Dietrich L, Bollmann M, Brandstädter K, Grossmann TN, et al. Targeting ß-catenin dependent Wnt signaling via peptidomimetic inhibitors in murine chondrocytes and OA cartilage. Osteoarthritis Cartilage 2018;26(6):818-23.
- Topol L, Chen W, Song H, Day TF, Yang Y. Sox9 inhibits Wnt signaling by promoting ß-catenin phosphorylation in the nucleus. J Biol Chem 2009;284(5):3323-33.
- Zhang Y, Huang X, Yuan Y. MicroRNA-410 promotes chondrogenic differentiation of human bone marrow mesenchymal stem cells through down-regulating Wnt3a. Am J Transl Res 2017;9(1):136-45.
- Akiyama H, Lyons JP, Mori-Akiyama Y, Yang X, Zhang R, Zhang Z, et al. Interactions between Sox9 and ß-catenin control chondrocyte differentiation. Genes Dev 2004;18(9):1072-87.
- Frisch J, Rey-Rico A, Venkatesan JK, Schmitt G, Madry H, Cucchiarini M. rAAV-mediated overexpression of sox9, TGF-ß and IGF-I in minipig bone marrow aspirates to enhance the chondrogenic processes for cartilage repair. Gene Ther 2016;23(3):247-55.
- Cucchiarini M, Orth P, Madry H. Direct rAAV SOX9 administration for durable articular cartilage repair with delayed terminal differentiation and hypertrophy in vivo. J Mol Med 2013;91(5):625-36.
- Li W, Xiong Y, Chen W, Wu L. Wnt/ß-catenin signaling may induce senescence of chondrocytes in osteoarthritis. Exp Ther Med 2020;20(3):2631-8.
- Wang W, He N, Feng C, Liu V, Zhang L, Wang F, et al. Human adipose-derived mesenchymal progenitor cells engraft into rabbit articular cartilage. Int J Mol Sci 2015;16(6):12076-91.
- Day TF, Guo X, Garrett-Beal L, Yang Y. Wnt/ß-catenin signaling in mesenchymal progenitors controls osteoblast and chondrocyte differentiation during vertebrate skeletogenesis. Dev Cell 2005;8(5):7