- *Corresponding Author:
- T. C. Wang
Department of Virology,
Changchun Veterinary Research Institute,
Chinese Academy of Agricultural Sciences,
Changchun,
Jilin 130122,
China
E-mail: q3504517@126.com
| This article was originally published in a special issue,“New Advancements in Biomedical and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2022:84(2) Spl Issue “48-58” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The complement system played critical roles in antimicrobial defense response, immune regulation and immunopathological damage. As an important negative regulator in this system, complement factor H provided selective advantage for tumor cell proliferation to escape immune surveillance, leading to avoid apoptosis. However, the influence of its expression on the pathological process and prognosis of liver cancer were still unclear. In this study, we analyzed the pattern of complement factor H expression in liver cancer in order to clarify its potential application value in the diagnosis and prognosis by bioinformatics analysis of data-set collected from The Cancer Genome Atlas database. By evaluating the clinical diagnostic value of complement factor H, we studied the correlation between complement factor H expression and clinicopathological parameters of liver cancer. Additionally, we found that patients with low complement factor H expression had poor overall survival and relapse-free survival, and confirmed that low complement factor H expression was an independent predictor of poor prognosis through risk regression analysis. Gene-set enrichment analysis identified E2 factor targets, growth 2 phase of mitosis checkpoint, spermatogenesis, mitotic spindle, deoxyribonucleic acid repair and wingless-related integration site/beta-catenin signaling were enriched with low complement factor H expression phenotype. Taking together, these findings suggested that complement factor H may be a useful biomarker for the diagnosis and prognosis of liver cancer.
Keywords
Liver cancer, complement factor H, biomarker, complement system, poor prognosis
Liver cancer, a malignant disease with high mortality, often leads to patients with poor prognosis due to high recurrence [1-3]. In recent years, the diagnosis and therapies of liver cancer are significantly improving, including adjuvant radiotherapy, surgical resection, biological therapy and other comprehensive treatments [4]. However, the relapse and metastasis of liver cancer are still steadily increasing [5]. With the research on the treatment of liver cancer, the screening of molecular targets has become a new strategy [6]. Thus, identification, discovery and search for more sensitive and specific new diagnostic and prognostic markers can help patients to make reasonable choices of therapies and to monitor regularly during treatment.
Complement system (also called complement activation pathway), an important component of innate immunity, was widely involved in host antimicrobial defense reactions, immune regulation and immunopathological damage [7-9]. It was able to be activated through three pathways, including classical, lectin and alternative pathway. When C3 convertase cleaved C3 into C3a and C3b, three complement pathways would converge into a final universal pathway and activated C3 leads to the formation of Membrane Attack Complex (MAC) to induce the disintegration of target cells such as tumor cells [10,11]. It had been reported that the complement system stimulated the inflammatory response to isolate microorganisms or toxic-molecules to attack the host by attracting neutrophils and macrophages to increase the levels of interferon’s and interleukins [12].
Complement Factor H (CFH), produced by urothelial tumor cells and macrophages, gives a selective growth advantage to tumor cells in vivo, avoiding apoptosis by escaping host immune surveillance [13,14]. However, CFH played a critical negative feedback role in controlling the alternative pathway of complement activation [15-17]. In addition, CFH prevented cells from being lysed by interfering with the complement cascade [18]. A recent study showed that CFH was able to be a biomarker for progression of cutaneous squamous cell carcinoma [19]. However, few studies had reported the role of CFH expression in clinical diagnosis and prognosis.
In this study, our team focused on the impact of CFH expression on clinical features, diagnosis and prognosis in liver cancer. Based on the clinical dataset of The Cancer Genome Atlas (TCGA), we analyzed the expression pattern of CFH at different stages and revealed its diagnostic value in liver cancer. Further, we suggested that the Overall Survival (OS) and Relapse- Free Survival (RFS) were significantly shortened in patients with low CFH expression. Indeed, its low expression was a risk factor for poor prognosis through Cox analysis. In summary, our findings indicated that CFH might be a useful biomarker for the diagnosis and prognosis of liver cancer in clinical applications.
Materials and Methods
Data collection and mining:
We obtained Ribonucleic Acid sequencing (RNAseq) of CFH and clinical information of liver cancer patient from TCGA database by using R software (version 4.0.1) and RNAseq was transformed to RNA-Seq by Expectation Maximization (RSEM) by estimating as log2 (x+1) normalized counts and used for subsequent analysis by selecting R software [20].
Gene-set enrichment analysis:
To explore the distribution of predefined genomes and determine the potential mechanism to influence the effect of CFH expression on the prognosis of Liver Hepatocellular Carcinoma (LIHC) patients, we opted for Gene Set Enrichment Analysis (GSEA) (version 4.0.3). This analysis was performed through the “h.all. v7.2. symbols.gmt” gene set in the molecular signatures database [21]. Gene-sets with a normal p value<0.05 was regarded as significantly enriched.
Statistical analysis:
R software was used for statistical analysis of all data. Data visualization was performed via using grammar of graphics (ggplot2) package. The boxplots was used to analyze the expression pattern of CFH. The chi-square test verified the correlation between the expression of CFH and clinicopathological parameters. Receiver Operating Characteristic (ROC) analysis was preformed through pROC package [22]. The ROC curve was used to evaluate the diagnostic value of CFH and the patients were divided into two groups (high and low expression) according to cut-off values [23]. Kaplan- Meier and log-rank tests were used to evaluate the effect of CFH expression on patient’s survival. Univariate and multivariate analysis were used to verify the correlation between CFH expression and OS and RFS, p<0.05 was expressed as a difference and considered statistically significant.
Results and Discussion
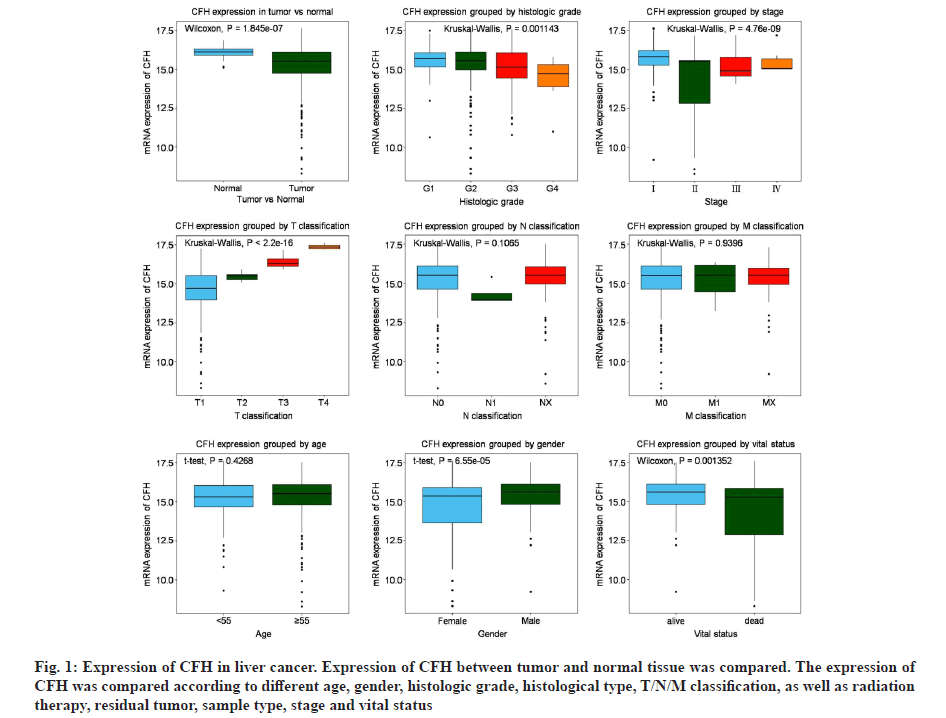
The clinical dataset of liver cancer patients were obtained from TCGA database. Table 1 lists the patient clinical characteristics, including age, gender, histological type, histologic grade, pathologic stage and Tumor/Nodes/Metastases (T/N/M) classification, as well as radiation therapy, residual tumor and vital status (Table 1). Subsequently, CFH expression analysis (fig. 1) showed that it was significantly higher in healthy tissues than in tumor tissues (p=1.845×10-7). Moreover, we also observed that CFH expression was negatively correlated with histological grades (p=0.001143), pathologic stage (p=4.760×10-9), gender (p=6.550×10-5), vital status (p=0.01352) and positively correlated with T classification (p<2.200×10-16), indicating that CFH expression was associated with tumor progression.
| Parameters | Variables | Numbers (%) |
|---|---|---|
| Age | NA | 1 (0) |
| ≥55 | 256 (67.90) | |
| <55 | 120 (32.10) | |
| Gender | Male | 255 (67.64) |
| Female | 122 (32.36) | |
| Histological type | Fibrolamellar carcinoma | 3 (0.8) |
| HCC | 367 (97.35) | |
| Hepatocholangiocarcinoma (mixed) | 7 (1.86) | |
| Histologic grade | NA | 5 (1.33) |
| G1 | 55 (14.59) | |
| G2 | 180 (47.75) | |
| G3 | 124 (32.89) | |
| G4 | 13 (3.45) | |
| Pathologic stage | NA | 22 (5.84) |
| Ⅰ | 175 (46.42) | |
| Ⅱ | 88 (23.34) | |
| Ⅲ | 86 (22.81) | |
| Ⅳ | 6 (1.59) | |
| M classification | M0 | 272 (72.15) |
| M1 | 4 (1.06) | |
| MX | 101 (26.79) | |
| N classification | NA | 1 (0) |
| N0 | 257 (68.17) | |
| N1 | 4 (1.06) | |
| NX | 115 (30.50) | |
| T classification | NA | 2 (0.53) |
| T1 | 185 (49.07) | |
| T2 | 95 (25.20) | |
| T3 | 81 (21.48) | |
| T4 | 14 (3.71) | |
| Radiation therapy | NA | 30 (7.96) |
| NO | 338 (89.66) | |
| Yes | 9 (2.39) | |
| Residual tumor | NA | 7 (1.86) |
| R0 | 330 (87.53) | |
| R1 | 17 (4.51) | |
| R2 | 1 (0) | |
| RX | 22 (5.84) | |
| RFS | NA | 33 (8.75) |
| No | 233 (61.80) | |
| Yes | 111 (29.44) | |
| Vital status | Dead | 191 (50.66) |
| Survival | 286 (75.86) | |
| CFH | NA | 6 (1.59) |
| High | 157 (41.64) | |
| Low | 214 (56.77) |
Note: NA: Not available
Table 1: Demographic and Clinical Characteristics of Tcga-Lihc Cohort
Fig. 1: Expression of CFH in liver cancer. Expression of CFH between tumor and normal tissue was compared. The expression of CFH was compared according to different age, gender, histologic grade, histological type, T/N/M classification, as well as radiation therapy, residual tumor, sample type, stage and vital status
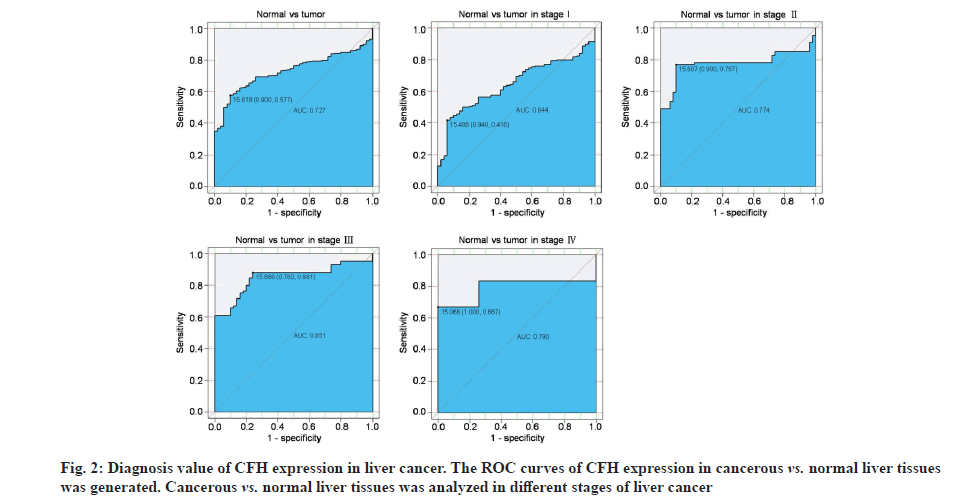
To evaluate the diagnostic capability of CFH expression, ROC curve was performed. We observed that CFH expression had modest diagnostic value (Area Under the Curve (AUC)=0.727; fig. 2) and it can also distinguish non-cancerous tissues from stage I disease (AUC=0.644), stage II disease (AUC=0.774), stage III disease (AUC=0.851) and stage IV (AUC=0.790). Additionally, we also observed that the low expression of CFH was related to the patient’s clinical characteristics (Table 2), including gender (p=0.002), histologic grade (p=0.025), pathologic stage (p=0.0001), T classification (p=0.0001), RFS (p=0.014) and worse survival (p=0.0001).
| CFH | ||||||||
|---|---|---|---|---|---|---|---|---|
| Parameters | Variables | Numbers | High | Probability (%) | Low | Probability (%) | X2 | p-value |
| Age | ≥55 | 256 | 117 | 72.22 | 139 | 64.95 | 2.242 | 0.134 |
| <55 | 120 | 45 | 27.78 | 75 | 35.05 | |||
| Gender | Male | 255 | 124 | 76.07 | 131 | 61.21 | 9.333 | 0.002 |
| Female | 122 | 39 | 23.93 | 83 | 38.79 | |||
| Histological type | Fibrolamellar carcinoma | 3 | 1 | 0.61 | 2 | 0.93 | 2.57 | 0.277 |
| HCC | 367 | 161 | 98.77 | 206 | 96.26 | |||
| Hepatocholangiocarcinoma (mixed) | 7 | 1 | 0.61 | 6 | 2.81 | |||
| Histologic grade | G1 | 55 | 29 | 18.01 | 26 | 12.32 | 9.383 | 0.025 |
| G2 | 180 | 86 | 53.42 | 94 | 44.55 | |||
| G3 | 124 | 43 | 26.71 | 81 | 38.89 | |||
| G4 | 13 | 3 | 1.86 | 10 | 4.74 | |||
| Pathologic stage | Ⅰ | 175 | 98 | 64.05 | 77 | 38.12 | 25.61 | 0.001 |
| Ⅱ | 88 | 22 | 14.38 | 66 | 32.67 | |||
| Ⅲ | 86 | 31 | 20.26 | 55 | 27.23 | |||
| Ⅳ | 6 | 2 | 1.31 | 4 | 1.98 | |||
| M classification | M0 | 272 | 120 | 73.62 | 152 | 71.03 | 0.448 | 0.799 |
| M1 | 4 | 2 | 1.23 | 2 | 0.93 | |||
| MX | 101 | 41 | 25.15 | 60 | 28.04 | |||
| N classification | N0 | 257 | 114 | 69.94 | 143 | 67.14 | 3.193 | 0.203 |
| N1 | 4 | 0 | 0 | 4 | 1.88 | |||
| NX | 115 | 49 | 30.06 | 66 | 30.99 | |||
| T classification | T1 | 185 | 44 | 27.33 | 141 | 65.89 | 23.66 | 0.001 |
| T2 | 95 | 22 | 13.66 | 73 | 34.11 | |||
| T3 | 81 | 81 | 50.31 | 0 | 0 | |||
| T4 | 14 | 14 | 8.7 | 0 | 0 | |||
| Radiation therapy | NO | 338 | 170 | 96.59 | 168 | 98.25 | 0.816 | 0.366 |
| Yes | 9 | 6 | 3.41 | 3 | 1.75 | |||
| Residual tumor | R0 | 330 | 140 | 86.96 | 190 | 90.91 | 6.985 | 0.072 |
| R1 | 17 | 12 | 7.45 | 5 | 2.39 | |||
| R2 | 1 | 1 | 0.62 | 0 | 0 | |||
| RX | 22 | 8 | 4.97 | 14 | 6.7 | |||
| RFS | No | 233 | 108 | 75 | 125 | 62.5 | 5.985 | 0.014 |
| Yes | 111 | 36 | 25 | 75 | 37.5 | |||
| Vital status | Dead | 191 | 25 | 15.34 | 66 | 30.84 | 12.15 | 0.001 |
| Survival | 286 | 138 | 84.66 | 148 | 69.16 | |||
Table 2: Correlation Between the Expressions of CFH and the Clinic Pathologic Characteristics in Liver Cancer
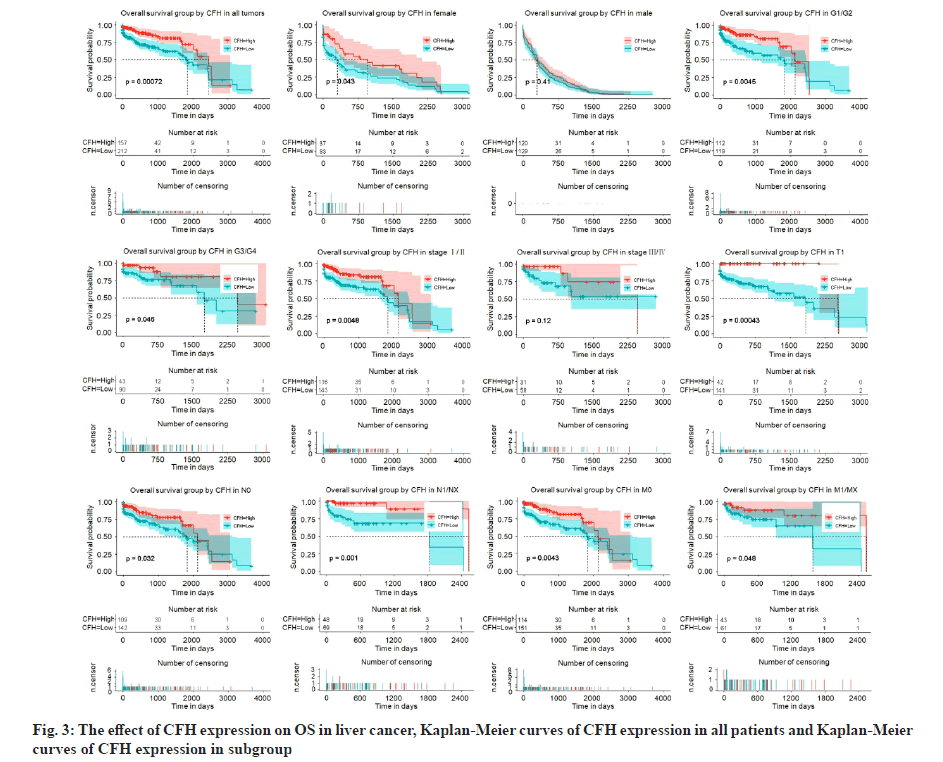
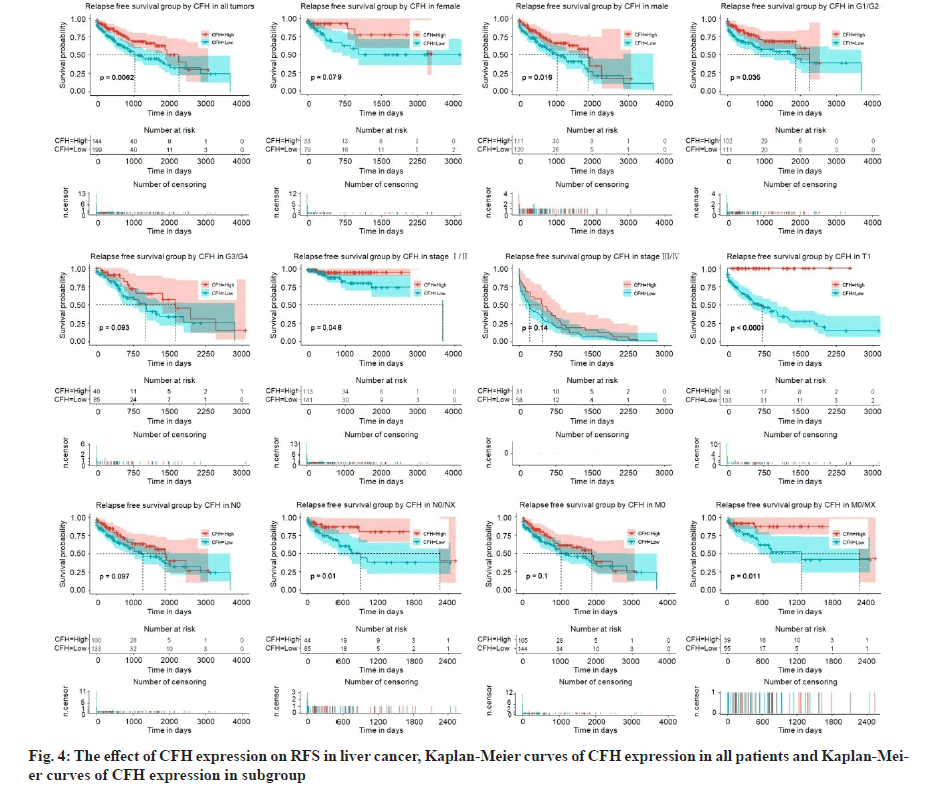
We had previously shown that CFH expression was associated with poor survival. To assess the effect of CFH expression on patient survival, we constructed Kaplan-Meier curves. We found that patients with low expression of CFH had lower OS levels (p=0.00072) and subgroups analysis also showed that low CFH expression decreased the OS in liver cancer cases of histologic grade, G1: Well differentiated (low grade)/ G2: Moderately differentiated (intermediate grade), G3: Poorly differentiated (high grade)/G4: Undifferentiated (high grade), stage I/II, T1, N0, N1/NX, M0 and M1/MX (fig. 3). Moreover, patients with low CFH expression had poor RFS (p=0.0062) and subgroups analysis also showed that low CFH expression decreased the RFS in liver cancer cases of histologic grade G1/G2, stage I/II, T1, N1/NX and M1/MX (fig. 4).
Low CFH expression is an independent risk factor for prognostic among patients with liver cancer. We selected potential variables that were significant in univariate analysis to conduct multivariable Cox analysis to assess the prognostic significance of CFH expression (Table 3 and Table 4). We found that low CFH is an independent risk factor for poor OS (Hazard Ratio (HR)=2.190, 95 % Confidence Interval (CI): 1.19-4.02, p=0.011 and RFS (HR=1.892, 95 % CI:1.21-2.37, p=0.038).
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| HR | CI95 | p value | HR | CI95 | p value | |
| Age | 0.881 | 0.57-1.37 | 0.572 | |||
| Gender | 0.998 | 0.80-1.24 | 0.994 | |||
| Histological type | 0.500 | 0.12-2.07 | 0.340 | |||
| Histologic grade | 0.957 | 0.72-1.27 | 0.759 | |||
| Pathologic stage | 1.758 | 1.40-2.20 | 0.001 | 1.736 | 1.37-2.20 | 0.001 |
| M classification | 0.962 | 0.75-1.24 | 0.764 | |||
| N classification | 0.93 | 0.72-1.19 | 0.555 | |||
| T classification | 1.286 | 1.00-1.65 | 0.050 | 1.090 | 0.77-1.55 | 0.632 |
| Radiation therapy | 1.706 | 0.63-4.60 | 0.997 | |||
| Residual tumor | 0.993 | 0.86-1.15 | 0.994 | |||
| CFH | 2.210 | 1.38-3.54 | 0.001 | 2.190 | 1.19-4.02 | 0.011 |
Table 3: Univariate and Multivariate Analysis of OS in Patients With Liver Cancer
| Univariate analysis | Multivariate analysis | |||||
|---|---|---|---|---|---|---|
| HR | CI95 | p value | HR | CI95 | p value | |
| Age | 0.900 | 0.61-1.34 | 0.600 | |||
| Gender | 1.599 | 1.02-2.51 | 0.042 | 2.134 | 1.34-3.41 | 0.001 |
| Histological type | 1.725 | 0.47-6.34 | 0.411 | |||
| Histologic grade | 1.240 | 0.97-1.58 | 0.086 | |||
| Pathologic stage | 4.373 | 3.44-5.56 | 0.000 | 5.014 | 3.75-6.70 | 0.001 |
| M classification | 0.874 | 0.69-1.10 | 0.252 | |||
| N classification | 0.924 | 0.74-1.15 | 0.477 | |||
| T classification | 1.357 | 1.08-1.71 | 0.009 | 1.630 | 1.07-2.49 | 0.023 |
| Radiation therapy | 3.036 | 0.80-13.7 | 0.099 | |||
| Residual tumor | 0.902 | 0.67-1.21 | 0.486 | |||
| CFH | 1.737 | 1.16-2.59 | 0.007 | 1.892 | 1.21-2.37 | 0.038 |
Table 4: Univariate and Multivariate Analysis of RFS in Patients With Liver Cancer
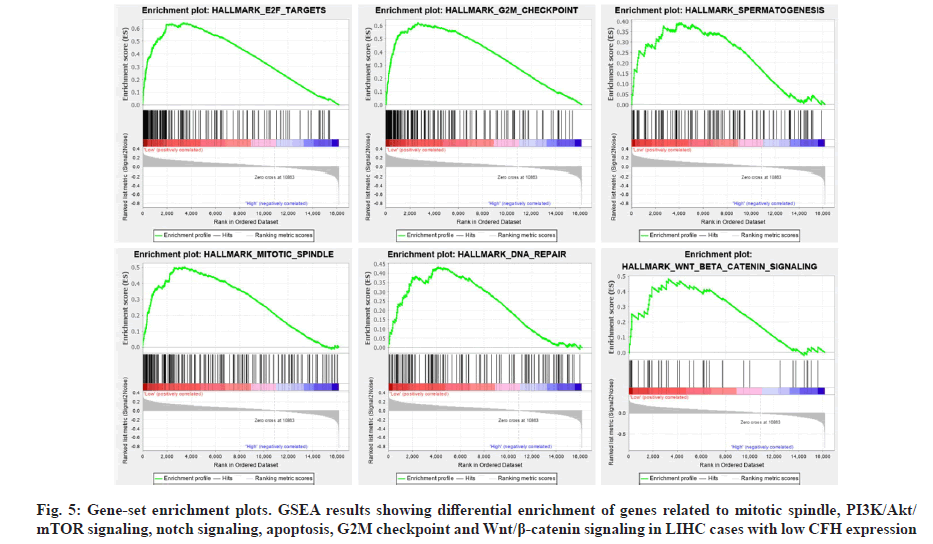
Identifying the activation of signaling pathways would facilitate a better understanding of molecular interactions, reactions and relationships, as well as disease process. To determine the signaling pathways activated in LIHC, we used GSEA to analyze the high and low CFH expression datasets. The results showed that E2 Factor (E2F) targets, Growth 2 phase of Mitosis (G2M) checkpoint, spermatogenesis, mitotic spindle, Deoxyribonucleic Acid (DNA) repair and Wingless- related integration site (Wnt)/beta (β)-catenin signaling were enriched to the low CFH expression phenotype (Table 5 and fig. 5).
| Name | ES | NES | NOM p-value |
|---|---|---|---|
| Hallmark_E2F_targets | 0.64 | 1.85 | 0.008 |
| Hallmark_G2M_checkpoint | 0.62 | 1.84 | 0.014 |
| Hallmark_spermatogenesis | 0.39 | 1.58 | 0.017 |
| Hallmark_ mitotic_spindle | 0.50 | 1.70 | 0.024 |
| Hallmark_ DNA_repair | 0.43 | 1.64 | 0.035 |
| Hallmark_Wnt_beta_catenin_signaling | 0.48 | 1.56 | 0.039 |
Table 5: Gene Set Enrichment Analysis in Low CFH Expression Phenotype Among Liver Cancer
In this study, we revealed that CFH was down-regulated in liver cancer and found that low CFH expression was relevant to histological grades, pathologic stage, T classification, patient’s gender and survival status. The ROC curve showed that CFH expression had excellent clinical diagnostic value. Through the survival curve, we observed that patients with low CFH expression had a worse OS and RFS. Univariate and multivariate Cox regression analysis confirmed that CFH is an independent predictor of poor prognosis in patients with liver cancer.
As an important negative regulator in the alternative pathway, CFH played a critical role in the activation of the alternative pathway, target cell binding and amplification [18,26-28]. It has been reported that CFH was considered to be a functional role for tumor cells to escape from complement-mediated cytotoxicity, including lung cancer [29], ovarian cancer [30], bladder cancer [31] and glioblastoma [32]. Recent studies have shown that CFH controls the stemness of liver cancer cells via Liver Suppressor Factor 1 (LSF-1) and CFH- deficient mice had spontaneous liver tumors due to T cell infiltration and neutropenia [33,34]. This means that CFH down-regulated, plays an important role in the development of liver tumors. Consistent with our findings, CFH was down-regulated in liver cancer and the expression of CFH gradually increased with the worsening of T classification (fig. 1). Interestingly, patients with low CFH expression can hardly survive to T3 and T4 (Table 2), suggesting that CFH was related to liver cancer progression. Thus, we speculate that liver cancer cells protect themselves from complement- mediated cell killing by affecting CFH, but the mechanism needs further study.
CFH also had other functions which might play important roles in tumor progression and tumorigenesis.Previous studies had shown that CFH combined C3b by competing with complement factor-B to inhibit the activity of C3 convertase [35]. Additionally, it regulated cells adhesion by binding to cell surface receptors (Cluster of Differentiation (CD) 11b (CD11b)/CD18) to promote their proliferation, including some endothelial cells and tumor cells [36]. The over-expression of Complement Factor H-Related protein 3 (CFHR3) promoted apoptosis of Hepatocellular Carcinoma (HCC) cells by inhibiting the Phosphoinositide 3-Kinase (PI3K)/protein kinase B (Akt)/mammalian Target of Rapamycin (mTOR) signaling pathway [37], which indicated that CFH was extremely important for the occurrence and progression of liver cancer. In fact, our results indicate that CFH expression is related to liver cancer progression and malignant tumors and the implicit mechanism may be linked to E2F targets, G2M checkpoint, spermatogenesis, mitotic spindle, DNA repair and Wnt/β-catenin signaling as GSEA identified.
Considering the association with immune response and anti-tumor therapy, obviously, that tumor cells were regulated by many new antigens that might be recognized by the immune system [38]. It had been reported that CFH was a barrier to monoclonal antibody therapy in ovarian cancer, because it protected tumor cells from being attacked by the immune system [39]. Additionally, it promoted the progression of skin squamous cell carcinoma by regulating immune surveillance, which indicated that it could be used as indicator of the disease’s progression and possible therapeutic targets [40]. Recently, abnormal CFH expression (mutation or deletion) has been associated with poor prognosis of many tumors, including gallbladder cancer [41] and lung adenocarcinoma [42]. In this study, our findings revealed that low CFH expression reduced OS and RFS in patients with liver cancer. Furthermore, we also found that it impacted patients OS at G1/G2, stage I/II, T1, N0, N1/NX, M0, M1/MX stage and RFS at G1/G2, stage I/II, T1, N1/NX and M1/MX stages. These indicated that CFH expression was specific in predicting the prognosis of patients, which was conducive to accurate clinical treatment of patients.
To our knowledge, this is the first report of the effect of CFH expression on the clinical features and poor prognostic of patients with liver cancer. We revealed that it has good clinical diagnostic value in patients with liver cancer and is a risk factor of poor prognosis. However, we need to further determine specific mechanisms between low CFH expression and poor prognosis in the future, so as to provide patients with more treatment options and supervision strategies.
Funding:
This work was supported by grants from 2016YFD0501001 (National Key Research and Development Program of China) and 2016YFC1201602 (National Key Research and Development Program of China).
Author’s contributions:
Chaoxiang Lv and Qiqi Zhang contributed equally to this work.
Acknowledgements:
Authors would like to thank Dr. Lichun Wang for providing R technical analysis and writing guidance.
Conflict of interests:
The authors declared that there were no conflicts of interest.
References
- Sia D, Villanueva A, Friedman SL, Llovet JM. Liver cancer cell of origin, molecular class and effects on patient prognosis. Gastroenterology 2017;152(4):745-61.
[Crossref] [Google Scholar] [PubMed]
- Sun JH, Luo Q, Liu LL, Song GB. Liver cancer stem cell markers: Progression and therapeutic implications. World J Gastroenterol 2016;22(13):3547-57.
[Crossref] [Google Scholar] [PubMed]
- Lee JH, Suh JH, Choi SY, Kang HJ, Lee HH, Ye BJ, et al. Tonicity-responsive enhancer-binding protein promotes hepatocellular carcinogenesis, recurrence and metastasis. Gut 2019;68(2):347-58.
[Crossref] [Google Scholar] [PubMed]
- Piron L, Cassinotto C, Guiu B. Prise en charge des tumeurs malignes du foie en radiologie interventionnelle. Presse Méd 2019;48(10):1156-68.
- Cheng Z, Li X, Ding J. Characteristics of liver cancer stem cells and clinical correlations. Cancer Lett 2016;379(2):230-8.
[Crossref] [Google Scholar] [PubMed]
- Cervello M, McCubrey JA, Cusimano A, Lampiasi N, Azzolina A, Montalto G. Targeted therapy for hepatocellular carcinoma: Novel agents on the horizon. Oncotarget 2012;3(3):236-60.
[Crossref] [Google Scholar] [PubMed]
- Reichhardt MP, Meri S. Intracellular complement activation-An alarm raising mechanism? Semin Immunol 2018;38:54-62.
[Crossref] [Google Scholar] [PubMed]
- Arbore G, Kemper C, Kolev M. Intracellular complement the complosome in immune cell regulation. Mol Immunol 2017;89:2-9.
[Crossref] [Google Scholar] [PubMed]
- Pereira WL, Reiche EM, Kallaur AP, Kaimen-Maciel DR. Epidemiological, clinical, and immunological characteristics of neuromyelitis optica: A review. J Neurol Sci 2015;355(1-2):7-17.
[Crossref] [Google Scholar] [PubMed]
- Irmscher S, Döring N, Halder LD, Jo EA, Kopka I, Dunker C, et al. Kallikrein cleaves C3 and activates complement. J Innate Immun 2018;10(2):94-105.
[Crossref] [Google Scholar] [PubMed]
- Khan MA, Assiri AM, Broering DC. Complement and macrophage crosstalk during process of angiogenesis in tumor progression. J Biomed Sci 2015;22(1):1-9.
[Crossref] [Google Scholar] [PubMed]
- Hoth JJ, Wells JD, Jones SE, Yoza BK, McCall CE. Complement mediates a primed inflammatory response after traumatic lung injury. J Trauma Acute Care Surg 2014;76(3):601-9.
[Crossref] [Google Scholar] [PubMed]
- Qi P, Wu B, Guo B, Zhang C, Xu K. The complement factor H (CFH) and its related protein 2 (CFHR2) mediating immune response in large yellow croaker Larimichthyscrocea. Dev Comp Immunol 2018;84:241-9.
[Crossref] [Google Scholar] [PubMed]
- Zipfel PF, Hallström T, Hammerschmidt S, Skerka C. The complement fitness factor H: Role in human diseases and for immune escape of pathogens, like pneumococci. Vaccine 2008;26:I67-74.
[Crossref] [Google Scholar] [PubMed]
- van der Maten E, van Selm S, Langereis JD, Bootsma HJ, van Opzeeland FJ, de Groot R, et al. Alternative pathway inhibition by exogenous factor H fails to attenuate inflammation and vascular leakage in experimental pneumococcal sepsis in mice. PLoS One 2016;11(2):e0149307.
- Tateishi K, Imaoka M, Matsushita M. Dual modulating functions of thrombomodulin in the alternative complement pathway. Biosci Trends 2016;10(3):231-4.
[Crossref] [Google Scholar] [PubMed]
- Simon N, Friedrich O, Kappes B. Quantification of human complement factor H binding to asexual malaria blood stages by an enzyme-linked immunosorbent assay. Vaccine 2018;36(12):1545-7.
[Crossref] [Google Scholar] [PubMed]
- Parente R, Clark SJ, Inforzato A, Day AJ. Complement factor H in host defense and immune evasion. Cell Mol Life Sci 2017;74(9):1605-24.
[Crossref] [Google Scholar] [PubMed]
- Riihilä PM, Nissinen LM, Ala-Aho R, Kallajoki M, Grénman R, Meri S, et al. Complement factor H: a biomarker for progression of cutaneous squamous cell carcinoma. J Invest Dermatol 2014;134(2):498-506.
[Crossref] [Google Scholar] [PubMed]
- Jiao Y, Fu Z, Li Y, Zhang W, Liu Y. Aberrant FAM64A mRNA expression is an independent predictor of poor survival in pancreatic cancer. PLoS One 2019;14(1):e0211291.
- Wagih O. ggseqlogo: A versatile R package for drawing sequence logos. Bioinformatics 2017;33(22):3645-7.
[Crossref] [Google Scholar] [PubMed]
- Robin X, Turck N, Hainard A, Tiberti N, Lisacek F, Sanchez JC, et al. pROC: An open-source package for R and S+ to analyze and compare ROC curves. BMC Bioinformatics 2011;12(1):1-8.
[Crossref] [Google Scholar] [PubMed]
- Jiao Y, Li Y, Fu Z, Hou L, Chen Q, Cai Y, et al. OGDHL expression as a prognostic biomarker for liver cancer patients. Dis Markers 2019;2019.
[Crossref] [Google Scholar] [PubMed]
- Aufhauser Jr DD, Sadot E, Murken DR, Eddinger K, Hoteit M, Abt PL, et al. Incidence of occult intrahepatic metastasis in hepatocellular carcinoma treated with transplantation corresponds to early recurrence rates after partial hepatectomy. Ann Surg 2018;267(5):922-8.
[Crossref] [Google Scholar] [PubMed]
- Jiao Y, Li Y, Lu Z, Liu Y. High trophinin-associated protein expression is an independent predictor of poor survival in liver cancer. Dig Dis Sci 2019;64(1):137-43.
[Crossref] [Google Scholar] [PubMed]
- Makou E, Herbert AP, Barlow PN. Functional anatomy of complement factor H. Biochemistry 2013;52(23):3949-62.
[Crossref] [Google Scholar] [PubMed]
- Nakamura H, Oku K, Ogata Y, Ohmura K, Yoshida Y, Kitano E, et al. Alternative pathway activation due to low level of complement factor H in primary antiphospholipid syndrome. Thromb Res 2018;164:63-8.
[Crossref] [Google Scholar] [PubMed]
- Pathak A, Bergstrand J, Sender V, Spelmink L, Aschtgen MS, Muschiol S, et al. Factor H binding proteins protect division septa on encapsulated Streptococcus pneumoniae against complement C3b deposition and amplification. Nat Commun 2018;9(1):1-6.
[Crossref] [Google Scholar] [PubMed]
- Yoon YH, Hwang HJ, Sung HJ, Heo SH, Kim DS, Hong SH, et al. Upregulation of complement factor H by SOCS-1/3–STAT4 in lung cancer. Cancers 2019;11(4):471.
- Honda KI, Asada R, Kageyama K, Fukuda T, Terada H, Yasui T, et al. Protein complex of fibrinogen gamma chain and complement factor H in ovarian cancer patient plasma. Anticancer Res 2017;37(6):2861-6.
[Crossref] [Google Scholar] [PubMed]
- Raitanen MP, Marttila T, Nurmi M, Ala-Opas MA, Nieminen P, Aine R, et al. Human complement factor H related protein test for monitoring bladder cancer. J Urol 2001;165(2):374-7.
[Crossref] [Google Scholar] [PubMed]
- DeCordova S, Abdelgany A, Murugaiah V, Pathan AA, Nayak A, Walker T, et al. Secretion of functionally active complement factor H related protein 5 (FHR5) by primary tumour cells derived from glioblastoma multiforme patients. Immunobiology 2019;224(5):625-31.
[Crossref] [Google Scholar] [PubMed]
- Seol HS, Lee SE, Song JS, Rhee JK, Singh SR, Chang S, et al. Complement proteins C7 and CFH control the stemness of liver cancer cells via LSF-1. Cancer Lett 2016;372(1):24-35.
[Crossref] [Google Scholar] [PubMed]
- Laskowski J, Renner B, Pickering MC, Serkova NJ, Smith-Jones PM, Clambey ET, et al. Complement factor H–deficient mice develop spontaneous hepatic tumors. J Clin Invest 2020;130(8):4039-54.
[Crossref] [Google Scholar] [PubMed]
- Gęca A, Mazurek U, Muc-Wierzgoń M, Nowakowska-Zajdel E, Niedworok E, Ziółko E, Kokot T. The role of complement factor H in the pathogenesis of Borrelia infection. Postepy Hig Med Dosw 2012;66:501-6.
- Losse J, Zipfel PF, Józsi M. Factor H and factor H-related protein 1 bind to human neutrophils via complement receptor 3, mediate attachment to Candida albicans and enhance neutrophil antimicrobial activity. J Immunol 2010;184(2):912-21.
[Crossref] [Google Scholar] [PubMed]
- Liu H, Zhang L, Wang P. Complement factor H‑related 3 overexpression affects hepatocellular carcinoma proliferation and apoptosis. Mol Med Rep 2019;20(3):2694-702.
[Crossref] [Google Scholar] [PubMed]
- Tang H, Qiao J, Fu YX. Immunotherapy and tumor microenvironment. Cancer Lett 2016;370(1):85-90.
[Crossref] [Google Scholar] [PubMed]
- Junnikkala S, Hakulinen J, Jarva H, Manuelian T, Bjørge L, Bützow R, et al. Secretion of soluble complement inhibitors factor H and factor H-like protein (FHL-1) by ovarian tumour cells. Br J Cancer 2002;87(10):1119-27.
[Crossref] [Google Scholar] [PubMed]
- Riihilä PM, Nissinen LM, Ala-Aho R, Kallajoki M, Grénman R, Meri S, et al. Complement factor H: A biomarker for progression of cutaneous squamous cell carcinoma. J Invest Dermatol 2014;134(2):498-506.
[Crossref] [Google Scholar] [PubMed]
- Cheng ZZ, Corey MJ, Parepalo M, Majno S, Hellwage J, Zipfel PF, et al. Complement factor H as a marker for detection of bladder cancer. Clin Chem 2005;51(5):856-63.
[Crossref] [Google Scholar] [PubMed]
- Cui T, Chen Y, Knösel T, Yang L, Zöller K, Galler K, et al. Human complement factor H is a novel diagnostic marker for lung adenocarcinoma. Int J Oncol 2011;39(1):161-8.
[Crossref] [Google Scholar] [PubMed]