- *Corresponding Author:
- Z. F. Wang
Department of Neurosurgery, China-Japan Union Hospital of Jilin University, Changchun, Jilin 130000, China
E-mail: wzf2013@jlu.edu.cn
| This article was originally published in a special issue, “Role of Biomedicine in Pharmaceutical Sciences” |
| Indian J Pharm Sci 2023:85(2) Spl Issue “124-136” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
We conducted a study to identify circular RNAs associated with invasive non-functioning pituitary adenomas and chemoresistance, as well as to construct a competitive endogenous RNA network. We performed circular RNA expression profiling of 10 specimens using RNA sequencing and identified 37 differentially expressed circular RNAs, which were validated using real-time quantitative polymerase chain reaction. We also detected 26 differentially expressed microRNAs and 388 differentially expressed messenger RNAs using microarray expression profiles downloaded from GSE191113. We predicted the interaction among differentially expressed circular RNAs, differentially expressed microRNAs and differentially expressed messenger RNAs and constructed a circular RNA-microRNA-messenger RNA regulatory network. Our comprehensive bioinformatic analysis revealed the regulatory pathways associated with PA invasion and chemoresistance. Enrichment analysis of differentially expressed circular RNAs revealed their association with non-functioning pituitary adenomas invasion and drug resistance. We established a protein-protein interaction network and identified four hub genes (suppressor of cytokine signaling 3, leptin receptor, interleukin 6 receptor and protein tyrosine phosphatase receptor-type C). Overall, our study provides valuable insights into the invasive and drug-resistance nature of non-functioning pituitary adenomas and could be useful in identifying potential biomarkers or therapeutic targets for invasive non-functioning pituitary adenomas
Keywords
Pituitary adenoma, hub genes, protein-protein interaction network, polymerase chain reaction, circular ribonucleic acids
Pituitary Adenomas (PAs) are the most prevalent intracranial neoplasms, accounting for up to 15 % of all intracranial tumors[1]. In recent years, because of the advances in diagnostic technologies with improved sensitivity, the detection rate of PA has increased[1,2]. According to the type of hormone secretion, PAs can be divided into functioning and Non-Functioning Pituitary Adenomas (NFPAs). NFPAs account for 14 %-54 % of PAs[3,4]. Although PAs are benign cancers, 25 %-55 % of PAs exhibit invasive biological behavior and can infiltrate the cavernous sinuses, dural membrane, skull and other surrounding brain tissue and encircle vital structures such as the internal carotid artery[5]. Surgical resection remains the first-line treatment for symptomatic patients with NFPAs; however, the high rate (~50 %) of incomplete resection owing to the hindrances imposed by invasive NFPAs is a major concern as it leads to reoperation or adjuvant therapy[6]. It is still worse as a significant proportion of aggressive pituitary tumors are chemoresistant[7]. Therefore, to improve the clinical diagnosis and treatment of PAs, studying its invasion and chemoresistance mechanism is crucial.
Circular Ribonucleic Acids (circRNAs) are a special class of RNA with a closed-loop structure and without 5' terminal cap and 3ʹ terminal poly (A) tail. They are mainly located in the cytoplasm or stored in exosomes, unaffected by RNA exonucleases and are stably expressed[8]. Because of their diverse biological functions, circRNAs have gained attention. Recent advances in sequencing technologies have led to efficient detection of circRNAs and therefore, increased understanding of their formation mechanism and biological functions. It has been confirmed that circRNAs can be obtained by reverse splicing messenger RNAs (mRNAs). They have been identified as the predominant transcript isoform in several human genes and shown to participate in gene expression in diverse cell types[9]. CircRNAs can bind and adsorb microRNAs (miRNAs); thus, they lower miRNA activity and indirectly upregulate miRNA-related target gene expression[10]. An increasing number of studies have shown that circRNAs may have an essential effect on biological processes, especially in oncogenesis, tumor progress[11-15]. For instance, hsa_circ_0001564 has been confirmed to regulate osteosarcoma proliferation and apoptosis by acting as miRNA sponge[14]. In addition, circRNA has been found to play an important role in drug resistance of various tumors such as urinary system tumors and pancreatic cancer[16,17]. However, there are few studies on invasiveness and drug resistance of PAs and circRNA.
In this study, we aimed to identify Differentially Expressed (DE) circRNAs (DEcircRNAs) in invasive PAs and explore the pathways through which they might affect the invasiveness or chemoresistance of PAs.
Materials and Methods
Tissue samples for RNA sequencing:
10 specimens were obtained from 10 patients with PAs, who were treated surgically at China-Japan Union Hospital (Changchun, China) between October 2007 and July 2014. Knosp classification grade III and IV and/or Hardy-Wilson classification grade IV PAs were considered invasive[18,19]. The type of tumor was determined based on pathologic diagnosis and preoperative pituitary hormone examination results. Based on the above criteria, the 10 NFPA tumor tissues were divided into two groups: 5 in the non-invasive group, indicated as N256, N261, N301, N209 and N318, and five in the invasive group, indicated as N181, N195, N255, N278 and N283. All samples were immediately flash-frozen and stored in liquid nitrogen or at -80°.
The Ethics Committee of China-Japan Union Hospital reviewed and approved this study (2014-004). Written informed consent was obtained from individual patients or their families before the procedure.
CircRNA sequencing (circRNA-seq):
The total RNA was isolated from patient samples using the RNeasy Mini kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The Deoxyribonucleic Acid (DNA) in the purification column was removed using Deoxyribonuclease (DNase). The total RNA was digested with Ribonuclease R (RNase R) (Epicenter Technologies, Madison, Wisconsin (WI)) at 37° for 1 h and then the RNA library was prepared. The NEBNext Ultra™ Directional RNA Library Prep kit (E7420L; New England Biolabs, Ipswich, Massachusetts (MA)) for Illumina was used to prepare the circRNA-seq library. Paired-end sequencing was performed using an Illumina HiSeq 3000 System (Illumina, Inc., San Diego, California (CA)) at Amogene Biotech (Xiamen, China).
RNA-seq data analysis:
We mapped the sequenced reads to the Human Genome version 19 (HG19), reference genome and then utilized the latest versions of CIRI2, find_circ, CIRCexplorer2, MapSplice, and circRNA_finder tools to predict circRNAs. To quantify overlapping circRNAs, the number of reads from the back-splicing junction for each circRNA was extracted and combined. To determine the abundance of circRNAs, we scaled the reads to Revolutions Per minute (RPM) value (the number of reads corresponding to the number of reads sequenced per million reads), at least, 10 % of the samples with RPM larger than 0.1 were retained for DE analysis. We used principal component analysis to explore transcriptional heterogeneity between the two groups of specimens. The edgeR package was used to identify DEcircRNAs between the invasive and non-invasive groups[20]. CircRNAs with a Fold Change (FC)≥2 and p<0.05 were considered as DEcircRNAs. Next, the results of DEcircRNAs from the five software tools were merged and analyzed. Finally, DEcircRNAs with consistent differential expression patterns among the five programs were retained.
Identification of DEmRNAs and DEmiRNAs:
The GSE191113 datasets for invasive NFPAs were downloaded from the Gene Expression Omnibus (GEO) database. This dataset was used to screen for DEmRNAs and DEmiRNAs. The DEseq2 package in R software was used to analyze mRNA and miRNA expression data[21]. DEmRNAs were selected using the criteria of log |FC|>1 and p<0.05. DEmiRNAs were selected using the criteria of log |FC|>0 and p<0.05.
Construction of circRNA-miRNA pairs:
We used the Cancer-Specific CircRNA Database (CSCD) to predict the target miRNAs of DEcircRNAs[22,23]. Subsequently, we took the intersection of the predicted miRNAs with the DEmiRNAs. Next, the circRNA-miRNA pairs were constructed.
Construction of miRNA-mRNA pairs:
The miRNAs obtained in the previous step were used to predict their downstream regulatory genes in the microRNA Target Prediction Database (miRDB) and TargetScan databases[24,25]. To increase the reliability of the prediction results, we intersected the DEmRNAs of GSE191113 with the predicted mRNAs and miRNA-mRNA pairs were then formed.
Construction of the circRNA-miRNA-mRNA network:
Based on the pairing of miRNA-mRNA and miRNA-circRNA, we constructed a circRNA-miRNA-mRNA network. This network was displayed using Cytoscape (ver. 3.9.0)[26].
Enrichment analyses:
The potential main functions of the DEmRNAs were analyzed using the Gene Ontology (GO) analysis. To predict the potential principal functional pathways of invasive NFPAs, the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was performed using the clusterProfiler package in R[27]. A p-value of 0.05 was considered to indicate significant GO or KEGG terms. We also performed Gene Set Enrichment Analysis (GSEA) of the KEGG pathway and the set of drug-resistant genes set from Enrichr to assess whether the corresponding pathway was in an activated or inhibited state[28,29].
Construction of a Protein-Protein Interaction (PPI) network and screening of hub genes:
Based on the obtained DEmRNAs, the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (https://string-db.org/) was used to construct a PPI network[30]. Visualization was performed using Cytoscape software (version 3.9.0). The Molecular Complex Detection (MCODE) app was used to select hub gene modules from the PPI network[31].
Validation of the identified circRNAs and mRNAs using quantitative Reverse Transcription-Polymerase Chain Reaction (RT-qPCR):
To test the validity of the RNA-seq and the results of the bioinformatic analysis, we performed RT-qPCR of all samples from both groups. Five circRNAs were selected for validation and we used the SYBR Green assay (ArrayStar Inc., Rockville, Maryland (MD)). The primers used for the RT-qPCR are listed in Table 1. We performed three replicate experiments using beta (β)-actin as the internal reference. We used the 2-ΔΔCT method to determine the relative expression of each circRNA.
| Primers | Sequences (from 5′ to 3′) |
|---|---|
| hsa_circ_0105129 Forward primer | GGAAAATGCCAGCCTAGTGT |
| hsa_circ_0105129 Reverse primer | CAACATTCAGTCTGCCAGCA |
| hsa_circ_0109731 Forward primer | AGTGAGACCTCCCAAAAGG |
| hsa_circ_0109731 Reverse primer | CTCCAAATCCAGAGCATCAG |
| hsa_circ_0093733 Forward primer | AGTGGACAGGGAAGAACA |
| hsa_circ_0093733 Reverse primer | CTGGCTCCAGGAGACTAA |
| hsa_circ_0031563 Forward primer | TAGATTAGCCCAAGTGGT |
| hsa_circ_0031563 Reverse primer | ACAGAGGTTTCCTTCATAG |
| hsa_circ_0133631 Forward primer | GAAACTGGAAACCCTATCA |
| hsa_circ_0133631 Reverse primer | GAGACAAGTGGCTCATCAC |
Table 1: Primers used in the present study.
Statistical analysis:
For the comparison of circRNA and mRNA expression levels between the groups, Student’s t-test (two-tailed) was used when the data followed a normal distribution; otherwise, the nonparametric Mann-Whitney U test was used. All statistical analyses were performed using Prism 7 (GraphPad Software, San Diego, CA). Differences were considered statistically significant atp<0.05.
Results and Discussion
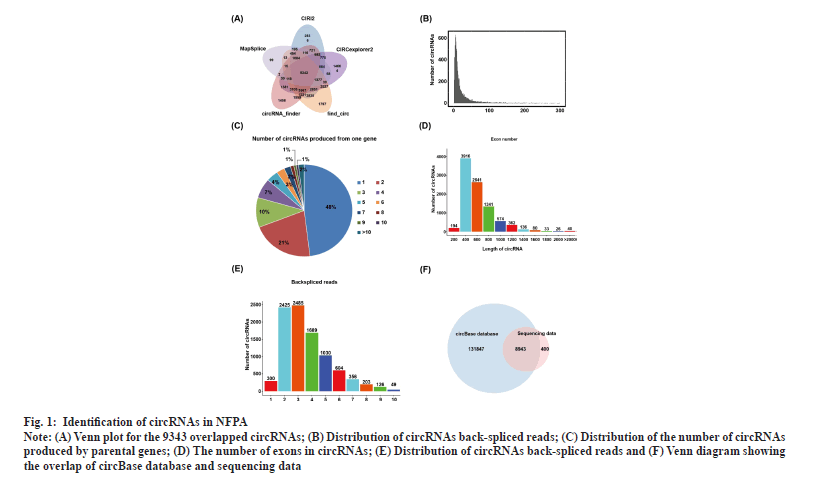
Identification of circRNAs in NFPAs was shown in fig. 1. RNA-seq of invasive and non-invasive NFPA samples was performed to explore the differential expression of circRNAs in invasive NFPAs. To comprehensively screen reliable circRNAs in NFPA samples, the sequencing dataset was analyzed using five software tools (find_circ, CIRCexplorer2, circRNA_finder, CIRI2 and MapSplice)[25,32-36]. In total, 9343 highly credible circRNAs were identified using all five tools and the number of circRNAs that contained at least two unique back-spliced reads was 9341, accounting for nearly 99.98 % of the total circRNAs (fig. 1A and fig. 1B). As one gene could generate multiple circRNAs through the alternative back-spliced mechanism, we also examined the contribution of alternative back-splicing to the diversity of circRNAs in PAs. The data revealed that ~52 % of circRNAs were derived from repeated parental genes (fig. 1C). The majority of circRNAs contained 2-6 exons (fig. 1D) and the length of circRNA ranged from 200 to 1200 base pair (bp) in NFPAs (fig. 1E). Alignment of these circRNAs with the circBase database containing 140 790 human circRNAs revealed that 8943 circRNAs were embedded in the circBase database and 400 were discovered in this study for the first time (fig. 1F).
Fig. 1: Identification of circRNAs in NFPA.
Note: (A) Venn plot for the 9343 overlapped circRNAs; (B) Distribution of circRNAs back-spliced reads; (C) Distribution of the number of circRNAs
produced by parental genes; (D) The number of exons in circRNAs; (E) Distribution of circRNAs back-spliced reads and (F) Venn diagram showing
the overlap of circBase database and sequencing data.
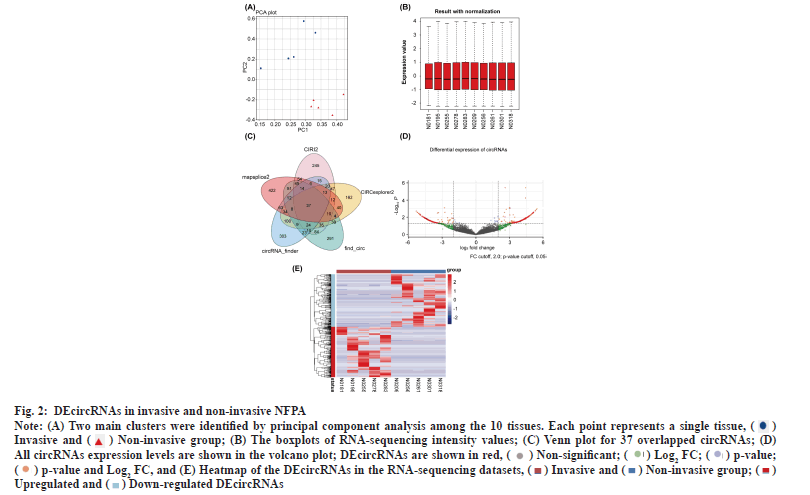
DEcircRNAs in invasive and non-invasive NFPAs was shown in fig. 2. Principal component analysis clearly distinguished the two groups based on the expression of circRNAs (fig. 2A). The boxplot shows that the overall differences in the test sample data were not significant and were comparable (fig. 2B). The differential analysis using edgeR followed by the intersection of the circRNAs obtained using the five software tools revealed 37 significantly DEcircRNAs (fig. 2C). Of these, 24 DEcircRNAs were upregulated and the rest were downregulated in invasive NFPAs (Table 2 and Table 3). The volcano plot displayed differences in circRNA expression between the invasive and non-invasive groups (fig. 2D). We developed a clustering heatmap based on DEcircRNAs (fig. 2E); it revealed a remarkable difference in circRNA expression between the groups.
Fig. 2: DEcircRNAs in invasive and non-invasive NFPA.
Note: (A) Two main clusters were identified by principal component analysis among the 10 tissues. Each point represents a single tissue,  Invasive and
Invasive and  (B) The boxplots of RNA-sequencing intensity values; (C) Venn plot for 37 overlapped circRNAs; (D)
All circRNAs expression levels are shown in the volcano plot; DEcircRNAs are shown in red,
(B) The boxplots of RNA-sequencing intensity values; (C) Venn plot for 37 overlapped circRNAs; (D)
All circRNAs expression levels are shown in the volcano plot; DEcircRNAs are shown in red, 
 and (E) Heatmap of the DEcircRNAs in the RNA-sequencing datasets,
and (E) Heatmap of the DEcircRNAs in the RNA-sequencing datasets,  Upregulated and
Upregulated and  .
.
| CircRNA ID | Regulation | Log2 FC | p | Chromosome | CircBase ID | Host gene | Length (bp) |
|---|---|---|---|---|---|---|---|
| chr1:215342542|215368435 | Up | 3.73611723 | 0.009804 | chr1 | hsa_circ_0007131 | KCNK2 | 488 |
| chr10:7262373|7327916 | Up | 3.267319548 | 7.01E-05 | chr10 | hsa_circ_0017628 | SFMBT2 | 894 |
| chr10:7285520|7327916 | Up | 2.346619647 | 1.00E-06 | chr10 | hsa_circ_0017636 | SFMBT2 | 684 |
| chr10:96005703|96006378 | Up | 2.045588475 | 0.012048 | chr10 | hsa_circ_0019225 | PLCE1 | 676 |
| chr11:15198657|15212360 | Up | 3.189563 | 0.026963 | chr11 | INSC | 291 | |
| chr12:78334099|78362482 | Up | 4.1226873 | 0.00426 | chr12 | hsa_circ_0003514 | NAV3 | 428 |
| chr13:70456415|70549934 | Up | 4.24249163 | 7.74E-05 | chr13 | hsa_circ_0100793 | KLHL1 | 730 |
| chr14:31818995|31833504 | Up | 5.041415654 | 0.000238 | chr14 | hsa_circ_0031563 | HEATR5A | 747 |
| chr14:63416851|63417277 | Up | 4.107522612 | 0.004575 | chr14 | hsa_circ_0102346 | KCNH5 | 427 |
| chr14:63468049|63483672 | Up | 4.466171524 | 0.001879 | chr14 | hsa_circ_0102352 | KCNH5 | 360 |
| chr15:49517402|49531564 | Up | 3.90406049 | 0.007017 | chr15 | hsa_circ_0007487 | GALK2 | 10884 |
| chr16:19234367|19236160 | Up | 3.189563 | 0.026963 | chr16 | hsa_circ_0105129 | SYT17 | 277 |
| chr17:3967655|3970536 | Up | 2.343851426 | 0.044861 | chr17 | hsa_circ_0106826 | ZZEF1 | 643 |
| chr18:6824761|6837413 | Up | 3.744993846 | 0.010354 | chr18 | hsa_circ_0108851 | ARHGAP28 | 421 |
| chr19:46832464|46834530 | Up | 2.944593427 | 0.000286 | chr19 | hsa_circ_0109731 | HIF3A | 390 |
| chr2:116066815|116101488 | Up | 3.050448406 | 0.032399 | chr2 | hsa_circ_0117102 | DPP10 | 211 |
| chr3:183454506|183480067 | Up | 2.574085179 | 0.048717 | chr3 | hsa_circ_0009131 | YEATS2 | 1135 |
| chr3:38263069|38271921 | Up | 3.877980446 | 0.00736 | chr3 | hsa_circ_0064871 | OXSR1 | 461 |
| chr4:95506716|95585186 | Up | 3.711987142 | 0.010202 | chr4 | hsa_circ_0127356 | PDLIM5 | 1445 |
| chr5:94204038|94248681 | Up | 3.425475647 | 0.018353 | chr5 | hsa_circ_0005540 | MCTP1 | 1086 |
| chr6:147581751|147599340 | Up | 3.546971783 | 0.014151 | chr6 | hsa_circ_0007762 | STXBP5 | 407 |
| chr6:150147400|150158599 | Up | 4.234874869 | 0.003264 | chr6 | hsa_circ_0078258 | LRP11 | 435 |
| chr7:146471363|146536996 | Up | 4.191932871 | 0.000105 | chr7 | hsa_circ_0133631 | CNTNAP2 | 305 |
| chr9:97209105|97218619 | Up | 2.867634137 | 0.025923 | chr9 | MFSD14B | 397 |
Table 2: CircRNAs Upregulated in NFPAs
| CircRNA ID | Regulation | Log2 FC | p | Chromosome | CircBase ID | Host gene | Length (bp) |
|---|---|---|---|---|---|---|---|
| chr1:190195212|190234185 | Down | -3.794029223 | 0.00864 | chr1 | hsa_circ_0111549 | FAM5C | 534 |
| chr1:57252848|57258476 | Down | -4.718839287 | 0.00088 | chr1 | hsa_circ_0113682 | C1orf168 | 944 |
| chr10:123842162|123848106 | Down | -3.218556434 | 0.02745 | chr10 | / | TACC2 | 5427 |
| chr10:55943204|55973808 | Down | -3.586121739 | 0.01302 | chr10 | hsa_circ_0093733 | PCDH15 | 626 |
| chr11:36103203|36120011 | Down | -2.825802835 | 0.04962 | chr11 | hsa_circ_0004453 | LDLRAD3 | 261 |
| chr13:51501543|51504895 | Down | -3.835628827 | 0.0073 | chr13 | hsa_circ_0005980 | RNASEH2B | 257 |
| chr14:102368056|102376027 | Down | -3.813743064 | 0.00833 | chr14 | hsa_circ_0101268 | PPP2R5C | 401 |
| chr17:53150261|53158544 | Down | -2.128649422 | 0.02003 | chr17 | hsa_circ_0044699 | STXBP4 | 478 |
| chr20:40979268|40980925 | Down | -3.621897262 | 0.01235 | chr20 | hsa_circ_0115238 | PTPRT | 305 |
| chr4:119252871|119259469 | Down | -4.716179632 | 0.00088 | chr4 | hsa_circ_0006472 | PRSS12 | 469 |
| chr4:17963526|17974508 | Down | -2.370031353 | 0.00063 | chr4 | hsa_circ_0069285 | LCORL | 276 |
| chr9:113703701|113735838 | Down | -2.458745423 | 0.03762 | chr9 | hsa_circ_0003611 | LPAR1 | 974 |
| chrX:132887509|132888203 | Down | -4.107270947 | 0.00464 | chrX | hsa_circ_0091581 | GPC3 | 695 |
Table 3: CircRNAs Downregulated in NFPAs
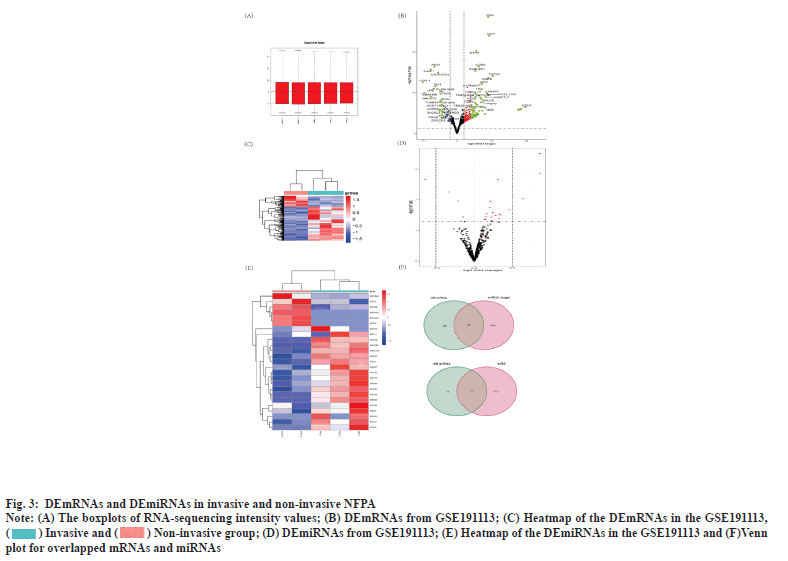
Identification of DEmRNA and DEmiRNA was shown in fig. 3. Plotting the distribution of the samples in the GSE191113 database using the boxplot demonstrated that the samples in the database had the same overall distribution (fig. 3A). Based on the aforementioned criteria, we screened 388 DEmRNAs from the GSE191113 dataset, of which 277 were upregulated and 111 downregulated (fig. 3B). Heatmaps demonstrated differences in mRNA expression between the groups (fig. 3C). We obtained 26 DEmiRNAs from GSE191113, 19 of which were up-regulated miRNAs and the rest were down-regulated (fig. 3D). Heatmaps demonstrated differences in mRNA expression between the groups (fig. 3E).
Fig. 3: DEmRNAs and DEmiRNAs in invasive and non-invasive NFPA.
Note: (A) The boxplots of RNA-sequencing intensity values; (B) DEmRNAs from GSE191113; (C) Heatmap of the DEmRNAs in the GSE191113,  ; (D) DEmiRNAs from GSE191113; (E) Heatmap of the DEmiRNAs in the GSE191113 and (F)Venn
plot for overlapped mRNAs and miRNAs.
; (D) DEmiRNAs from GSE191113; (E) Heatmap of the DEmiRNAs in the GSE191113 and (F)Venn
plot for overlapped mRNAs and miRNAs.
Construction of circRNA-miRNA and miRNA-mRNA pairing was shown in fig. 3F. We obtained 9 miRNAs by taking the intersection of the miRNAs that can be bound by DEcircRNAs predicted from CSCD with DEmiRNAs. Subsequently, we predicted the mRNAs that can be bound by the above 9 miRNAs using miRDB and TargetScan, and took the intersection with DEmRNAs to finally obtain 60 mRNAs.
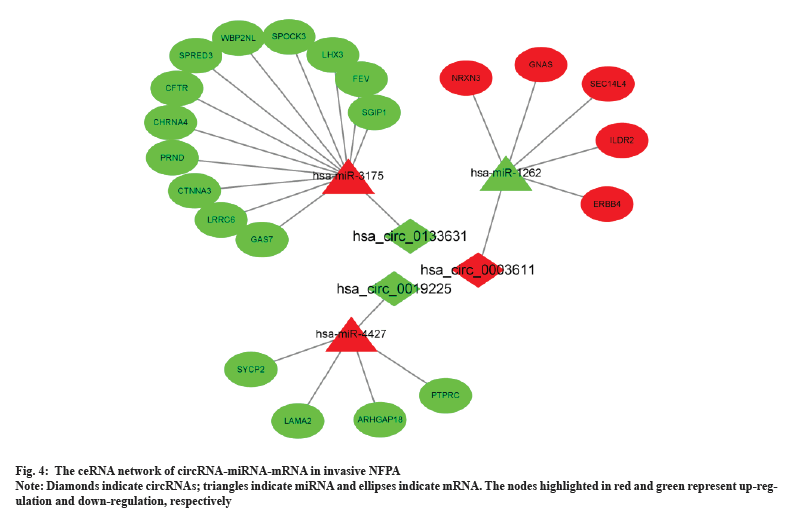
A circRNA-miRNA-mRNA network was constructed by combining miRNA-mRNA and miRNA-circRNA pairs as shown in fig. 4. The network comprised 3 circRNA nodes, 3 miRNA nodes, 21 DEmRNA nodes and 24 edges.
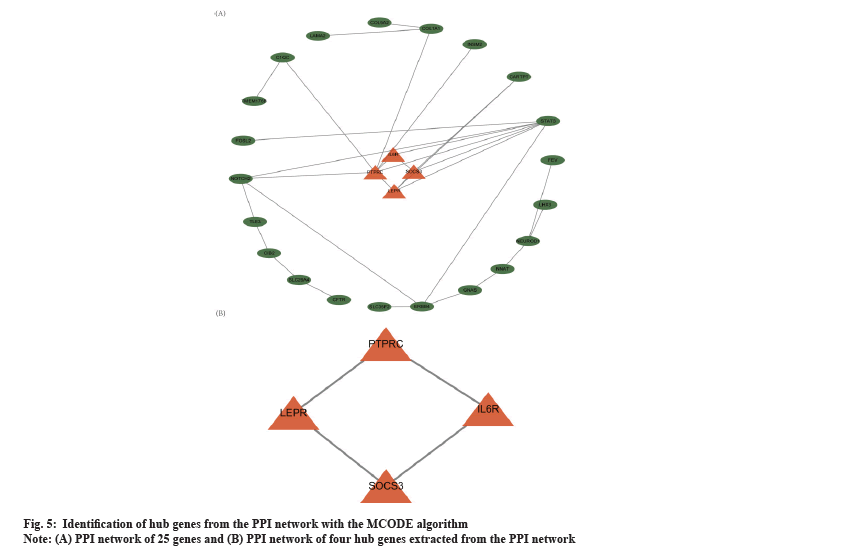
PPI network analysis was shown in fig. 5. Based on the overlapped 60 DEmRNAs, we constructed the PPI network using STRING. The PPI network had 25 nodes and 31 edges (fig. 5A). The MCODE app identified a prominent cluster at k-core of 2. This cluster contained four genes (Suppressor of Cytokine Signaling 3 (SOCS3), Leptin Receptor (LEPR), Interleukin 6 Receptor (IL6R) and Protein Tyrosine Phosphatase Receptor-type C (PTPRC)) that were considered as hub genes (fig. 5B). Finally, we obtained one circRNA-miRNA-mRNA regulatory modules (hsa_circ_0019225/hsa-miR-4427/PTPRC).
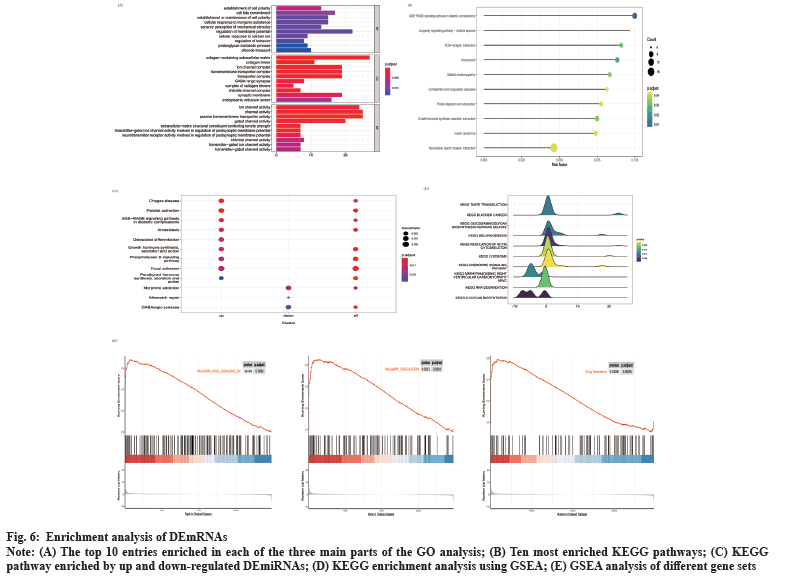
Enrichment analysis of DEmRNAs was shown in fig. 6. We performed GO and KEGG enrichment analysis for DEmRNAs. These DEmRNAs were enriched in 91 GO terms related to biological processes, cellular components and molecular functions. In fig. 6A, the top 10 GO significant terms for each classification are shown and it can be seen that the regulation of ion channels is closely related to the invasion of NFPA. A total of 49 pathways were enriched by KEGG analysis and the top 10 pathways are shown in fig. 6B. These pathways include Extracellular Matrix (ECM)-receptor interactions, complement and coagulation cascades and neuroactive ligand-receptor interaction pathways, suggesting that these pathways may also be involved in the regulation of NFPA invasiveness. We also performed KEGG enrichment analysis for the up and down-regulated differential genes, respectively, and the results are shown in fig. 6C. Among them, platelet activation, phospholipase D signaling pathway and focal adhesion pathways were activated, while mismatch repair pathway was inhibited. In addition, we performed GSEA analysis of the KEGG pathway and found that pathways such as the regulation of the actin cytoskeleton, glycosaminoglycan biosynthesis acetyl heparan sulfate and chemokine signaling pathways were activated, while pathways such as RNA degradation were inhibited (fig. 6D). By performing GSEA analysis on different gene sets, we found that HALLMARK_KRAS_SIGNALING_DN and HALLMARK_COAGULATION were also activated (fig. 6E). More interestingly, we found that the drug resistance gene set was also in the activated state (fig. 6F), indicating that the aggressive NFPA is also drug resistant.
Fig. 6: Enrichment analysis of DEmRNAs.
Note: (A) The top 10 entries enriched in each of the three main parts of the GO analysis; (B) Ten most enriched KEGG pathways; (C) KEGG
pathway enriched by up and down-regulated DEmiRNAs; (D) KEGG enrichment analysis using GSEA; (E) GSEA analysis of different gene sets.
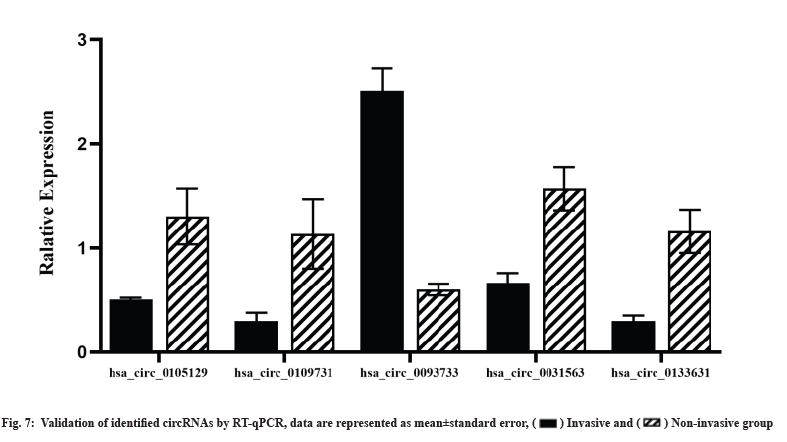
Validation of identified circRNAs using RT-qPCR was shown in fig. 7. We selected the five circRNAs for RT-qPCR validation in the original sequencing specimens to confirm the sequencing results. The expression profile of circRNAs obtained using RT-qPCR was concordant with the RNA-seq results, validating the sequencing results (fig. 7).
Invasive NFPA remains a clinical challenge because of its extensive infiltration of surrounding structures, difficulty in complete resection, poor drug therapy, high postoperative recurrence rate and poor prognosis[7,36,37]. Thus, there is an urgent need to further explore the mechanisms underlying PA invasion and chemoresistance. Here, we constructed a competitive endogenous RNA (ceRNA) network to clarify the downstream mechanisms of circRNAs in invasive NFPA. We demonstrated the actual variations of mRNAs and miRNAs in invasive PAs and identified significant DEmRNAs and DEmiRNAs by analyzing the GSE191113 database. In the present study, we constructed the first circRNA-miRNA-mRNA and PPI networks to identify four hub genes, namely, SOCS3, LEPR, IL6R and PTPRC, as the core circRNAs involved in the regulatory network. The findings were validated using RT-qPCR. Searching for pathways enriched by DEmRNAs in invasive NFPAs by GO, KEGG analysis. Also, the overall differences in gene expression between invasive and non-invasive NFPA were analyzed using GSEA.
Recent studies have reported that circRNAs are closely associated with the invasiveness and chemoresistance of tumors[15-17,38]. Previous studies have performed gene enrichment analysis based on circRNA parental genes and equated circRNA-regulated genes to their parental genes without considering the changes in mRNAs in the actual situation. To the best of our knowledge, this is the first study to identify DEcircRNAs using high-throughput sequencing of invasive NFPA samples, we identified 37 DEcircRNAs, of which 24 were upregulated and 13 were downregulated. In our study, four upregulated circRNAs (hsa_circ_0105129, hsa_circ_0109731, hsa_circ_0031563 and hsa_circ_0133631) and 1 downregulated circRNA (hsa_circ_0093733) were confirmed using RT-qPCR; this result was concordant with our RNA-seq results. The bioinformatic analysis revealed differences in circRNA expression profiles between the groups suggesting that DEcircRNAs are involved in the modulation of NFPA invasion and drug resistance mechanisms.
To clarify the downstream mechanism of circRNAs in invasive NFPA, it is essential to construct a ceRNA network. To determine the actual variation of mRNAs and miRNAs in invasive PAs and screen for significant DEmRNAs, we analyzed the GSE191113 database and constructed circRNA-miRNA-mRNA and PPI networks; the analysis enabled us to elucidate the regulatory mechanism of circRNAs in the invasiveness and chemoresistance of NFPAs for the first time. In our study, 3 circRNAs, including the one circRNAs validated using RT-qPCR, were included in the ceRNA network. These results provide evidence that circRNA plays an important regulatory role in the interaction between miRNAs and target genes, regulating many target genes by acting as a miRNA sponge to adsorb a large number of miRNAs. For example, hsa_circ_0003611 might be associated with miRNAs such as hsa-miR-1262, hsa-miR-4483, hsa-miR-1293, hsa-miR-127-5p and hsa-miR-1306-5p. Many of the target genes such as Neurexin 3 (NRXN3), Guanine Nucleotide Binding Protein, Alpha Stimulating activity polypeptide (GNAS), Immunoglobulin Like Domain Containing Receptor 2 (ILDR2) and Erb-B2 Receptor Tyrosine Kinase 4 (ERBB4) regulated by these miRNAs in the reconstructed ceRNA network are related to PAs. NRXN3 was shown to be associated with tumor aggressiveness in studies of gliomas and glioblastoma patients with high NRXN3 expression were found to have a poor prognosis[39]. By analyzing the Cancer Cell Line Encyclopedia (CCLE)-Genomics of Drug Sensitivity in Cancer (GDSC) dataset, Krushkal et al. found a significant correlation between increased GNAS expression and tumor resistance to chemotherapeutic agents[40]. Furthermore, GNAS activation significantly promoted growth and progression in a mouse model of Small Cell Lung Cancer (SCLC)[41]. ERBB4 is a member of the epidermal growth factor receptor subfamily and its high expression in colorectal cancer has been found to promote tumor progression[42]. These findings also indirectly suggest that hsa_circ_0003611 might regulate the proliferation, invasiveness and chemoresistance of PAs by acting on these genes.
To further investigate the potential regulatory mechanisms in the aggressiveness and drug resistance of NFPAs, we conducted functional and pathway enrichment analyses of DEmiRNAs. In the GO term enrichment analysis, the most closely associated with invasive and drug-resistant NFPAs was “ion channel transport”, such as calcium ions, chloride ions, etc. Our results were consistent with previous studies. Wu et al. found that DE genes in NFPAs were also enriched in ion channel transport, suggesting that ion channel regulation is involved in the regulation of PA aggressiveness[43]. It is now increasingly clear that the regulation of membrane ion channels is involved in the regulation of drug resistance. Increasing Calcium ion (Ca2+) inward flow by upregulating Transient Receptor Potential Cation Channel subfamily C member 5 (TRPC5) expressions can activate multiple signaling pathways such as Calcium ions/Calmodulins stimulate protein Kinase Kinases β (CaMKKβ)/Adenosine Monophosphate (AMP)-activated Protein Kinase (AMPK)/mammalian Target of Rapamycin (mTOR) pathway to promote and/or maintain chemoresistance[44]. It had been discovered that blocking the chloride channel protein-coding gene Transmembrane Member 16A (TMEM16A) or one of its downstream pathways, such as Extracellular Signal-Regulated Kinase 1 (ERK1) or 2, could overcome the cisplatin resistance seen in tumors with high TMEM16A expression[45]. In conclusion, ion channels are involved in the regulation of tumor invasion and drug resistance.
Furthermore, the KEGG pathway enrichment analysis revealed that pathways such as "Phospholipase D signaling pathway", "Focal adhesion" and "Mismatch repair" may be associated with the aggressiveness and drug resistance of NFPA. Among those pathways, "Phospholipase D signaling pathway" and "Focal adhesion" pathway were considered to be activated, while the “Mismatch repair” pathway was inhibited. Although there were no studies on the regulation of PA aggressiveness by the phospholipase D signaling pathway, activation of this pathway had been shown to promote tumor cell proliferation in a variety of other tumors, such as breast and prostate cancers[46,47]. Through KEGG analysis of the parental genes of DEcircRNAs in invasive PA, Wang et al. also found that they were enriched in the “Focal adhesion” pathway[15]. As a key gene in the “Focal adhesion” pathway, the expression level of Focal Adhesion Kinase (FAK) was significantly positively correlated with the invasiveness of PAs[48]. The "Mismatch repair" pathway had likewise been associated with pituitary tumour aggressiveness, with studies finding that downregulation of mismatch repair gene MutS Homolog (MSH) 6/MSH2 expression promoted pituitary tumour growth[49]. The surprising finding was that platelet activation was not only seen in the results of the KEGG analysis, but also in the GSEA analysis. A correlation between platelet activation pathways and the development of NFPA has been previously reported[50]. Wu et al. also discovered that Tumor Necrosis Factor alpha (TNF-α) expression was upregulated in aggressive NFPA and that this increased production of TNF-α could facilitate platelet activation by increasing p-integrin. The ability of immune cells to assault tumor cells was decreased when activated platelets adhered to them[51]. The above mechanisms may partially explain the aggressive mechanism of PAs. More interestingly, through GSEA, our study found for the first time that drug resistance genes set were activated in invasive NFPA, suggesting that invasive NFPA was indeed associated with drug resistance. It had been demonstrated that platelets in cancer patients undergoing chemotherapy might influence the efficacy of medication therapy for a variety of cancer types[52]. Therefore, although there is no direct evidence of platelet activation and drug resistance of pituitary tumors, our analysis results suggest a correlation between platelet activation and NFPA resistance.
To further discern the core circRNAs involved in the regulatory network, we constructed a PPI network and screened four hub genes, namely SOCS3, LEPR, IL6R and PTPRC. The PTPRC is the only one of these hub genes that contributes to the formation of the ceRNA network, which is the focus of this section. PTPRC was found to be significantly reduced in aggressive PAs and to be enriched in the "cell adhesion" pathway by Cao et al. using computational bioinformatics analysis of DNA microarray expression profiles. This finding suggested that PTPRC was significantly associated with the aggressiveness of PAs and could be used as a biomarker for aggressive pituitary tumours[53]. No studies have been reported on the association of PTPRC with drug resistance in pituitary tumours, although studies have been done in other tumours. For example, Niu et al. found that PTPRC expression was significantly reduced in drug-resistant ovarian cancer, suggesting that low expression of PTPRC promotes drug resistance and the same had been observed in breast cancer[54,55]. Thus a correlation between PTPRC and drug resistance in invasive NFPAs can be established through this study. Here, we discovered only one circRNA-miRNA hub gene axe, suggesting competitive regulation of one circRNAs with one gene in the invasive NFPA. The above regulatory axe was derived from bioinformatics analysis and their actual regulatory role in invasive NFPA still needed to be further investigated.
There are some limitations in this study. First, due to the limited number of patients, the sample size is small. Second, to obtain more circRNAs and discover new circRNAs, we performed circRNA seq instead of whole transcriptome sequencing. For the subsequent analysis of circRNA-mRNA associations, we had to resort to GEO microarray data. Finally, we only performed preliminary RT-qPCR validation of the sequencing results of circRNA and further validation of circRNA function in in vivo and in vitro experiments is still needed.
For the first time, we reported the expression of circRNAs in invasive NFPA tissues determined by high-throughput sequencing. We selected DEmRNAs and DEmiRNAs from publicly available microarray datasets and combined them with bioinformatics analysis results to construct a circRNA-related ceRNA network. The circRNA-miRNA hub gene regulator axe revealed one important circRNA that may be involved in the regulation of NFPA invasion or chemoresistance, providing new perspectives on the invasion mechanism of NFPA and suggesting possible therapeutic targets for further study.
Funding:
This work was supported by the Natural Science Foundation of Jilin Province (20200201581JC) and the High Technology Research and Development Program of Jilin Province of China (2014G074).
Author’s contributions:
Ping Chen and Xingli Zhao contributed equally to this work and they are co-first authors. All authors had full access to the data in the study and take responsibility for the integrity of the data and the accuracy of the analysis. Conceptualization: Zhanfeng Wang and Xingli Zhao; Methodology: Ping Chen, Xiaonan Wu and Guosheng Hu; Investigation: Jialin Li, Rao Fu and Xingli Zhao; Formal analysis: Ping Chen and Guosheng Hu; Resources: Zhanfeng Wang and Xingli Zhao; Writing-Original draft: Ping Chen and Xingli Zhao; Writing-Review and Editing: Zhanfeng Wang, Ping Chen and Xingli Zhao; Visualization: Ping Chen and Guosheng Hu; Supervision: Zhanfeng Wang and Funding acquisition: Zhanfeng Wang.
Acknowledgements:
We would like to thank Editage (www.editage.cn) for English language editing.
Conflict of interests:
The authors declared no conflict of interest.
References
- Melmed S. Pituitary-tumor endocrinopathies. N Engl J Med 2020;382(10):937-50.
[Crossref] [Google scholar] [PubMed]
- Mahajan A, Bronen RA, Mian AY, Omay SB, Spencer DD, Inzucchi SE. Diagnosis and management of pituitary disease with focus on the role of magnetic resonance imaging. Endocrine 2020;68:489-501.
[Crossref] [Google scholar] [PubMed]
- Al-Dahmani K, Mohammad S, Imran F, Theriault C, Doucette S, Zwicker D, et al. Sellar masses: An epidemiological study. Can J Neurol Sci 2016;43(2):291-7.
[Crossref] [Google scholar] [PubMed]
- Raappana A, Koivukangas J, Ebeling T, Pirilä T. Incidence of pituitary adenomas in Northern Finland in 1992-2007. J Clin Endocrinol Metab 2010;95(9):4268-75.
[Crossref] [Google scholar] [PubMed]
- Di Ieva A, Rotondo F, Syro LV, Cusimano MD, Kovacs K. Aggressive pituitary adenomas-diagnosis and emerging treatments. Nat Rev Endocrinol 2014;10(7):423-35.
[Crossref] [Google scholar] [PubMed]
- Lamberts SW, Hofland LJ. Future treatment strategies of aggressive pituitary tumors. Pituitary 2009;12(3):261-4.
[Crossref] [Google scholar] [PubMed]
- Raverot G, Ilie MD, Lasolle H, Amodru V, Trouillas J, Castinetti F, et al. Aggressive pituitary tumours and pituitary carcinomas. Nat Rev Endocrinol 2021;17(11):671-84.
[Crossref] [Google scholar] [PubMed]
- Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J, et al. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res 2015;25(8):981-4.
[Crossref] [Google scholar] [PubMed]
- Salzman J, Gawad C, Wang PL, Lacayo N, Brown PO. Circular RNAs are the predominant transcript isoform from hundreds of human genes in diverse cell types. PLoS One 2012;7(2):e30733.
[Crossref] [Google scholar] [PubMed]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, et al. Natural RNA circles function as efficient microRNA sponges. Nature 2013;495(7441):384-8.
[Crossref] [Google scholar] [PubMed]
- Cheng J, Nie D, Li B, Gui S, Li C, Zhang Y, et al. CircNFIX promotes progression of pituitary adenoma via CCNB1 by sponging miR-34a-5p. Mol Cell Endocrinol 2021;525:111140.
[Crossref] [Google scholar] [PubMed]
- Guan YJ, Ma JY, Song W. Identification of circRNA-miRNA-mRNA regulatory network in gastric cancer by analysis of microarray data. Cancer Cell Int 2019;19(1):1-9.
[Crossref] [Google scholar] [PubMed]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 2013;495(7441):333-8.
[Crossref] [Google scholar] [PubMed]
- Song YZ, Li JF. Circular RNA hsa_circ_0001564 regulates osteosarcoma proliferation and apoptosis by acting miRNA sponge. Biochem Biophys Res Commun 2018;495(3):2369-75.
[Crossref] [Google scholar] [PubMed]
- Wang J, Wang D, Wan D, Ma Q, Liu Q, Li J, et al. Circular RNA in invasive and recurrent clinical nonfunctioning pituitary adenomas: Expression profiles and bioinformatic analysis. World Neurosurg 2018;117:e371-86.
[Crossref] [Google scholar] [PubMed]
- Hu C, Xia R, Zhang X, Li T, Ye Y, Li G, et al. circFARP1 enables cancer-associated fibroblasts to promote gemcitabine resistance in pancreatic cancer via the LIF/STAT3 axis. Mol Cancer 2022;21(1):1-21.
[Crossref] [Google scholar] [PubMed]
- Long G, Ma S, Shi R, Sun Y, Hu Z, Chen K. Circular RNAs and drug resistance in genitourinary cancers: A literature review. Cancers 2022;14(4):1-20.
[Crossref] [Google scholar] [PubMed]
- Knosp E, Steiner E, Kitz K, Matula C. Pituitary adenomas with invasion of the cavernous sinus space: A magnetic resonance imaging classification compared with surgical findings. Neurosurgery 1993;33(4):610-7.
[Crossref] [Google scholar] [PubMed]
- Wilson CB. A decade of pituitary microsurgery: The Herbert Olivecrona lecture. J Neurosurg 1984;61(5):814-33.
[Crossref] [Google scholar] [PubMed]
- Robinson MD, McCarthy DJ, Smyth GK. EdgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26(1):139-40.
[Crossref] [Google scholar] [PubMed]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15(12):1-21.
[Crossref] [Google scholar] [PubMed]
- Xia S, Feng J, Chen K, Ma Y, Gong J, Cai F, et al. CSCD: A database for cancer-specific circular RNAs. Nucleic Acids Res 2018;46(D1):D925-9.
[Crossref] [Google scholar] [PubMed]
- Feng J, Chen W, Dong X, Wang J, Mei X, Deng J, et al. CSCD2: An integrated interactional database of cancer-specific circular RNAs. Nucleic Acids Res 2022;50(D1):D1179-83.
[Crossref] [Google scholar] [PubMed]
- Fromm B, Billipp T, Peck LE, Johansen M, Tarver JE, King BL, et al. A uniform system for the annotation of vertebrate microRNA genes and the evolution of the human microRNAome. Annu Rev Genet 2015;49:213-42.
[Crossref] [Google scholar] [PubMed]
- Wong N, Wang X. miRDB: An online resource for microRNA target prediction and functional annotations. Nucleic Acids Res 2015;43(D1):D146-52.
[Crossref] [Google scholar] [PubMed]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13(11):2498-504.
[Crossref] [Google scholar] [PubMed]
- Yu G, Wang LG, Han Y, He QY. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS 2012;16(5):284-7.
[Crossref] [Google scholar] [PubMed]
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci 2005;102(43):15545-50.
[Crossref] [Google scholar] [PubMed]
- Xie Z, Bailey A, Kuleshov MV, Clarke DJ, Evangelista JE, Jenkins SL, et al. Gene set knowledge discovery with Enrichr. Curr Protoc 2021;1(3):e90.
[Crossref] [Google scholar] [PubMed]
- Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res 2015;43(D1):D447-52.
[Crossref] [Google scholar] [PubMed]
- Bandettini WP, Kellman P, Mancini C, Booker OJ, Vasu S, Leung SW, et al. MultiContrast Delayed Enhancement (MCODE) improves detection of subendocardial myocardial infarction by late gadolinium enhancement cardiovascular magnetic resonance: A clinical validation study. J Cardiovasc Magn Reson 2012;14(1):1-10.
[Crossref] [Google scholar] [PubMed]
- Gao Y, Zhang J, Zhao F. Circular RNA identification based on multiple seed matching. Brief Bioinform 2018 Sep;19(5):803-10.
[Crossref] [Google scholar] [PubMed]
- Wang K, Singh D, Zeng Z, Coleman SJ, Huang Y, Savich GL, et al. MapSplice: Accurate mapping of RNA-seq reads for splice junction discovery. Nucleic Acids Res 2010;38(18):e178.
[Crossref] [Google scholar] [PubMed]
- Westholm JO, Miura P, Olson S, Shenker S, Joseph B, Sanfilippo P, et al. Genome-wide analysis of drosophila circular RNAs reveals their structural and sequence properties and age-dependent neural accumulation. Cell Rep 2014;9(5):1966-80.
[Crossref] [Google scholar] [PubMed]
- Zhang XO, Dong R, Zhang Y, Zhang JL, Luo Z, Zhang J, et al. Diverse alternative back-splicing and alternative splicing landscape of circular RNAs. Genome Res 2016;26(9):1277-87.
[Crossref] [Google scholar] [PubMed]
- Heaney A. Management of aggressive pituitary adenomas and pituitary carcinomas. J Neurooncol 2014;117:459-68.
[Crossref] [Google scholar] [PubMed]
- Hui P, Xu X, Xu L, Hui G, Wu S, Lan Q. Expression of MMP14 in invasive pituitary adenomas: Relationship to invasion and angiogenesis. Int J Clin Exp Pathol 2015;8(4):3556.
[Google scholar] [PubMed]
- Hu Y, Zhang N, Zhang S, Zhou P, Lv L, Richard SA, et al. Differential circular RNA expression profiles of invasive and non-invasive non-functioning pituitary adenomas: A microarray analysis. Medicine 2019;98(26):e16148.
[Crossref] [Google scholar] [PubMed]
- Yang Q, Wang R, Wei B, Peng C, Wang L, Hu G, et al. Gene and microRNA signatures are associated with the development and survival of glioblastoma patients. DNA Cell Biol 2019;38(7):688-99.
[Crossref] [Google scholar] [PubMed]
- Krushkal J, Vural S, Jensen TL, Wright G, Zhao Y. Increased copy number of imprinted genes in the chromosomal region 20q11-q13. 32 is associated with resistance to antitumor agents in cancer cell lines. Clin Epigenetics 2022;14(1):161.
[Crossref] [Google scholar] [PubMed]
- Coles GL, Cristea S, Webber JT, Levin RS, Moss SM, He A, et al. Unbiased proteomic profiling uncovers a targetable GNAS/PKA/PP2A axis in small cell lung cancer stem cells. Cancer Cell 2020;38(1):129-43.
[Crossref] [Google scholar] [PubMed]
- Williams CS, Bernard JK, Demory Beckler M, Almohazey D, Washington MK, Smith JJ, et al. ERBB4 is over-expressed in human colon cancer and enhances cellular transformation. Carcinogenesis 2015;36(7):710-8.
[Crossref] [Google scholar] [PubMed]
- Wu S, Gu Y, Huang Y, Wong TC, Ding H, Liu T, et al. Novel biomarkers for non-functioning invasive pituitary adenomas were identified by using analysis of microRNAs expression profile. Biochem Genet 2017;55:253-67.
[Crossref] [Google scholar] [PubMed]
- Bong AH, Monteith GR. Calcium signaling and the therapeutic targeting of cancer cells. Biochim Biophys Acta Mol Cell Res 2018;1865(11):1786-94.
[Crossref] [Google scholar] [PubMed]
- Fujimoto M, Inoue T, Kito H, Niwa S, Suzuki T, Muraki K, et al. Transcriptional repression of HER2 by ANO1 Cl-channel inhibition in human breast cancer cells with resistance to trastuzumab. Biochem Biophys Res Commun 2017;482(1):188-94.
[Crossref] [Google scholar] [PubMed]
- Ye Q, Kantonen S, Henkels KM, Gomez-Cambronero J. A new signaling pathway (JAK-Fes-phospholipase D) that is enhanced in highly proliferative breast cancer cells. J Biol Chem 2013;288(14):9881-91.
[Crossref] [Google scholar] [PubMed]
- Noble AR, Maitland NJ, Berney DM, Rumsby MG. Phospholipase D inhibitors reduce human prostate cancer cell proliferation and colony formation. Br J Cancer 2018;118(2):189-99.
[Crossref] [Google scholar] [PubMed]
- Wang F, Shu K, Lei T, Xue D. The expression of integrinβ1 and FAK in pituitary adenomas and their correlation with invasiveness. J Huazhong Univ Sci Technolog Med Sci 2008;28:572-5.
[Crossref] [Google scholar] [PubMed]
- Uraki S, Ariyasu H, Doi A, Kawai S, Takeshima K, Morita S, et al. Reduced expression of mismatch repair genes MSH6/MSH2 directly promotes pituitary tumor growth via the ATR-Chk1 pathway. J Clin Endocrinol Metab 2018;103(3):1171-9.
[Crossref] [Google scholar] [PubMed]
- Cheng T, Wang Y, Lu M, Zhan X, Zhou T, Li B, et al. Quantitative analysis of proteome in non-functional pituitary adenomas: Clinical relevance and potential benefits for the patients. Front Endocrinol 2019;10:1-19.
[Crossref] [Google scholar] [PubMed]
- Wu JL, Qiao JY, Duan QH. Significance of TNF-α and IL-6 expression in invasive pituitary adenomas. Genet Mol Res 2016;15(1):1-9.
[Crossref] [Google scholar] [PubMed]
- Haemmerle M, Stone RL, Menter DG, Afshar-Kharghan V, Sood AK. The platelet lifeline to cancer: Challenges and opportunities. Cancer Cell 2018;33(6):965-83.
[Crossref] [Google scholar] [PubMed]
- Cao C, Wang W, Ma C, Jiang P. Computational analysis identifies invasion-associated genes in pituitary adenomas. Mol Med Rep 2015;12(2):1977-82.
[Crossref] [Google scholar] [PubMed]
- Niu N, Shen W, Zhong Y, Bast Jr RC, Jazaeri A, Sood AK, et al. Expression of B7-H4 and IDO1 is associated with drug resistance and poor prognosis in high-grade serous ovarian carcinomas. Hum Pathol 2021;113:20-7.
[Crossref] [Google scholar] [PubMed]
- Perou CM, Sørlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, et al. Molecular portraits of human breast tumours. Nature 2000;406(6797):747-52.
[Crossref] [Google scholar] [PubMed]
 .
.



