- *Corresponding Author:
- Jing Wang
Hepatobilary Department, The Affiliated Traditional Chinese Medicine Hospital of Southwest Medical University, Luzhou, Sichuan 646000, China
E-mail: lywj68@swmu.edu.cn
| This article was originally published in a special issue, “Drug Development in Biomedical and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2023:85(5) Spl Issue “119-128” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Cordyceps sinensis is a widely utilized medicinal substance in clinical practice, notably recognized for its applications in various cancer treatments. Although several studies have demonstrated its therapeutic effects on liver cancer, a comprehensive analysis of the underlying molecular mechanisms is still lacking. In this study, we systematically gathered 197 components and identified 343 action targets of Cordyceps from traditional Chinese medicine systems pharmacology and high-throughput experiment- and reference-guided databases, as well as 1482 therapeutic targets associated with hepatocellular carcinoma from various sources, including Online Mendelian Inheritance in Man, DrugBank and other databases. Utilizing this extensive dataset, we conducted a comprehensive analysis that revealed tumor necrosis factor, cysteine-aspartic acid protease 3, B-cell lymphoma 2, interleukin 6, vascular endothelial growth factor A and peroxisome proliferator-activated receptor as the central targets responsible for the efficacy of Cordyceps sinensis in liver cancer therapy. Further enrichment analysis unveiled potential mechanisms of Cordyceps action in liver cancer treatment, highlighting the tumor necrosis factor-nuclear factor kappa B signalling pathway and wingless-related integration site/catenin beta 1 signalling pathway as areas warranting further exploration. Finally, we utilized molecular docking to visualize the binding models of the relevant components and core targets. In summary, our study systematically and effectively analysed the potential active ingredients, action targets and relevant pathways associated with Cordyceps sinensis in the treatment of hepatocellular carcinoma. These findings shed light on the underlying mechanisms of action of Cordyceps sinensis, thus establishing a robust theoretical foundation for future experimental investigations.
Keywords
Cordyceps sinensis, network pharmacology, molecular docking, traditional Chinese medicine
Liver cancer stands as one of the most prevalent malignant neoplasms globally, encompassing Hepatocellular Carcinoma (HCC), Intrahepatic Cholangiocarcinoma (ICC), and combined hepatocellular-cholangiocarcinoma. Among these, HCC constitutes 75 %-85 % of all primary liver cancer, while ICC accounting for 10 %-15 %[1]. Across the world, the incidence of liver cancer remains notably elevated in Asian nations and Pacific islands, particularly within developing countries[2,3]. In China, nearly 500 000 new cases of primary liver cancer are diagnosed annually, constituting around 50 % of all newly reported instances of primary liver cancer globally. This heightened occurrence primarily stems from deficient early diagnosis and management of Hepatitis B Virus (HBV) infection[4,5]. Concurrently, the prodromal manifestations of liver cancer often evade attention, resulting in detections predominantly manifesting in the intermediate to advanced stages. Present therapeutic modalities are considerably constrained, profoundly compromising the quality of patients life and exacerbating their burdens[2].
Cordyceps sinensis, documented as far back in the early 15th century, is a widely employed medicinal substance in clinical practice, particularly notable for its applications in various tumor treatments. It emerges as a complex entity formed through the symbiotic relationship between the Cordyceps fungus and the remains of its host insect larva. Relevant research has indicated that it actively engages in intricate signalling pathways, both directly and indirectly, thereby exerting diverse pharmacological effects such as anti-infective, antioxidant, immunoregulatory and anticancer properties[6]. There are also studies indicating that Cordyceps sinensis exhibits protective effects against chemical-induced liver injury, immune-mediated liver damage, as well as alcoholic and non-alcoholic liver injuries.
Significantly, Cordyceps sinensis exhibits a clearly defined anticancer effect. Research conducted by Lee et al. has unveiled that Cordyceps is capable of enhancing the sensitivity of liver cancer human hepatoma cell line (Hep3B) to Tumor Necrosis Factor (TNF)-Related Apoptosis-Inducing Ligand (TRAIL) by modulating the c-Jun N-terminal Kinase (c-JNK) signalling pathway, thereby inducing apoptosis in liver cancer cells[7]. Research discovered that Cordyceps sinensis also exhibited inhibitory effects on the production of nitric oxide and Reactive Oxygen Species (ROS). Additionally, it led to an elevation in mitochondrial membrane potential, Superoxide Dismutase (SOD) and catalase levels, while concurrently decreasing the content of Glutathione (GSH)[8]. Furthermore, investigations conducted by Zhang et al. have demonstrated the ability of Cordyceps to augment the responsiveness of the immune system toward tumor cells, thereby significantly inhibiting the growth of tumours within Hepatoma 22 (H-22) tumor-bearing mice[9,10].
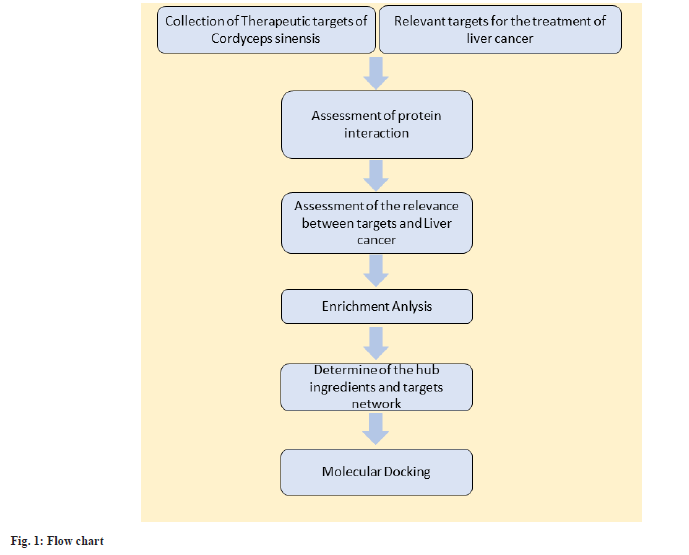
However, a comprehensive systematic analysis of Cordyceps sinensis in the context of liver cancer treatment remains lacking. Network pharmacology is a method to analyse the roles of drug components and their target interactions in disease therapeutics. It facilitates the systematic assessment of how drugs exert their therapeutic effects through intricate signalling pathways, making it particularly suitable for studies involving multiple components in disease treatment. Such investigations hold pivotal significance in unravelling the mechanisms of action of Traditional Chinese Medicine (TCM)[11]. Consequently, this study aims to employ network pharmacology method combined with molecular docking to systematically reveal the underlying mechanism of Cordyceps sinensis in treating liver cancer (fig. 1).
Materials and Methods
Potential targets of Cordyceps sinensis:
The ingredients and targets of Cordyceps sinensis were sourced from the TCM Systems Pharmacology (TCMSP)[12] and High-throughput Experiment- and Reference-guided database of TCM (HERB)[13]. The search keyword used was “Dong Chong Xia Cao”. In TCMSP, the selection criteria included an Oral Bioavailability (OB) rate >30 % and a drug-likeness score exceeding 0.18[14]. Conversely, in the HERB database, all ingredients and targets were included as they were derived from previously validated research studies.
Compilation of therapeutic targets for liver cancer:
Several databases like Online Mendelian Inheritance in Man (OMIM)[15], DrugBank[16], MalaCards[17], GeneCards[18], DisGeNET[19] and Therapeutic Target Database (TTD)[20] offer contemporary insights into human genes, granting unhindered access to pertinent information regarding targets for established diseases. In this study, liver cancer serves as the primary search term, employed to retrieve pertinent targets from the aforementioned databases. The gene sets characterized by differential expression, along with genes obtained from each database, are amalgamated, purged of redundancies, and subsequently categorized as genes associated with liver cancer.
Exploration of interrelation networks and core modules:
Protein-Protein Interaction (PPI) stands as a pivotal facet governing the interactions between proteins, with central modules offering elucidation into crucial functional units within PPI networks, thereby aiding the identification of protein complexes and novel pathways. In this study, the PPI network was scrutinized via the Search Tool for the Retrieval of Interacting Genes (STRING) platform for online analysis. Subsequently, the outcomes were imported into Cytoscape (version 3.10.0)[18], and the CytoNCA plugin was employed to meticulously dissect and appraise the intricacies of the PPI network[19]. Simultaneously, in order to further ascertain the core genes, Variant Election (VarElect) was also harnessed to analyze the correlation between targets and diseases. Furthermore, Molecular Complex Detection (MCODE) was employed to delve into the identification of core modules.
Enrichment analysis:
Utilizing Gene Ontology (GO) and the Kyoto Encyclopedia of Genes and Genomes (KEGG) empowers the systematic scrutiny of data and the potential for unearthing novel insights[21]. Therefore, within the comprehensive exploration of the therapeutic mechanism of Cordyceps sinensis concerning liver cancer, we engaged in meticulous GO and KEGG analysis concerning the potential targets and functional modules. By uploading the prospective targets onto Metascape[21], we conducted a rigorous GO analysis encompassing biological processes, cellular components, molecular functions and KEGG pathway enrichment. Significance was accorded to elements bearing a value of p<0.01, denoting pronounced enrichment.
Molecular docking:
In the context of this study, the application of molecular docking was instrumental in evaluating the interplay between Cordyceps sinensis and the pivotal targets related to liver cancer. At first, the ligand structures of each ingredient were downloaded from PubChem. Subsequently, the corresponding structures of hub targets were meticulously obtained from the Protein Data Bank (PDB) database[22]. The process of molecular docking will be orchestrated through the utilization of AutoDock[23] in conjunction with (PyMOL)[24].
Results and Discussion
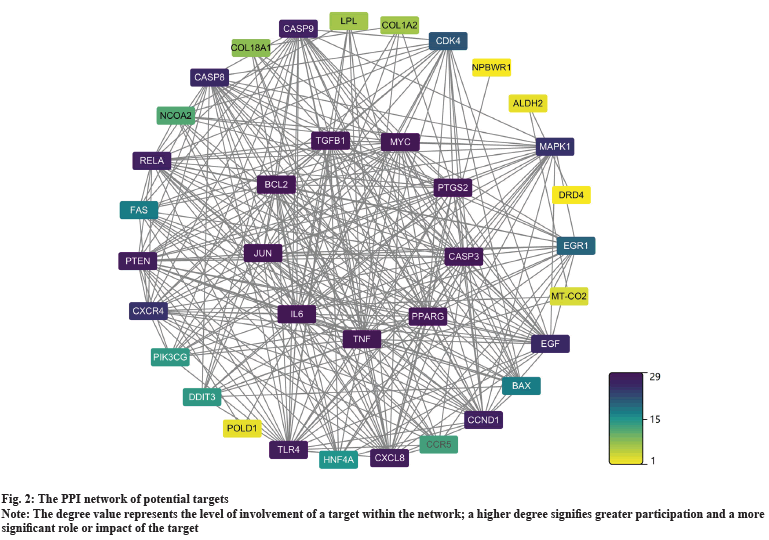
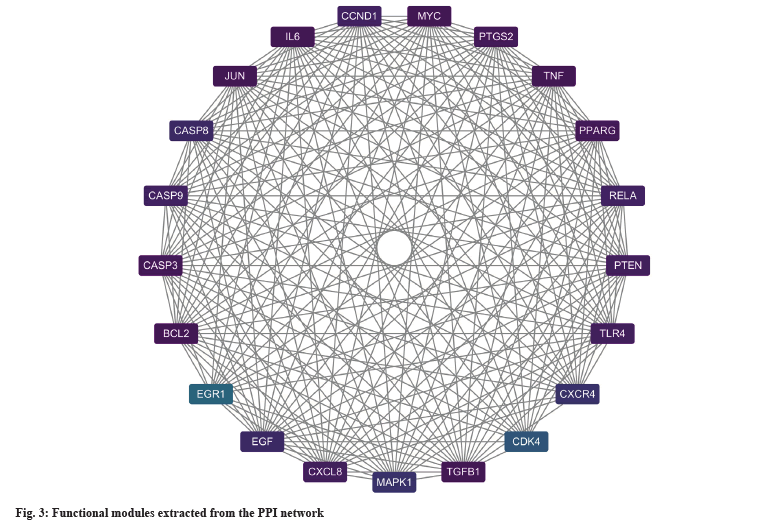
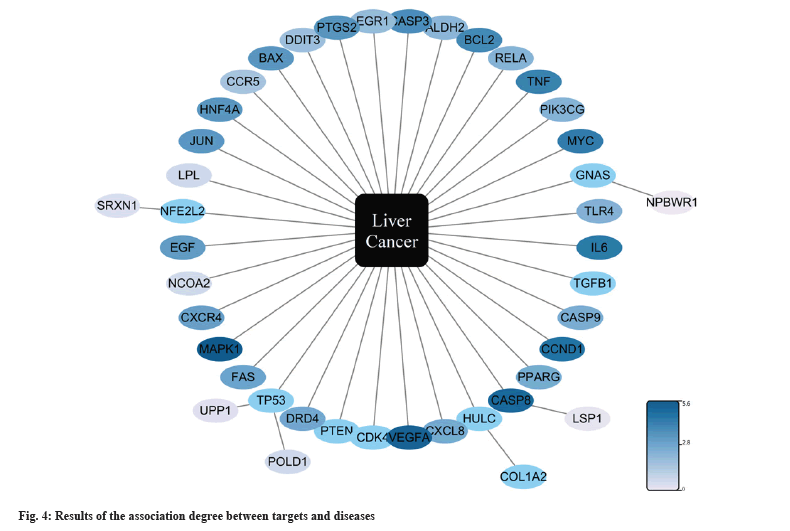
Using “Dong Chong Xia Cao” as the pivotal keyword, a comprehensive search was conducted within the TCMSP and HERB databases. Following meticulous curation and deduplication, a cumulative compilation of 197 ingredients and 343 targets was assembled. Simultaneously, employing “liver cancer” as the primary search term, research across the OMIM, DrugBank, MalaCards, GeneCards, DisGeNET, and TTD databases recruited 508, 229, 50, 434, 1186, and 75 liver cancer-related targets respectively. Upon elimination of duplicates, a total of 1482 targets pertinent to liver cancer therapy were obtained. Finally, the intersections between two sets in a total of 40 targets were defined as potential therapy targets. As shown in fig. 2, we conducted an analysis of the interactions among these 40 targets. Based on their degree values, we identified TNF, Caspase 3 (CASP3), B-Cell Lymphoma 2 (BCL2), Interleukin-6 (IL-6), Vascular Endothelial Growth Factor-A (VEGF-A), and Prostaglandin-endoperoxide Synthase 2 (PTGS2) as central targets within this interaction network. Furthermore, the exploration revealed a highly interconnected functional module (fig. 3). In addition, an analysis of the relevance between targets and disease treatment was conducted, unveiling that among them, 32 targets are directly implicated in disease prognosis, while rest of the targets are indirectly associated with disease prognosis. Notably, TNF, CASP3, BCL2, IL6, VEGF-A, and other targets exhibited the highest degrees of correlation (fig. 4).
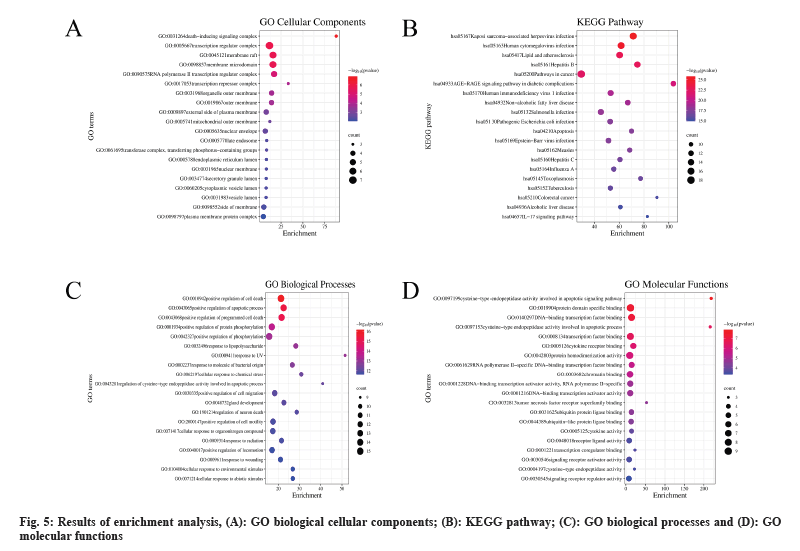
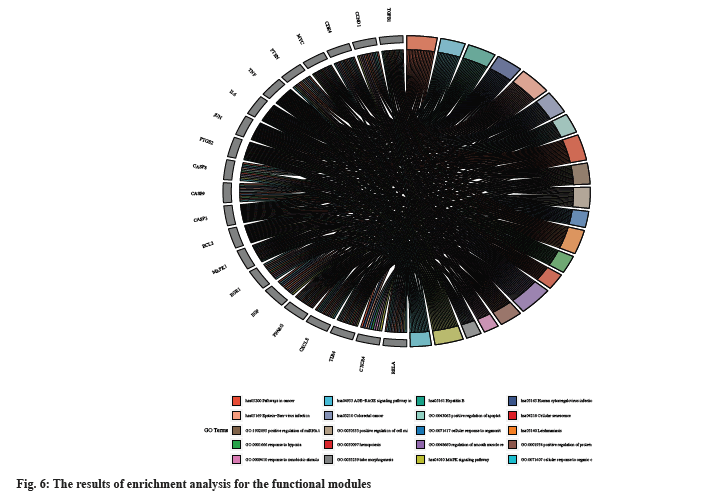
In our study, we continued with an enrichment analysis to explore the main pathways involved in the treatment of liver cancer using Cordyceps (fig. 5). Our analysis of biological processes revealed a strong focus on pathways related to cellular apoptosis. KEGG pathway analysis showed that Cordyceps sinensis was involved in regulating pathways like hepatitis B, cancer, Advanced Glycation Endproducts-Receptor for Advanced Glycation Endproducts (AGE-RAGE) signaling pathway, non-alcoholic fatty liver disease, hepatitis C, alcoholic liver disease, and IL-17 signaling pathway, Nucleotide-binding Oligomerization Domain (NOD)-like receptor signaling pathway, and TNF signaling pathway (fig. 6).
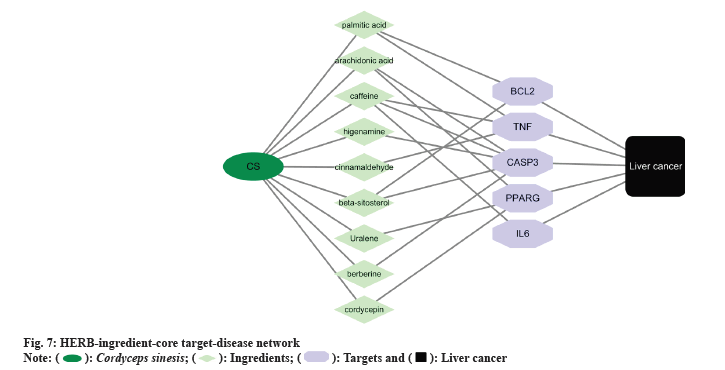
We established HERB-ingredient-core target-disease network (fig. 7) by integrating the effective ingredients of Cordyceps sinensis with the highly associated core targets in liver cancer. This network consists by 18 nodes and 26 edges, succinctly encapsulating the intricate relationships between the key ingredients and core targets of our analysis.
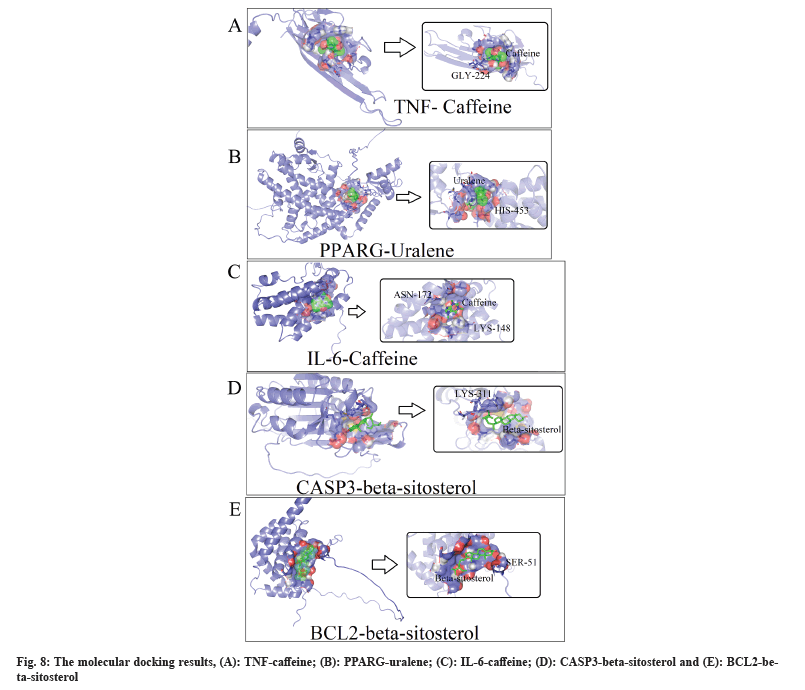
To validate our findings, we conducted molecular docking to visualize the binding method of key ingredients and core targets (fig. 8). Besides, in all molecular docking models, stable hydrogen bonds were formed, and the strength of their binding energies is illustrated in Table 1.
| Protein | Gene name | Ligands | Pubchem ID | Binding energy |
|---|---|---|---|---|
| Caspase-3 | CASP3 | Arachidonic acid | 444899 | -5.6 |
| Caspase-3 | CASP3 | Beta-sitosterol | 222284 | -7.3 |
| Caspase-3 | CASP3 | Berberine | 2353 | -6.8 |
| Caspase-3 | CASP3 | Caffeine | 2519 | -5.1 |
| Caspase-3 | CASP3 | Higenamine | 114840 | -6.8 |
| Apoptosis regulator Bcl-2 | BCL2 | Beta-sitosterol | 222284 | -6.4 |
| Apoptosis regulator Bcl-2 | BCL2 | Palmitic acid | 985 | -4.4 |
| Tumor necrosis factor | TNF | Caffeine | 2519 | -5.6 |
| Tumor necrosis factor | TNF | Cinnamaldehyde | 637511 | -5.3 |
| Tumor necrosis factor | TNF | Palmitic acid | 985 | -4.2 |
| Interleukin-6 | IL-6 | Caffeine | 2519 | -5 |
| Peroxisome proliferator-activated receptor gamma | PPARG | Crachidonic acid | 444899 | -5.1 |
| Peroxisome proliferator-activated receptor gamma | PPARG | Cordycepin | 6303 | -6.3 |
| Peroxisome proliferator-activated receptor gamma | PPARG | Uralene | 192409 | -7.3 |
Note: Binding energy is a crucial parameter in molecular docking, evaluating the strength of interaction between a receptor and a ligand. A lower value indicates a tighter binding affinity.
Table 1: The results of binding energy of each molecular docking.
The research conducted by Chen et al. indicates that the subunits of TNF brings changes in the microenvironment, inducing the generation of an inflammatory microenvironment by Allograft Inflammatory Factor 1 (AIF1)+Colony Stimulating Factor 1 Receptor (CSF1R)+Mesenchymal Stem Cells (MSCs) and promoting the onset of liver cancer[21]. Concurrently, other studies suggest that the regulation of the TNF-Nuclear Factor Kappa B (NFκB) signaling pathway is crucial for compensatory proliferation of liver cells, with excessive TNF expression diminishing this compensatory response, thereby fostering the development of HCC[22]. Research conducted in mice demonstrates that by targeting a pivotal component of the TNF-mediated NFκB signaling pathway, tumor growth is decelerated, highlighting TNF as a promotive factor in liver cancer, with the NFκB signaling pathway being one of its essential routes[23]. Based on a review of analogous studies, anti-TNF treatment stands as a promising avenue in the advancement of liver cancer therapy[24]. CASP3 has emerged as a prominent target in recent liver cancer-related investigations. Research has unveiled that CASP3 cleaves Sterol Regulatory Element-Binding Protein 2 (SREBP2) in the endoplasmic reticulum, facilitating cholesterol biosynthesis and leading to resistance against anti-liver cancer treatment by triggering the activation of the sonic hedgehog signaling pathway[25]. In various studies, the decline of BCL2 due to various reasons has shown to promote apoptosis in liver cancer cells[26,27]. Certain research findings concerning Peroxisome Proliferator-Activated Receptor Gamma (PPARG) suggests that its upregulation can activate the Wingless-related integration site/Catenin Beta-1 (WNT/CTNNB1) signaling pathway, contributing to the occurrence and progression of liver cancer[28,29].
IL-6 is a pleiotropic cytokine with a four-helix bundle structure that plays multiple roles in the body. Since its discovery, it has been recognized as a participant in the regulation of various pathways, including inflammation and immunity. Studies have indicated that the activation of IL-6 triggers the activation of the IL-6/Signal Transducer and Activator of Transcription 3 (STAT3) signaling pathway. Conversely, anti-IL-6 therapy has an inhibitory effect on this pathway, revealing a novel mechanism in macrophage polarization regulation with significant potential for future liver cancer treatment[30]. A comprehensive review underscores the vital role of IL-6 in maintaining hepatocellular homeostasis. Sustained activation of the IL-6 signaling pathway can be detrimental to the liver and may ultimately lead to the development of hepatic tumors[31].
VEGF-A, is conventionally known for its roles in inducing endothelial cell proliferation, promoting cell migration, inhibiting apoptosis, and increasing vascular permeability. Research has indicated that the overexpression of bone morphogenetic protein 9 can stimulate VEGF-A secretion, consequently activating the Hypoxia-Inducible Factor 1-alpha (HIF-1α)/VEGF-A signaling pathway, ultimately promoting the development of blood vessels in liver cancer. Furthermore, targeted therapy against VEGF-A represents a crucial avenue in the treatment of advanced liver cancer[32,33]. Moreover, non-alcoholic fatty liver disease is considered a significant risk factor for liver cancer. During the transition from non-alcoholic fatty liver disease to liver cancer, the activation of VEGF-A through various pathways is believed to facilitate the development of liver cancer[34].
However, this study has some limitations. Foremost among these is that the primary objective of this investigation was to gain a deeper understanding of its mechanism through the application of network pharmacology and molecular docking techniques. Despite yielding results of considerable exploratory significance, further progression necessitates the implementation of clinical randomized controlled trials to validate the accuracy of the responsive targets and pathways.
In the virtual analysis conducted in this study, Cordyceps sinensis exhibited therapeutic potential against liver cancer. This effect primarily originates from its intricate influence on the target proteins through a complex interplay of its constituents, notably its regulation of targets such as BCL2, IL6, CASP3, and TNF, as well as its modulation of pertinent pathways. These findings have illuminated the underlying mechanism of action of Cordyceps sinensis, thus establishing a solid theoretical foundation for future experimental endeavors.
Acknowledgements:
This study was supported by Luzhou “Jiucheng Talents, Scientific and Technological Innovation Team” (Number: [2021] No. 162), traditional Chinese medicine talent growth platform construction project of Sichuan Provincial Administration of Traditional Chinese Medicine, National Traditional Chinese Medicine Clinical Research Base Construction Unit, research project of the Affiliated Traditional Chinese Medicine Hospital, Southwest Medical University (Number: [2020] No. 33), and joint project of Southwest Medical University-Affiliated Traditional Chinese Medicine Hospital of Southwest Medical University (Number: [2020] No.6, 2020XYLH-037), basic clinical research on the prevention and treatment of primary liver cancer based on the academic thought of professor Sun Tongjiao, a famous national traditional Chinese medicine (Number: [2022] 2022YFS0619).
Authors’ contributions:
Yaping Mu and Jing Wang contributed equally to this work. Yaping Mu, Ding Zheng, and Qinghua Peng conceived, designed, and planned the study. Yaping Mu acquired and analysed the data. Yaping Mu and Fei Ye interpreted the results. Yaping Mu and Jing Wang drafted the manuscript and Encheng Wang and Jing Wang contributed to the critical revision of the manuscript. All authors read and approved the final manuscript.
Conflicts of interests:
The authors declare no conflict of interests.
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68(6):394-424.
[Crossref] [Google Scholar] [PubMed]
- Rumgay H, Arnold M, Ferlay J, Lesi O, Cabasag CJ, Vignat J, et al. Global burden of primary liver cancer in 2020 and predictions to 2040. J Hepatol 2022;77(6):1598-606.
[Crossref] [Google Scholar] [PubMed]
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71(3):209-49.
[Crossref] [Google Scholar] [PubMed]
- Cao W, Chen HD, Yu YW, Li N, Chen WQ. Changing profiles of cancer burden worldwide and in China: A secondary analysis of the global cancer statistics 2020. Chin Med J 2021;134(7):783-91.
[Crossref] [Google Scholar] [PubMed]
- Chen JG, Zhang SW. Liver cancer epidemic in China: Past, present and future. Semin Cancer Biol 2011;21(1):59-69.
[Crossref] [Google Scholar] [PubMed]
- Olatunji OJ, Tang J, Tola A, Auberon F, Oluwaniyi O, Ouyang Z. The genus Cordyceps: An extensive review of its traditional uses, phytochemistry and pharmacology. Fitoterapia 2018;129:293-316.
[Crossref] [Google Scholar] [PubMed]
- Lee HH, Jeong JW, Lee JH, Kim GY, Cheong J, Jeong YK, et al. Cordycepin increases sensitivity of Hep3B human hepatocellular carcinoma cells to TRAIL-mediated apoptosis by inactivating the JNK signaling pathway. Oncol Rep 2013;30(3):1257-64.
[Crossref] [Google Scholar] [PubMed]
- Xiao Y, Zhang X, Huang Q. Protective effects of Cordyceps sinensis exopolysaccharide-selenium nanoparticles on H2O2-induced oxidative stress in HepG2 cells. Int J Biol Macromol 2022;213:339-51.
[Crossref] [Google Scholar] [PubMed]
- Zhang W, Li J, Qiu S, Chen J, Zheng Y. Effects of the exopolysaccharide fraction (EPSF) from a cultivated Cordyceps sinensis on immunocytes of H22 tumor bearing mice. Fitoterapia 2008;79(3):168-73.
[Crossref] [Google Scholar] [PubMed]
- Zhang W, Yang J, Chen J, Hou Y, Han X. Immunomodulatory and antitumour effects of an exopolysaccharide fraction from cultivated Cordyceps sinensis (Chinese caterpillar fungus) on tumour-bearing mice. Biotechnol Appl Biochem 2005;42(1):9-15.
[Crossref] [Google Scholar] [PubMed]
- Liang YI, Liang BO, Chen W, Wu XR, Liu-Huo WS, Zhao LZ. Potential mechanism of dingji fumai decoction against atrial fibrillation based on network pharmacology, molecular docking, and experimental verification integration strategy. Front Cardiovasc Med 2021;8:1-16.
[Crossref] [Google Scholar] [PubMed]
- Ru J, Li P, Wang J, Zhou W, Li B, Huang C, et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J Cheminform 2014;6:1-6.
[Crossref] [Google Scholar] [PubMed]
- Fang S, Dong L, Liu L, Guo J, Zhao L, Zhang J, et al. HERB: A high-throughput experiment-and reference-guided database of traditional Chinese medicine. Nucleic Acids Res 2021;49(D1):D1197-206.
[Crossref] [Google Scholar] [PubMed]
- Liang B, Liang Y, Li R, Zhang H, Gu N. Integrating systematic pharmacology-based strategy and experimental validation to explore the synergistic pharmacological mechanisms of Guanxin V in treating ventricular remodeling. Bioorg Chem 2021;115:1-11.
[Crossref] [Google Scholar] [PubMed]
- Amberger JS, Hamosh A. Searching Online Mendelian Inheritance in Man (OMIM): A knowledgebase of human genes and genetic phenotypes. Current protocols in bioinformatics. 2017;58(1):1-2.
[Crossref] [Google Scholar] [PubMed]
- Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res 2018;46(D1):D1074-82.
[Crossref] [Google Scholar] [PubMed]
- Rappaport N, Twik M, Plaschkes I, Nudel R, Iny Stein T, Levitt J, et al. MalaCards: An amalgamated human disease compendium with diverse clinical and genetic annotation and structured search. Nucleic Acids Res 2017;45(D1):D877-87.
[Crossref] [Google Scholar] [PubMed]
- Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, et al. The GeneCards suite: From gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics 2016;54(1):1-30.
[Crossref] [Google Scholar] [PubMed]
- Piñero J, Bravo À, Queralt-Rosinach N, Gutiérrez-Sacristán A, Deu-Pons J, Centeno E, et al. DisGeNET: A comprehensive platform integrating information on human disease-associated genes and variants. Nucleic Acids Res 2017;45(D1):D833-39.
[Crossref] [Google Scholar] [PubMed]
- Zhou Y, Zhang Y, Lian X, Li F, Wang C, Zhu F, et al. Therapeutic target database update 2022: Facilitating drug discovery with enriched comparative data of targeted agents. Nucleic Acids Res 2022;50(D1):D1398-407.
[Crossref] [Google Scholar] [PubMed]
- Zong C, Meng Y, Ye F, Yang X, Li R, Jiang J, et al. AIF1+CSF1R+MSCs, induced by TNF‐α, act to generate an inflammatory microenvironment and promote hepatocarcinogenesis. Hepatology 2023;78(2):434-51.
[Crossref] [Google Scholar] [PubMed]
- Grivennikov SI, Karin M. Inflammatory cytokines in cancer: Tumour necrosis factor and interleukin 6 take the stage. Ann Rheum Dis 2011;70(1):i104-8.
[Crossref] [Google Scholar] [PubMed]
- Pikarsky E, Porat RM, Stein I, Abramovitch R, Amit S, Kasem S, et al. NF-κB functions as a tumour promoter in inflammation-associated cancer. Nature 2004;431(7007):461-6.
[Crossref] [Google Scholar] [PubMed]
- Tiegs G, Horst AK. TNF in the liver: Targeting a central player in inflammation. Semin Immunopathol 2022;44(4):445-59.
[Crossref] [Google Scholar] [PubMed]
- Mok EH, Leung CO, Zhou L, Lei MM, Leung HW, Tong M, et al. Caspase-3-induced activation of srebp2 drives drug resistance via promotion of cholesterol biosynthesis in hepatocellular carcinoma. Cancer Res 2022;82(17):3102-15.
[Crossref] [Google Scholar] [PubMed]
- Liu D, Chen C, Cui M, Zhang H. miR-140-3p inhibits colorectal cancer progression and its liver metastasis by targeting BCL9 and BCL2. Cancer Med 2021;10(10):3358-72.
[Crossref] [Google Scholar] [PubMed]
- Teng Y, Li Z, Liu J, Teng L, Li H. Proliferation inhibition and apoptosis of liver cancer cells treated by blue light irradiation. Med Oncol 2023;40(8):227.
[Crossref] [Google Scholar] [PubMed]
- To JC, Chiu AP, Tschida BR, Lo LH, Chiu CH, Li XX, et al. ZBTB20 regulates WNT/CTNNB1 signalling pathway by suppressing PPARG during hepatocellular carcinoma tumourigenesis. JHEP Rep 2021;3(2):1-10.
[Crossref] [Google Scholar] [PubMed]
- Zuo Q, He J, Zhang S, Wang H, Jin G, Jin H, Cheng Z, et al. Pparγ coactivator‐1α suppresses metastasis of hepatocellular carcinoma by inhibiting Warburg effect by PPARγ-Dependent WNT/Β‐Catenin/Pyruvate dehydrogenase kinase isozyme 1 axis. Hepatology 2021;73(2):644-60.
[Crossref] [Google Scholar] [PubMed]
- Zhou YF, Song SS, Tian MX, Tang Z, Wang H, Fang Y, et al. Cystathionine β-synthase mediated PRRX2/IL-6/STAT3 inactivation suppresses Tregs infiltration and induces apoptosis to inhibit HCC carcinogenesis. J Immunother Cancer 2021;9(8):1-14.
[Crossref] [Google Scholar] [PubMed]
- Schmidt-Arras D, Rose-John S. IL-6 pathway in the liver: From physiopathology to therapy. J Hepatol 2016;64(6):1403-15.
[Crossref] [Google Scholar] [PubMed]
- Chen H, Nio K, Tang H, Yamashita T, Okada H, Li Y, et al. BMP9-ID1 signaling activates HIF-1α and VEGFA expression to promote tumor angiogenesis in hepatocellular carcinoma. Int J Mol Sci 2022;23(3):1-14.
[Crossref] [Google Scholar] [PubMed]
- Wei H, Xu Z, Chen L, Wei Q, Huang Z, Liu G, et al. Long non-coding RNA PAARH promotes hepatocellular carcinoma progression and angiogenesis via upregulating HOTTIP and activating HIF-1α/VEGF signaling. Cell Death Dis 2022;13(2):1-13.
[Crossref] [Google Scholar] [PubMed]
- Shen H, Yu H, Li QY, Wei YT, Fu J, Dong H, et al. Hepatocyte-derived VEGFA accelerates the progression of non-alcoholic fatty liver disease to hepatocellular carcinoma via activating hepatic stellate cells. Acta Pharmacol Sin 2022;43(11):2917-28.
[Crossref] [Google Scholar] [PubMed]