- *Corresponding Author:
- Xiaotao Chen
Department of Stomatology, People’s Hospital of Xinjiang Uygur Autonomous Region, Urumqi, Xinjiang 830001, China
E-mail: zhushuaijun@fjmu.edu.cn
| This article was originally published in a special issue, “Drug Development in Biomedical and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2023:85(5) Spl Issue “253-261” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
We aimed to develop novel methods for treating periodontitis by identifying the main targets and pathways of curcumin in treating periodontitis. Potential targets of curcumin and differential genes were obtained from multiple drug databases and gene expression omnibus database chip data, respectively. Common targets of curcumin and periodontitis were screened, a protein-protein interaction network map of these common targets was constructed, and the critical targets were identified. These targets were subjected to gene ontology and Kyoto encyclopedia of genes and genomes enrichment analysis to construct a "curcumin-target-pathway" network. Finally, molecular docking was performed on curcumin and its key targets. We retrieved 115 curcumin-related targets from multiple drug databases. We further identified 376 differential genes and 413 curcumin targets. Among the 2587 targets for periodontitis diseases, 72 common curcumin-periodontitis targets were identified. Notably, AKT serine/threonine kinase 1, tumor protein P53, signal transducer and activator of transcription 3, matrix metallopeptidase 9, caspase-3, epidermal growth factor receptor, epidermal growth factor, cyclin D1, mitogen-activated protein kinase 14, and peroxisome proliferator activated receptor gamma were the key targets of curcumin in periodontitis treatment. Approximately 805 processes, including cellular response to chemical stress, and 39 signaling pathways, including forkhead box O1 signaling pathway, were enriched. All key targets, except tumor protein P53, had a high affinity for curcumin. Curcumin interacts with forkhead box O1, advanced glycosylation end-product specific receptor, mitogen-activated protein kinase, phosphoinositide 3 kinase-protein kinase B, and tumor necrosis factor signaling pathways in diabetic complications via key targets, including AKT serine/threonine kinase 1, mitogen-activated protein kinase 14, epidermal growth factor and epidermal growth factor receptor. Therefore, curcumin may reduce periodontal tissue inflammation and alveolar bone loss by inhibiting inflammatory cytokine expression and promoting osteoclast apoptosis, thereby facilitating periodontitis treatment.
Keywords
Curcumin, periodontitis, network pharmacology, diabetes, signaling pathway, lipopolysaccharide
Periodontitis is an inflammatory disease that affects the periodontal tissue and is characterized by the formation of periodontal pockets and the progressive destruction of the alveolar bone. It is one of the most common oral diseases[1]. Currently, periodontitis is primarily treated using basic treatment strategies, and adjuvant drug therapy controls inflammation and prevents further destruction of periodontal tissue[2]. However, antibiotic use is limited owing to side effects and the development of drug resistance. Therefore, traditional Chinese medicine has been widely preferred owing to its minimal side effects and clinical efficacy.
Curcumin is the main active component of the traditional Chinese medicine Curcuma longa. Curcumin has various pharmacological characteristics, including antioxidant, antiinflammatory, anti-cancer and antibacterial effects[3]. Animal models of periodontitis have indicated that curcumin can inhibit Lipopolysaccharide (LPS)- induced activation of the Nuclear Factor Kappa B (NF-κB) signaling pathway, reduce the expression of Interleukin (IL)-1beta (β) and Tumor Necrosis Factor-Alpha (TNF-α), and significantly alleviate periodontal inflammation and alveolar bone loss[4]. In clinical studies, 1 % curcumin solution has been used for repeated subgingival irrigation, and curcumin sustained-release tablets have been used in the periodontal pockets of patients with chronic periodontitis. The findings from these studies suggested that curcumin could effectively inhibit periodontal inflammation and improve periodontal indexes[5,6]. We hypothesized that curcumin likely has a therapeutic effect on periodontitis by inhibiting the NF-κB, Janus Kinase (JAK)-Signal Transducer and Activator of Transcription (STAT), and Mitogen- Activated Protein Kinase (MAPK) signaling pathways[7].
Treatment of diseases using traditional Chinese medicine involves a multi-target, multi-pathway process. Therefore, big data must be used to mine the targets and signaling pathways to further explore the action mechanism of curcumin in the treatment of periodontitis. Therefore, we used Gene Expression Omnibus (GEO) database mining and network pharmacology analysis to screen and predict the potential targets and signaling pathways of curcumin in the treatment of periodontitis. We believe our findings would provide novel insights for the development and clinical application of traditional Chinese medicine in periodontitis treatment.
Materials and Methods
The databases used in this study are listed in Table 1.
| Database | Website address |
|---|---|
| TCMSP | https://tcmspw.com/tcmsp.php |
| STITCH | http://stitch.embl.de/ |
| Swiss Target Prediction | http://www.swisstargetprediction.ch/ |
| Uniprot | https://www.uniprot.org/ |
| GEO | https://www.ncbi.nlm.nih.gov/geo/ |
| Gene Cards | https://www.genecards.org/ |
| DisGeNET | https://www.disgenet.org/ |
| Drug bank | https://go.drugbank.com/ |
| String | https://string-db.org/ |
| Alpha Fold | https://alphafold.ebi.ac.uk/ |
| PubChem | https://www.ncbi.nlm.nih,gov/home/chemicals/ |
Table 1: Basic Information Of The Database Used In This Article
Collection and screening of curcumin targets: "Curcumin" was used as the keyword in TCMSP, STITCH and Swiss Target Prediction databases to search and identify putative targets of curcumin. After repeated targets were removed, the protein target information was standardized using the UniProt database[8]. Using the same search term, microarray data numbered GSE10896, which contained 24 groups of data about the curcumin experimental and control groups, were obtained from the GEO database. The GEO2R tool in the GEO database was used for the analysis, and the results were saved after grouping. In R language (version 4.1.0), p<0.05, logFC >1, or logFC <−1 were set as the cutoff values to obtain differentially expressed genes, and the volcano map was plotted. The "limma" and "ggplot" packages were used to plot the differential gene heat map using GSE10896 microarray data. The obtained targets of curcumin and differential genes were merged, and redundant genes were removed. The predicted targets related to the pharmacological effects of curcumin were finalized.
Collection and screening of periodontitis targets: The periodontitis disease targets were obtained from the DisGeNET, Drugbank and GeneCards disease database using the search term "periodontitis." A Venn diagram was plotted for the periodontitis disease and predicted curcumin targets using the "ggvenn" package (R language) to identify the common targets of curcumin and periodontitis.
Protein-Protein Interaction (PPI) Network Construction:
The common targets of curcumin-periodontitis were imported to the STRING database, and the species was restricted to humans. A PPI network map was generated, and the PPI network was constructed using Cyto scape software (version 3.8.2).
The Network Analyzer plug-in was used to calculate several parameters, including the degree, clustering coefficient, and betweenness centrality. A degree ≥15 was used as the standard to screen key targets, and the marks were set to different shapes and sizes according to the degree value. The PPI network diagram was reconstructed, and the top 10 key targets were screened.
Gene Ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis:
The common targets of curcumin-periodontitis were imported into R software. GO and KEGG functional enrichment analysis of curcumin-periodontitis common target genes were performed through the "cluster profiler" package. Enrichment data of Biological Processes (BP), Cellular Components (CC) and Molecular Functions (MF), as well as major signaling pathways, were collected. The first 30 results were exported as a graph through the "ggplot2" package.
Construction of the curcumin-common targetpathway network:
The common targets and pathways between curcumin and periodontitis were imported into Cytoscape software and evaluated using the Network Analyzer plug-in. The signs were set to different shapes and sizes according to the degree value to correspond to curcumin, targets and pathways, to construct the curcumin-common target-pathway network diagram.
Molecular docking of curcumin and key targets: The Three-Dimensional (3D) structures of 10 key target proteins, which were regarded as protein receptors, screened by the PPI network were obtained from the Alpha Fold database[9]. The 3D structure of curcumin was obtained as a small-molecule ligand from the PubChem database and converted into PDBQT format[10]. AutoDock Vina (version 1.1.2) was used for molecular docking, and PyMOL (version 2.5.2) software was used for visualization.
Results and Discussion
FWe identified 4, 13 and 100 curcumin targets in the TCMSP, STITCH and Swiss Target Prediction databases, respectively. A total of 115 curcuminrelated targets were obtained after the combination. The GSE10896 microarray data from the GEO database contained experimental and control data (n=12 each) of curcumin intervention in human monocytes U937 cells. Differential expression analysis using the GEO2R web tool and filtering criteria of p<0.05 and logFC >1 or logFC <-1 resulted in 376 differentially expressed genes (Differentially Expressed Genes (DEGs); 232 downregulated and 144 upregulated genes), which were visualized using a volcano plot (fig. 1). According to the grouping information, a heat map of the DEGs was plotted (fig. 2). After deduplication, the 115 targets obtained from the three drug databases were combined with 376 differential genes obtained from chip data in the GEO database, and 413 predicted targets related to the pharmacological effects of curcumin were finalized.
A total of 2587 periodontitis disease targets were retrieved from the DisGeNET (113), drug bank (35) and gene cards (2439) disease database. A Venn diagram of periodontitis disease and predicted curcumin targets were plotted using the R program package, and 72 common curcumin-periodontitis targets were identified as shown in fig. 3.
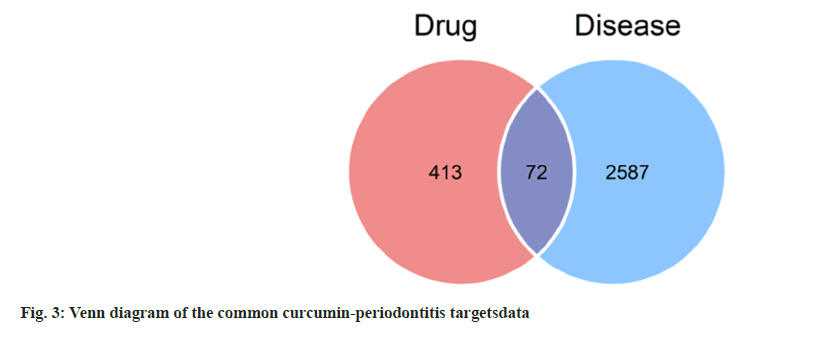
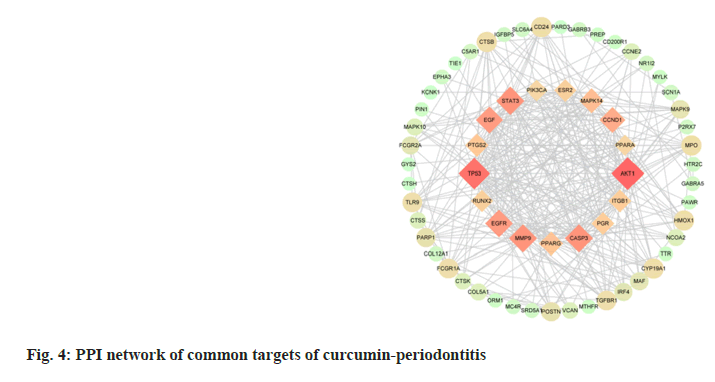
The 72 common targets of curcumin-periodontitis were imported to the STRING database to create a PPI network diagram. The closer the relationship between two nodes, the greater the degree value and the more crucial its position in the network. We identified 17 key targets, which are represented by diamonds (fig. 4). Among them, the top 10 targets were AKT Serine/Threonine Kinase 1 (AKT1), Tumor Protein P53 (TP53), STAT3, Matrix Metallopeptidase 9 (MMP9), Caspase-3 (CASP3), Epidermal Growth Factor Receptor (EGFR), EGF, Cyclin D1 (CCND1), Mitogen Activated Protein Kinase 14 (MAPK14), and Peroxisome Proliferator Activated Receptor Gamma (PPARG) as shown in Table 2.
| Target | Protein name | Degree | Clustering coefficient | Betweenness centrality | Binding energy (kcal/mol) |
|---|---|---|---|---|---|
| AKT1 | RAC-alpha serine/threonine-protein kinase | 36 | 0.34603 | 0.12891 | -5.438 |
| TP53 | Cellular tumor antigen p53 | 34 | 0.36542 | 0.20582 | -4.385 |
| STAT3 | Signal transducer and activator of transcription 3 | 28 | 0.47884 | 0.09712 | -6.634 |
| MMP9 | Matrix metalloproteinase-9 | 28 | 0.41534 | 0.1199 | -6.511 |
| CASP3 | Caspase-3 | 28 | 0.5 | 0.03712 | -6.778 |
| EGFR | Epidermal growth factor receptor | 27 | 0.46724 | 0.05377 | -6.778 |
| EGF | Epidermal growth factor | 26 | 0.52615 | 0.05383 | -6.027 |
| CCND1 | G1/S-specific cyclin-D1 | 24 | 0.57246 | 0.01986 | -5.826 |
| MAPK14 | Mitogen-activated protein kinase-14 | 21 | 0.64762 | 0.00953 | -7.685 |
| PPARG | Peroxisome proliferator-activated receptor gamma | 20 | 0.54211 | 0.05492 | -6.046 |
Table 2: Basic Information On The Key Targets Of Curcumin In The Treatment Of Periodontitis
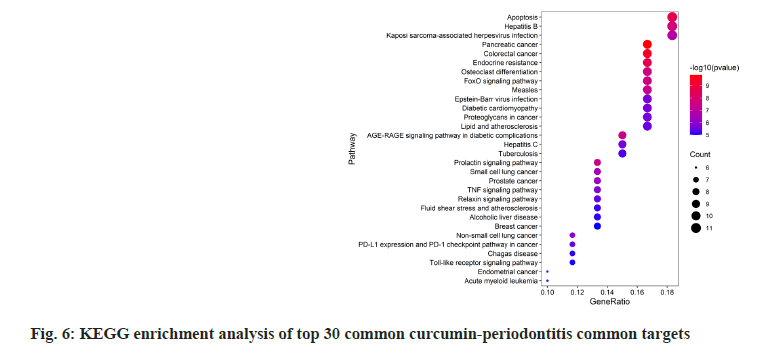
GO and KEGG enrichment analyses on 72 common curcumin-periodontitis targets helped identify 805 terms and 39 signaling pathways (p<0.05). The top 30 results were selected as the output (fig. 5 and fig. 6). We identified 16 enriched genes with count values >10, which were mainly associated with BP, such as response to nutrient levels, response to extracellular stimulus, response to oxygen levels, cellular response to chemical stress, and rhythmic processes (fig. 6). KEGG analysis suggested that the enriched genes belonged to the following top five pathways; Forkhead Box O1 (FoxO) (10), Advanced Glycosylation End-Product Specific Receptor (AGERAGE) signaling pathway in diabetic complications (9), MAPK (9), Phosphoinositide 3-Kinase (PI3K)- AKT (9), and TNF signaling pathways (8) as shown in fig. 6.
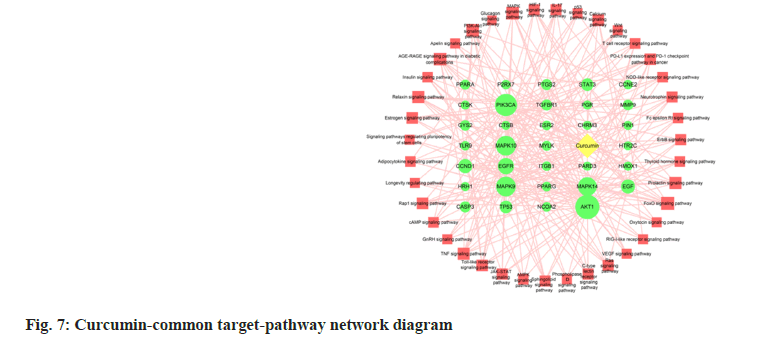
A curcumin-periodontitis-pathway network diagram was constructed using the common targets of drugs and diseases, and the KEGG-enriched pathways (fig. 7). The interaction network included 39 KEGG signaling pathways, 33 common drug-disease targets enriched in pathways (only the common targets with p<0.05 were included), and 260 edges. The top 10 common targets were AKT1, PIK3CA, MAPA9, MAPK10, MAPK14, EGFR, EGF, CCND1, STAT3 and TP53.
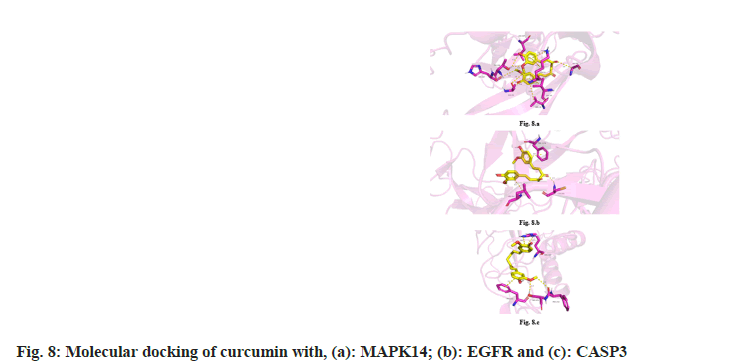
Molecular docking of the 10 key targets obtained from the PPI network with curcumin revealed that the binding energies of nine key targets, except TP53, were less than −5 kcal/mol. This suggests that these targets could bind spontaneously and stably to curcumin (Table 2). The binding energies of MAPK14, EGFR and CASP3 were the lowest at −7.685 kcal/mol, −7.383 kcal/mol and −6.778 kcal/ mol, respectively, indicating their excellent binding affinity to curcumin as shown in fig. 8.
The incidence of periodontitis is increasing annually and is currently the primary cause of tooth loss in adults, thereby severely affecting oral function[11,12]. Currently, the main drugs used to treat periodontitis are antibiotics, such as nitro imidazole, tetracycline and macrolides[13]. Curcumin, the main active component of turmeric, is effective in treating periodontitis[14]. However, we lack extensive reports on the targets and pathways through which curcumin exerts beneficial effects in periodontitis treatment. Therefore, we elucidated the potential action mechanism of curcumin in treating periodontitis using big data mining and network pharmacology. A total of 413 curcumin targets were obtained from the drug and GEO databases. Among the 2537 periodontitis targets, 73 common curcuminperiodontitis targets were identified. The top 10 key targets were AKT1, TP53, STAT3, MMP9, CASP3, EGFR, EGF, CCND1, MAPK14 and PPARG.
AKT is considered to be the main kinase regulating cell growth and survival, and is involved in regulating the PI3K signaling pathway. AKT1 activation can induce the proliferation and differentiation of mouse macrophages and simultaneously change the microenvironment from pro-inflammatory to anti-inflammatory by regulating NF-κB expression, thereby weakening the inflammatory alveolar bone resorption caused by periodontal tissue ligation in mice[15].
STAT3 can promote the expression of various proinflammatory products, including IL-6, IL-8, IL- 10 and TNF-α. These products bind to stromal cell receptors and activate the STAT3 signaling pathway, which plays a central role in inducing and maintaining the inflammatory microenvironment[16]. Curcumin can decrease MMP9 expression in polymorph nuclear leukocytes and inhibit extracellular matrix decomposition during inflammation[17]. In an inflammatory environment, the inhibition of CASP3 expression reduces epithelial cell apoptosis and contributes to mucosal homeostasis[18]. Serum EGF levels are associated with several tumors and inflammatory diseases. EGF and EGFR protein combinations can promote cell proliferation and differentiation through the MAPK/ERK pathway and participate in cell survival through the PI3KAKT- mammalian target of rapamycin pathway[19]. MAPK14, also known as p38α, is involved in signaling cascades that control cellular responses to cytokines and stress. It is closely related to many chronic inflammatory diseases and contributes to the production of proinflammatory cytokines. p38α inhibitors can be used to treat these diseases[20]. However, TP53, CCND1 and PPARG are considered key targets related to tumors or immunity and there is a lack of research on their role in periodontal disease or inflammation.
Among the 39 signaling pathways, the relationship between FoxO, AGE-RAGE signaling pathway in diabetic complications, MAPK, PI3K-AKT, and TNF signaling pathways and key targets are particularly important and were listed as key signaling pathways. The periodontal tissue of patients with periodontitis produces higher amounts of reactive oxygen species leading to oxidative stress and eventually periodontal tissue damage. Antioxidants can inhibit the PI3K/ AKT signaling pathway, which is crucial for inhibiting inflammation and apoptosis. The inhibition of this pathway leads to the upregulation of FoxO1 transcriptional activity and promotes osteoclast apoptosis[21,22]. Diabetes, an important risk factor for periodontitis, may aggravate the progression of periodontal pathogenesis. A large number of AGEs accumulate in the periodontal tissues of patients with periodontitis and diabetes. AGEs can increase the expression of IL-6 and Intercellular Adhesion Molecule 1 (ICAM1) in human gingival fibroblasts by binding to their receptor RAGE and MAPK pathways. This may aggravate periodontitis[22]. MAPK are critical regulators of cell survival, proliferation, apoptosis and differentiation. Activation of the MAPK signaling pathway can enhance inflammation, immune cell infiltration, M1 macrophage polarization, osteoclast formation and alveolar bone loss during periodontitis development and plays a vital role in alveolar bone loss and periodontitis aggravation[23]. The link between IL-17 and osteoclast proliferation highlights its crucial role in inflammatory bone destruction diseases. Some studies have reported its association with periodontitis. IL-17 can induce osteoclast production, disrupt bone homeostasis and lead to extensive bone destruction[24]. Simultaneously, IL-17 can stimulate human periodontal fibroblasts to secrete MMP9, IL-6, IL-8 and macrophages in periodontal tissue to release TNF-α and IL-1β[25], thus promoting periodontal inflammation, which has a strong synergistic effect with IL-17.
The upregulation of both cytokines can further stimulate the release of inflammatory mediators from fibroblasts and epithelial cells. TNF-α is a more potent inducer of matrix metalloproteinases (MMPs) than IL-17, but not of MMP9[25]. In addition, the activity of the JAK/STAT pathway may increase with an increase in proinflammatory cytokine levels[26]. Currently, the action mechanism of curcumin on periodontitis mainly focuses on the PI3KAKT signaling pathway. Curcumin can promote osteogenic differentiation of human Periodontal Ligament Stem Cells (hPDLSCs) by activating the PI3K-AKT signaling pathway[27]. However, we lack studies on other anti-inflammatory or pro-osteoclast apoptosis-related pathways, such as FoxO, the AGERAGE signaling pathway in diabetic complications, and MAPK and TNF signaling pathways. Through the analysis of ten key targets and five key pathways of curcumin, we provide a novel direction for future research.
In this study, GO and KEGG enrichment analyses were used to screen 73 common targets of curcumin for the treatment of periodontitis. GO analysis helped identify 16 terms with over 10 genes mainly enriched in BP, such as the response to nutrient levels, extracellular stimulation, oxygen content, cell response to chemical stimulation and rhythmic processes. The MF mainly include phosphatase binding, Deoxyribonucleic Acid (DNA)-binding transcription factor binding, protein serine/threonine kinase activity, protein phosphatase binding, and Ribonucleic Acid (RNA) polymerase II-specific DNA-binding transcription factor binding. CC included collagen-containing extracellular matrix, membrane rafts, membrane micro domain, vesicle lumen and apical part of the cell. KEGG pathway analysis of the common curcumin-periodontitis targets mainly focused on five key pathways, including FoxO signaling pathway. According to the curcumin-common target-pathway network diagram, we identified 10 targets, including six key targets, EGF, STAT3, MAPK14, EGFR, CCND1, and AKT1, in the FoxO signaling pathway and nine targets in the AGE-RAGE signaling pathway in diabetic complications. Four key targets, STAT3, MAPK14, AKT1 and CASP3, were involved in this process, and there were six, five, and five key targets involved in the MAPK, PI3K-AKT and TNF signaling pathways, respectively. AKT1 was involved in all five key pathways and MAPK14 was involved in the other four key pathways, except the PI3K-AKT signaling pathway. In contrast, EGF and EGFR were involved in the other three key pathways, except the AGERAGE and TNF signaling pathways, in diabetic complications.
The docking results indicated that the binding energy of the other nine key targets, except TP53, was <−5 kcal/mol, indicating that curcumin had a good affinity for these target proteins.
In summary, curcumin reduces periodontal tissue inflammation by inhibiting the expression of inflammatory factors and promoting osteoclast apoptosis through critical targets, such as AKT1, MAPK14, EGF, EGFR, FoxO, AGE-RAGE, MAPK, PI3K-AKT, and TNF signaling pathways in diabetic complications and alveolar bone loss in the treatment of periodontitis. However, experiments or clinical trials are required to validate the specific mechanism and therapeutic effects of curcumin. Nonetheless, we elucidated the characteristics of multiple targets and pathways by which curcumin mediates the treatment of periodontitis, thereby providing a possible direction for future research.
Acknowledgment:
The authors would like to thanks the Clinical Research Center Laboratory (The People's Hospital of Xinjiang Uygur Autonomous Region, Uygur, Xinjiang, China) for their technical support.
Funding:
This study was supported by Xinjiang Uygur Autonomous Region Regional Collaborative Innovation Special Fund (No. 2021E02071) and National Nature Science Foundation Project (No. 82260195).
Authors’ contributions:
All authors participated in the design of the experiment. Hailun Long performed the experiments, data analysis and wrote the first draft. Hailun Long wrote the revised manuscript and assisted in performing the experiments. Xiaomei Nan and Hongyan Zheng assisted in performing the experiments and processing pictures. Xiaotao Chen provides technical support and modifies the manuscript. Obtain permission from each person .All authors read and approved the final manuscript. All the authors identified in the manuscript as a source for unpublished data and agree on the final version of the manuscript to publish.
Conflict of interests:
The authors declared no conflict of interests.
References
- Slots J. Periodontitis: Facts, fallacies and the future. Periodontology 2017;75(1):7-23.
[Crossref] [Google Scholar] [PubMed]
- Kwon T, Lamster IB, Levin L. Current concepts in the management of periodontitis. Int Dental J 2021;71(6):462-76.
[Crossref] [Google Scholar] [PubMed]
- Hatamipour M, Ramezani M, Tabassi SA, Johnston TP, Sahebkar A. Demethoxycurcumin: A naturally occurring curcumin analogue for treating non-cancerous diseases. J Cell Physiol 2019;234(11):19320-30.
[Crossref] [Google Scholar] [PubMed]
- Xiao CJ, Yu XJ, Xie JL, Liu S, Li S. Protective effect and related mechanisms of curcumin in rat experimental periodontitis. Head Face Med 2018;14(1):1-8.
[Crossref] [Google Scholar] [PubMed]
- Chatterjee A, Debnath K, Rao NK. A comparative evaluation of the efficacy of curcumin and chlorhexidine mouth rinses on clinical inflammatory parameters of gingivitis: A double-blinded randomized controlled clinical study. J Indian Soc Periodontol 2017;21(2):132.
[Crossref] [Google Scholar] [PubMed]
- Singh A, Sridhar R, Shrihatti R, Mandloy A. Evaluation of turmeric chip compared with chlorhexidine chip as a local drug delivery agent in the treatment of chronic periodontitis: A split mouth randomized controlled clinical trial. J Altern Complement Med 2018;24(1):76-84.
[Crossref] [Google Scholar] [PubMed]
- Li Y, Jiao J, Qi Y, Yu W, Yang S, Zhang J, et al. Curcumin: A review of experimental studies and mechanisms related to periodontitis treatment. J Periodontal Res 2021;56(5):837-47.
[Crossref] [Google Scholar] [PubMed]
- UniProt Consortium. UniProt: The universal protein knowledgebase. Nucl Acids Res 2018;46(5):2699.
[Crossref] [Google Scholar] [PubMed]
- Burley SK, Bhikadiya C, Bi C, Bittrich S, Chen L, Crichlow GV, et al. RCSB Protein Data Bank: powerful new tools for exploring 3D structures of biological macromolecules for basic and applied research and education in fundamental biology, biomedicine, biotechnology, bioengineering and energy sciences. Nucl Acids Res 2021;49(D1):D437-51.
[Crossref] [Google Scholar] [PubMed]
- Syriopoulou A, Markopoulos I, Tzakos AG, Mavromoustakos T. Ligand–receptor interactions and drug design. Methods Mol Biol 2021;2266:89-104.
- Reynolds I, Duane B. Periodontal disease has an impact on patients' quality of life. Evid Based Dentistry 2018;19(1):14-5.
[Crossref] [Google Scholar] [PubMed]
- Tonetti MS, Jepsen S, Jin L, Otomo-Corgel J. Impact of the global burden of periodontal diseases on health, nutrition and wellbeing of mankind: A call for global action. J Clin Periodontol 2017;44(5):456-62.
[Crossref] [Google Scholar] [PubMed]
- Devji T. Mechanical and antibiotic periodontal therapies may be no different in terms of tooth loss in patients with chronic periodontitis. J Am Dental Assoc 2017;148(9):e124.
- Guimaraes MR, de Aquino SG, Coimbra LS, Spolidorio LC, Kirkwood KL, Rossa Jr C. Curcumin modulates the immune response associated with LPS-induced periodontal disease in rats. Innate Immun 2012;18(1):155-63.
[Crossref] [Google Scholar] [PubMed]
- Wu X, Wang Y, Chen H, Wang Y, Gu Y. Phosphatase and tensin homologue determine inflammatory status by differentially regulating the expression of Akt1 and Akt2 in macrophage alternative polarization of periodontitis. J Clin Periodontol 2023;50(2):220-31.
[Crossref] [Google Scholar] [PubMed]
- Ji Z, He L, Regev A, Struhl K. Inflammatory regulatory network mediated by the joint action of NF-kB, STAT3 and AP-1 factors is involved in many human cancers. Proc Natl Acad Sci 2019;116(19):9453-62.
[Crossref] [Google Scholar] [PubMed]
- Guru SR, Kothiwale SV, Saroch N, Guru RC. Comparative evaluation of inhibitory effect of curcumin and doxycycline on matrix metalloproteinase-9 activity in chronic periodontitis. Indian J Dental Res 2017;28(5):560-5.
[Crossref] [Google Scholar] [PubMed]
- Kuo WT, Shen L, Zuo L, Shashikanth N, Ong ML, Wu L, et al. Inflammation-induced occludin downregulation limits epithelial apoptosis by suppressing caspase-3 expression. Gastroenterology 2019;157(5):1323-37.
[Crossref] [Google Scholar] [PubMed]
- Dankner M, Rose AA, Rajkumar S, Siegel PM, Watson IR. Classifying BRAF alterations in cancer: New rational therapeutic strategies for actionable mutations. Oncogene 2018;37(24):3183-99.
[Crossref] [Google Scholar] [PubMed]
- Madkour MM, Anbar HS, El-Gamal MI. Current status and future prospects of p38α/MAPK14 kinase and its inhibitors. Eur J Med Chem 2021;213:113216.
[Crossref] [Google Scholar] [PubMed]
- Feng YL, Jiang XT, Ma FF, Han J, Tang XL. Resveratrol prevents osteoporosis by upregulating FoxO1 transcriptional activity. Int J Mol Med 2018;41(1):202-12.
[Crossref] [Google Scholar] [PubMed]
- Nonaka K, Kajiura Y, Bando M, Sakamoto E, Inagaki Y, Lew JH, et al. Advanced glycation end-products increase IL-6 and ICAM-1 expression via RAGE, MAPK and NF-κB pathways in human gingival fibroblasts. J Periodontal Res 2018;53(3):334-44.
[Crossref] [Google Scholar] [PubMed]
- Wang L, Zheng J, Pathak JL, Chen Y, Liang D, Yang L, et al. SLIT2 overexpression in periodontitis intensifies inflammation and alveolar bone loss, possibly via the activation of MAPK pathway. Front Cell Dev Biol 2020;8:593.
[Crossref] [Google Scholar] [PubMed]
- Sato K, Suematsu A, Okamoto K, Yamaguchi A, Morishita Y, Kadono Y, et al. Th17 functions as an osteoclastogenic helper T cell subset that links T cell activation and bone destruction. J Exp Med 2006;203(12):2673-82.
[Crossref] [Google Scholar] [PubMed]
- Beklen A, Ainola M, Hukkanen M, Gürgan C, Sorsa T, Konttinen YT. MMPs, IL-1 and TNF are regulated by IL-17 in periodontitis. J Dental Res 2007;86(4):347-51.
[Crossref] [Google Scholar] [PubMed]
- Jaiswal S, Natarajan P, Silver AJ, Gibson CJ, Bick AG, Shvartz E, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med 2017;377(2):111-21.
[Crossref] [Google Scholar] [PubMed]
- Xiong Y, Zhao B, Zhang W, Jia L, Zhang Y, Xu X. Curcumin promotes osteogenic differentiation of periodontal ligament stem cells through the PI3K/AKT/Nrf2 signaling pathway. Iranian J Basic Med Sci 2020;23(7):954.
[Crossref] [Google Scholar] [PubMed]
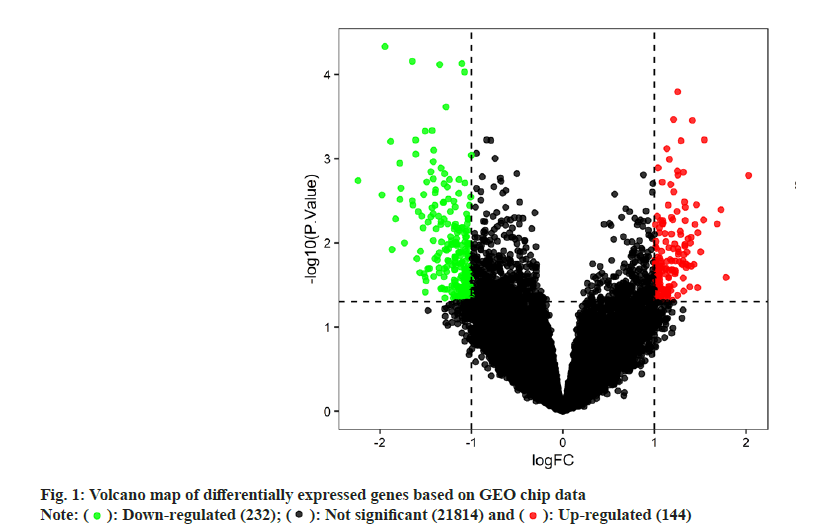
 ): Down-regulated (232); (
): Down-regulated (232); (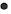 ): Not significant (21814) and (
): Not significant (21814) and ( ): Up-regulated (144)
): Up-regulated (144)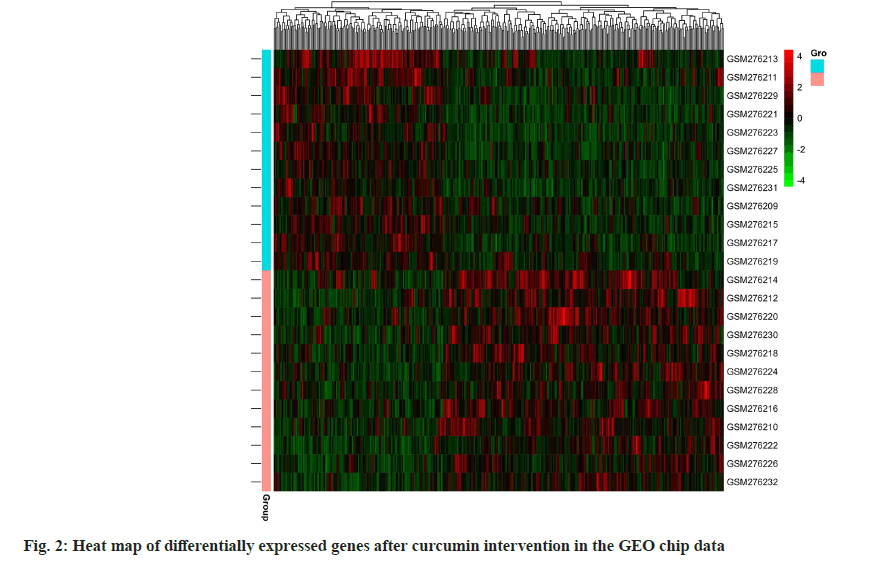
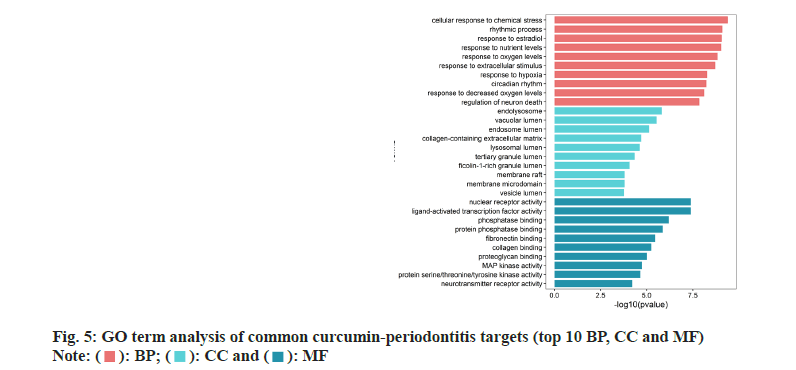
 ): BP; (
): BP; ( ): CC and (
): CC and ( ): MF
): MF