- *Corresponding Author:
- Dan Xu
Department of Clinical Laboratory, Xianyang Central Hospital, Xianyang, Shaanxi Province 712000, China
E-mail: 1165652549@qq.com
| This article was originally published in a special issue,“Clinical Advancements in Life Sciences and Pharmaceutical Research” |
| Indian J Pharm Sci 2024:86(5) Spl Issue “34-41” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
An integrated as well as an innovative deferentially expressed gene profile for hepatocellular carcinoma patients treated with sorafenib was established and provided several beneficial clinical hints for the drug usage. The messenger ribonucleic acid sequencing data and micro ribonucleic acid sequencing data from 114 hepatocellular carcinoma patients treated with sorafenib and 226 hepatocellular carcinoma control group patients who did not receive any drug therapy were studied by differential expression, functional enrichment and protein-protein interaction analysis. Screening target genes were tested in hepatocellular carcinoma patients treated with sorafenib and control group. 466 differentially expressed genes were identified in patients treated with sorafenib group compared with the control group. Among these deferentially expressed genes, 57 demonstrated increased expression level while 409 displayed decreased expression pattern. 12 micro ribonucleic acids showed differential expression patterns. With further examination, 5-hydroxytryptamine receptor 2C and thyrotropin releasing hormone, shown to be major targets for sorafenib. The potential target genes were also examined; 5-hydroxytryptamine receptor 2C concentration in control group hepatocellular carcinoma patients was approximately 61.43±10.5 ng.ml-1. However, after sorafenib treatment 5-hydroxytryptamine receptor 2C concentration decreased (25.34±6.31 ng.ml-1) (p<0.001). At the same time, thyrotropin releasing hormone concentration was 54.17±13.17 ng.ml-1 control group patients which decreased significantly in sorafenib group hepatocellular carcinoma patients (22.51±6.76 ng.ml-1 and p<0.001). 5-Hydroxytryptamine receptor 2C and thyrotropin releasing hormone can be considered as potential markers of sorafenib treatment. This study comprehensively studied the targets for sorafenib through a deep analysis of differentially expressed gene map and offered valuable references for clinical usage of the drug in hepatocellular carcinoma patients.
Keywords
Hepatocellular carcinoma, sorafenib, protein-protein interaction
Hepatocellular Carcinoma (HCC) has been considered as the most frequently occurring malignant tumor of liver, accounting for >70 % of entire liver cancer cases[1,2]. Hepatitis B Virus (HBV) and Hepatitis C Virus (HCV) have been long thought as major factors in the initiation as well as development of HCC[3]. The pathogenesis of HCC induced by virus involves many mechanisms[4-6].
Sorafenib, which is commercial name as nexavar is a chemical compound that has widely been applied in the clinical treatment of HCC patients[7-9]. In its mechanism, sorafenib acts as an multi-kinase blocker which massively affects the tumor cell growth as well as angiogenesis and accelerates the apoptosis of tumor cell[10-12]. As a small polytyrosine kinase inhibitor, sorafenib has been demonstrated as a primary inhibitor specially for Platelet Derived Growth Factor (PDGF), Vascular Endothelial Growth Factor (VEGF) as well as Rapidly Accelerated Fibrosarcoma (RAF) kinase[13-16].
As an effector for this signaling cassette, it is reasonable to assume that sorafenib is not an unique regulator in HCC patients. Indeed, sorafenib not only interferes with HCC tumor cell manner but also interacts with other cancer cells including breast, pancreatic and colon cancer[17]. Several side effects including diarrhea, weight loss and skin reactions on the hands and feet have been reported in patients treated with sorafenib[18]. In addition, a significant proportion of HCC patients develop resistance towards sorafenib[19]. The key target genes regulated by sorafenib are still unclear and the related molecular mechanisms remain poorly understood, which greatly restrict its clinical application.
In order to solve these problems, a deep differential expression analysis of sorafenib was established in this study. The major target genes and precise molecular pathways of sorafenib were comprehensively explored. These results provide potential guidance for precise drug administration of sorafenib which also provide great help to HCC patients.
Materials and Methods
Database analysis:
Data source: The genome Ribonucleic Acid (RNA) and (mi) micro RNA sequencing data was obtained from The Cancer Genome Atlas (TCGA) (https://TCGA-data.nci.nih.gov/TCGA/). The 226 HCC patients’ samples involved in the study included patients who did not administer any chemotherapy drugs and 114 HCC patients who administrated sorafenib. The clinical characteristics of the Clonal Hematopoiesis of Indeterminate Potential (CHIP) have been presented in Table 1.
| Features | Group | Patients | |
|---|---|---|---|
| Number (n) | % | ||
| Gender | Male | 236 | 69.41 |
| Female | 104 | 30.59 | |
| Age | ≤61 | 164 | 48.24 |
| ≥61 | 176 | 51.76 | |
| Clinical stage | I | 162 | 47.65 |
| II | 77 | 22.65 | |
| III | 77 | 22.65 | |
| IV | 4 | 1.18 | |
| Virus status | Unknown | 20 | 5.88 |
| Survival | 264 | 77.65 | |
| Death | 76 | 22.35 | |
| Adjacent liver tissue inflammation degree type | No | 104 | 30.59 |
| Mild | 93 | 27.35 | |
| Severe | 18 | 5.29 | |
| Unknown | 125 | 36.76 | |
| Vascular tumor status | No | 195 | 57.35 |
| Low | 84 | 24.71 | |
| Advanced | 13 | 3.82 | |
| Chemical drug use | Sorafenib | 114 | 33.53 |
| No drug | 226 | 66.47 | |
Table 1: Clinicopathological Features of 340 HCC Patients in TCCA
Differential expression analysis: The empirical analysis of digital gene expression data in R (edgeR) software package in R language was established to examine analyze differentially expressed messenger (m) RNA and miRNAs[20].
Protein-Protein Interaction (PPI): Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) (https://string-db.org/) version 11.0 was used to analyze the functional connections and interactions of potential proteins where confidence score≥0.4 was finally selected. The visual PPI network was built using Cytoscape (https://cytoscape.org/) version 3.7.2.
Functional enrichment analysis: In this study, the ClusterProfiler package in R language was established for Gene Ontology (GO) analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis.
Cell-type Identification by Estimating Relative Subsets of RNA Transcripts (CIBERSORT) analysis: It is an universal method which is used for quantifying the Cellular Components (CC) from cancer tissue gene expression profiles. Compared to the previous work, CIBERSORT has implemented a machine learning method called support vector regression, which improves the deconvolution performance by combining feature selection and powerful mathematical optimization techniques. In benchmark experiments, CIBERSORT is more accurate than other methods in separating, closely related cell subpopulations and mixtures with unknown cell types (such as solid tissue). Therefore, CIBERSORT is a useful method for high-throughput characterization of various cell types from complex tissues, such as tumor infiltrating lymphocytes. In order to study the tumor infiltrating lymphocytes, the feature of matrix input by CIBERSORT is called Leukocyte gene signature Matrix (LM22), which contains 547 genes and can distinguish 22 human hematopoietic cell phenotypes, including 7 T cell types, naive and memory B cells, plasma cells, Natural Killer (NK) cells and myeloid cell subsets.
General information:
83 HCC patients diagnosed pathologically were randomly selected, 4 of whom were excluded and totally 79 patients were included in the experiment. Among the 79 patients, 49 were male while 30 were female and were randomly assigned into two groups, HCC patients treated with sorafenib (n=48) and HCC patients not treated with sorafenib (n=31). The concentration of key target genes, 5-Hydroxytryptamine Receptor 2C (HTR2C) and Thyrotropin Releasing Hormone (TRH) were measured using Enzyme-Linked Immunosorbant Assay (ELISA) (Abcam company) double antibody sandwich method.
Informed consent from the patients and their families was taken which was approved by the hospital ethical review committee (Approval No: 2023TJZL670983).
Inclusion criteria: Individuals within the age of (40-80) y; patients who signed the informed consent form; patients whose clinical data was complete and patients meeting the clinical diagnostic criteria for HCC. It included the presence of HBV and/or HCV antigen (positive) in clinical diagnosis; if the mass diameter of the liver 1~2 cm or ≥2 cm; patients diagnosed with HCC in Computed Tomography (CT) and Magnetic Resonance Imaging (MRI); if the serum Alpha-Fetoprotein (AFP)≥400 μg/l for 1 mo or ≥200 μg/l for 2 mo and other causes of AFP elevation were included in the study.
Exclusion criteria: Presence of other malignancies other than HCC; presence of other chronic liver diseases; patients with germ line embryogenic tumors; patients with active liver disease; patients having secondary liver cancer and the patients who did not sign the consent from were excluded from the study.
Statistical analysis:
Excel 2013 software was used to establish the database. Statistical Analysis Software (SAS) version 9.4 and Statistical Package for Social Sciences (SPSS) version 19.0 statistical software were used to analyze the data. Continuous variables were tested for normal distribution and the variables conforming to normal distribution were expressed as mean±standard deviation (x±s) and t-test was used to compare the data where p<0.05 was considered as statistically significant difference.
Results and Discussion
The basic clinical data of patients was compared. HCC patients of the two groups, sorafenib-treated and non-sorafenib (control group) groups were evenly distributed, covering all clinical stages of HCC; they also included various types in terms of viral status and inflammation of adjacent liver tissues (Table 1).
Differentially expressed mRNAs and Differentially Expressed miRNAs (DEM) were analyzed. Sorafenibtreated HCC patients exhibited 466 Differentially Expressed Genes (DEG), including 57 up-regulated genes and 409 down-regulated genes, compared to HCC patients not treated with any chemical drugs (fig. 1A). Sorafenib-treated patients, on the other hand exhibited 12 DEM, including 7 up-regulated miRNAs and 5 down-regulated miRNAs (fig. 1B).
miRNA target gene identification was studied. The microRNA Target Prediction Database (miRDB) predicted the target genes of these 12 DEMs and identified a total of 7267 target genes. Among these, the 7267 genes interacted with 466 DEG, resulting in 58 genes specifically expressed in HCC patients treated with sorafenib.
Cluster 1 contains HTR2C, TRH, Angiotensin II Receptor Type 2 (AGTR2) and Melanin Concentrating Hormone Receptor 2 (MCHR2) genes, while Cluster2 contains Mucin 15 (MUC15), MUC17 and Polypeptide N-Acetylgalactosaminyltransferase Like 6 (GALNTL6) genes (fig. 2A); 10 of the 58 interacting genes were screened out. HTR2C has the largest nodal degree, with a nodal degree of 6 and TRH is the 2nd most important gene (fig. 2B).
GO and KEGG functional enrichment analysis was carried out. For the 58 genes specifically expressed in HCC patients treated with sorafenib, GO analysis (including Biological Process (BP), Molecular Function (MF) and CC) was carried out; further, KEGG pathway enrichment analysis was also conducted.
As shown in fig. 3A, the pathways like interleukin-1 mediated signaling pathway etc., were significantly up-regulated while T-cell activation pathways were down-regulated (fig. 3B). At the same time, toll-like receptor signaling pathway was obviously activated and T helper 17 (Th17) cell differentiation processes while others were deactivated (fig. 3C and fig. 3D).
Infiltrating immune cells were analyzed. Since the immune cells related signaling pathways were primarily enriched in HCC patients treated with sorafenib, we analyzed the proportion of 22 types of immune cells in each patient in sorafenib group and compared the difference in the proportion of 22 types of infiltrating immune cells based on the control group. The proportion of immature B cells, resting memory Cluster of Differentiation 4 (CD4+) and (CD8+) T cells, activated memory CD4+, follicular Th cells, gamma delta T cells, (M) Macrophage 1, resting dendritic like cells and activated dendritic like cells in the sorafenib treatment group was significantly lower than that in the control group. The proportion of resting NK cells, M0 macrophages, M2 macrophages, and resting mast cells in sorafenib treatment group was higher than that in the control group (fig. 4).
HTR2C and TRH were used as important markers to detect the therapeutic effect of sorafenib in clinical practice. HTR2C concentration in sorafenib-treated group of HCC patients was significantly lower than that in control group which was detected by ELISA double-antibody sandwich method (t=8.16 and p<0.001). TRH concentrations in HCC patients treated with sorafenib were also significantly lower than those in control group (t=7.03 and p<0.001) (Table 2).
| Group | HTR2C (ng.ml-1) | TRH (ng.ml-1) |
|---|---|---|
| Non-sorafenib (control) | 61.43±10.5 | 54.17±13.17 |
| Sorafenib | 25.34±6.31 | 22.51±6.76 |
| t | 8.16 | 7.03 |
| p | <0.001 | <0.001 |
Table 2: Comparison of HTR2C and TRH Concentrations Between Two Different Groups (X?±S)
Currently, the incidence of HCC is increasing yearly worldwide[21]. At present, sorafenib seems to be a well-known treatment option for HCC. However, it can even lead to hypertension, abdominal pain and eventually discontinuation of treatment. All of this seem to explain the fact that the target genes in sorafenib treatment for HCC are currently unknown. Therefore, it is significant that this study establishes a comprehensive gene specific expression profile in HCC patients treated with sorafenib alone.
Here, 57 up-regulated and 409 down-regulated genes and 12 DEMs were explored compared to the HCC patients in the control group. Using PPI networks, 2 most important target genes namely, HTR2C and TRH were further deleted. TRH represents the 30-35 long nucleotide 5’ transfer (t) RNA half 5’ tRNA-halves. TRH gene abundance is associated with the expression of tRNA-cleaving ribonuclease angiopoietin[22]. TRH has previously been reported to use immunochemical methods to study the malignant nature of metastatic HCC in tumor samples from patients with metastatic HCC[23]. At present, the research on HTR2C mainly focuses on non-small cell lung cancer and specific epilepsy, but has not been carried out in HCC research[24]. However, there are reports suggesting a close link between HTR2C and Thyroid Hormone (TH) signaling, which increases the possibility that TH regulation disorders affect the liver microenvironment and HCC formation[25]. In addition to the target genes, 12 potential miRNA candidates were identified in this study. Subsequently, we studied the molecular biological pathways between these miRNAs and 2 major genes which are the key for better evaluation of the efficacy of sorafenib in HCC treatment. Since both genes have shown to be down-regulated in treated patients, sorafenib may inhibit 2 key genes through 12 major miRNAs.
At present, the clinical application limitations of sorafenib include unclear associated molecular signals and difficulty in evaluating efficacy. The clinical efficacy of HCC patients now mainly depends on imaging methods especially in the early stages of chemical drug administration, the clinical treatment effect of HCC patients does not progress significantly, and it is difficult to evaluate some cases through imaging methods. Often, this period is a critical time point for HCC patient treatment, which can easily lead to erroneous judgment of results[26,27]. In this study, the two key genes deleted were tested clinically, and the results can confirm that the genes, HTR2C and TRH were significantly different between the sorafenib-treated group and the non-sorafenib (control) group. These two genes can be used as clinical markers of sorafenib treatment effect; ELISA method is a commonly used technique in clinical laboratory at present. It has simple operation, ELISA assay for HTR2C and TRH concentrations will be an important complementary tool for imaging evaluation of sorafenib therapy for HCC and quantitative evaluation of the results will help HCC patients to choose the appropriate treatment more quickly and accurately.
Here, we aimed to utilize the two specific genes, HTR2C and TRH to evaluate the efficiency of sorafenib. At the same time, the dosage responses of sorafenib for all the individuals are variable. Thus, the results of the study might give some suggestions for dose usage recommendation. Moreover, application of the drug could generate numerous of side effects, like diarrhea and weight loss, etc.[18]. To deeply explore these specific expressed genes, researchers might be able to pin-point the central sources of side effects and provide some possibilities of safety administration.
In summary, given the increasing dependence of sorafenib on HCC therapy and the limited understanding of sorafenib’s function, this study summarizes the differential expression profiles of potential target genes and miRNAs in sorafenibtreated HCC patients compared with control group HCC patients. Several specific target genes and miRNAs have established specific expression patterns, which opens up new insights into sorafenib’s therapeutic approach in clinical applications. In order to make HTR2C and TRH as specific markers of sorafenib efficacy widely used in clinical trials, future multicenter studies with large sample sizes are needed.
Author’s contributions:
Zhichao Sun, Ye Zhao and Dan Xu have equally contributed for this study.
Funding:
This study was supported by the Tianjin Key Medical Discipline (Specialty) Construction Project.
Conflict of interests:
The authors declared no conflict of interests.
References
- Forner A, Reig M, Bruix J. Hepatocellular cancer. Lancet 2018;391(10127):1301-14.
[Crossref] [Google Scholar] [PubMed]
- Colquhoun SD. Hepatocellular carcinoma: The current role of surgical intervention. Crit Rev Oncog 2016;21(1):93-103.
[Crossref] [Google Scholar] [PubMed]
- Hartke J, Johnson M, Ghabril M. The diagnosis and treatment of hepatocellular carcinoma. Semin Diagn Pathol 2017;34 (2):153-9.
[Crossref] [Google Scholar] [PubMed]
- El-Serag HB. Hepatocellular carcinoma. New English J Med 2011;365(12):1118-27.
- Grandhi MS, Kim AK, Ronnekleiv-Kelly SM, Kamel IR, Ghasebeh MA, Pawlik TM. Hepatocellular cancer: From diagnosis to treatment. Surg Oncol 2016;25(2):74-85.
[Crossref] [Google Scholar] [PubMed]
- Kudo M. Targeted and immune therapies for hepatocellular carcinoma: Predictions for 2019 and beyond. World J Gastroenterol 2019;25(7):789-807.
[Crossref] [Google Scholar] [PubMed]
- Golabi P, Rhea L, Henry L, Younossi ZM. Hepatocellular cancer and non-alcoholic fatty liver disease. Hepatol Int 2019;13(6):688-94.
[Crossref] [Google Scholar] [PubMed]
- Lurje I, Czigany Z, Bednarsch J, Roderburg C, Isfort P, Neumann UP, et al. Treatment strategies for hepatocellular carcinoma-a multidisciplinary approach. Int J Mol Sci 2019;22(6):1465-70.
[Crossref] [Google Scholar] [PubMed]
- Khemlina G, Ikeda S, Kurzrock R. The biology of hepatocellular cancer: Implications for genetic and immune therapies. Mol Cancer Res 2017;16(1):149-52.
[Crossref] [Google Scholar] [PubMed]
- Santos AL, Cardoso H, Silva M, Moreira A, Macedo G. Hepatocellular cancer treatment with Sorafenib: A remarkable case of eight-years remission without significant toxicity. Ann Hepatol 2019;24(1):34-40.
[Crossref] [Google Scholar] [PubMed]
- Bertacco A, Vitale A, Mescoli C, Cillo U. Sorafenib treatment has the potential to downstage advanced hepatocellular carcinoma before liver resection. Pers Med 2020;17(2):83-7.
[Crossref] [Google Scholar] [PubMed]
- Feng S, Zhou J, Li Z, Appelman HD, Zhao L, Zhu J, et al. Sorafenib encapsulated in nanocarrier functionalized with glycan-3 specific peptide for targeted therapy of hepatocellular carcinoma. Colloids Surf B Biointerfaces 2019;184(1):1-11.
[Crossref] [Google Scholar] [PubMed]
- Mammatas LH, Zandvliet AS, Rovithi M, Honeywell RJ, Swart EL, Peters GJ, et al. Sorafenib administered using a high dose, pulsatile region in patients with advanced solid malices: A phase I exposure escalation study. Cancer Chemother Pharmacol 2019;85(5):931-40.
[Crossref] [Google Scholar] [PubMed]
- Pearson H, Marshall LV, Carceller F. Sorafenib in pediatrics and pathocellular cancer from a clinical perspective. Pediatric Hematol Oncol 2020;37(5):412-23.
[Crossref] [Google Scholar] [PubMed]
- Labeur TA, Achterbergh R, Takkenberg B, van Delden O, Mathôt R, Klümpen HJ. Sorafenib for patients with hepatocellular carcinoma and child Pugh B liver cirrhosis: Lessons learned from a terminated study. Oncologist 2020;25(9):1274-79.
[Crossref] [Google Scholar] [PubMed]
- Hu J, Wang E, Liu L, Wang Q, Xia D, Bai W, et al. Sorafenib may enhance antitumour efficacy in hepatocellular carcinoma patients by modulating the proportions and functions of natural killer cells. Invest New Drugs 2020;38(5):1247-56.
[Crossref] [Google Scholar] [PubMed]
- Zhuang BW, Li W, Xie XH, Hu HT, Lu MD, Xie XY. Sorafenib vs. hepatic arterial infusion chemotherapy for advanced hepatocellular carcinoma: A systematic review and meta-analysis. Jpn J Clin Oncol 2019;49(9):845-55.
[Crossref] [Google Scholar] [PubMed]
- Pathania YS. Hand foot skin reaction induced by sorafenib in a hepatic cancer patient. Postgrad Med J 2020;96(1138):495-501.
[Crossref]
- Raoul JL, Adhoute X, Penaranda G, Perrier H, Castellani P, Oules V, et al. Sorafenib: Experience and better management of side effects improve overall survival in hepatocellular carcinoma patients: A real life retrospective analysis. Liver Cancer 2019;8(6):457-67.
[Crossref] [Google Scholar] [PubMed]
- Robinson MD, McCarthy DJ, Smyth GK. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010;26(1):139-40.
[Crossref] [Google Scholar] [PubMed]
- Villanueva A. Hepatocellular carcinoma. New English J Med 2019;380(15):1450-62.
- Wang J, Yang T, Chen H, Li H, Zheng S. Oncogene RPA1 promotes promotion of hepatocellular carcinoma via CDK4/Cyclin-D pathway. Biochem Biophys Res Commun 2018;498(3):424-30.
[Crossref] [Google Scholar] [PubMed]
- Selitsky SR, Baran-Gale J, Honda M, Yamane D, Masaki T, Fannin EE, et al. Small tRNA derived RNAs are increasing and more abundant than microRNAs in chronic hepatitis B and C. Sci Rep 2015;5(1):7675-80.
[Crossref] [Google Scholar] [PubMed]
- Ganesan P, Kulik LM. Hepatocellular carcinoma: New developments. Clin Liver Dis 2023;27(1):85-102.
[Crossref] [Google Scholar] [PubMed]
- Manka P, Coombes JD, Boosman R, Gauthier K, Papa S, Syn WK. Thyroid hormone in the regulation of hepatocellular carcinoma and its microenvironment. Cancer Lett 2018;419(1):175-86.
[Crossref] [Google Scholar] [PubMed]
- Brown ZJ, Tsilimigras DI, Ruff SM, Mohseni A, Kamel IR, Cloyd JM, et al. Management of hepatocellular carcinoma: A review. JAMA Surg 2023;158(4):410-20.
[Crossref] [Google Scholar] [PubMed]
- Wang Y, Deng B. Hepatocellular carcinoma: Molecular mechanism, targeted therapy, and biomarkers. Cancer Metastasis Rev 2023;42(3):629-52.
[Crossref] [Google Scholar] [PubMed]
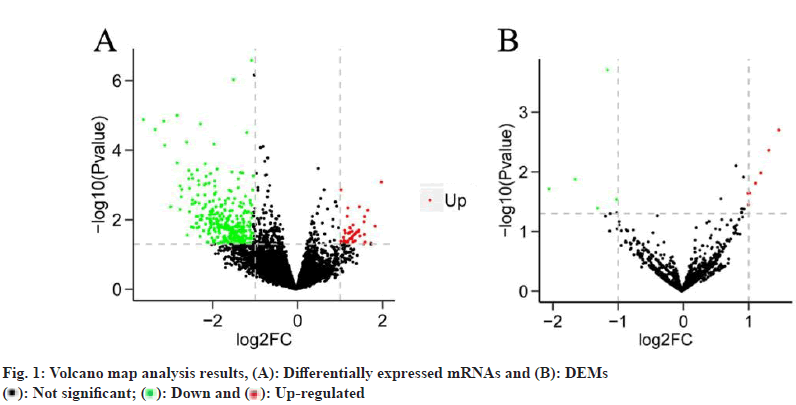
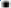 ): Not significant; (
): Not significant; ( ): Down and (
): Down and ( ): Up-regulated
): Up-regulated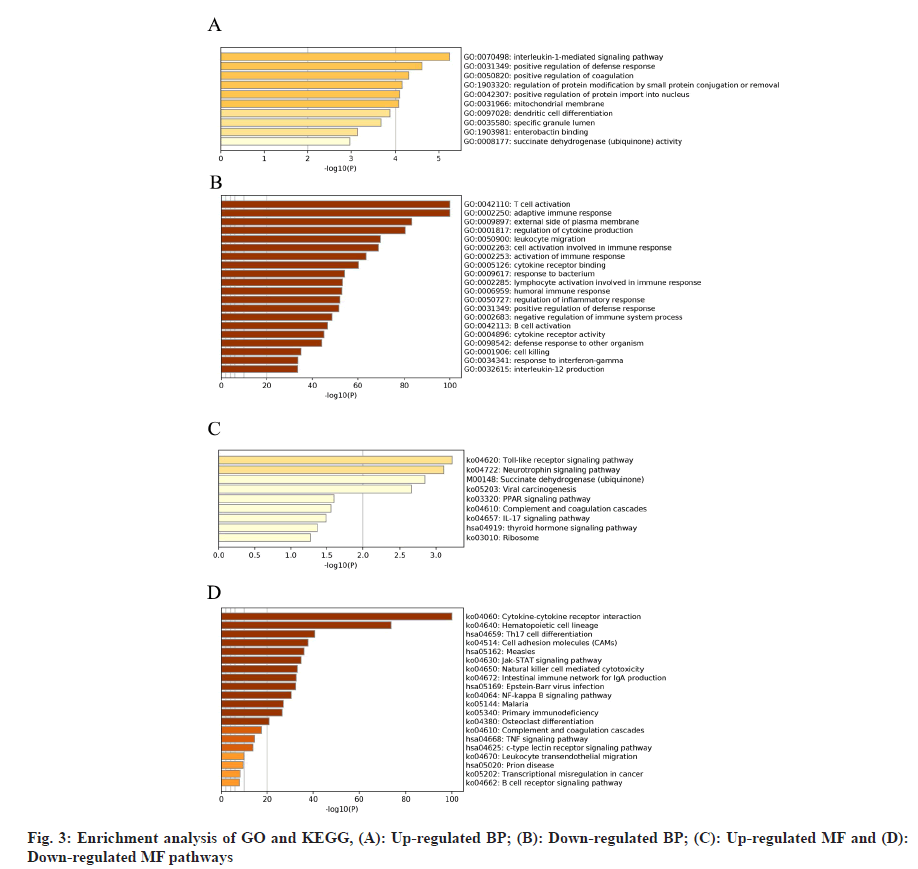
 ): High and (
): High and ( ): Low risk score, (*), (**), (***) and (****) are p<0.05, p<0.01, p<0.001 and p<0.0001, respectively while (ns): Not significant
): Low risk score, (*), (**), (***) and (****) are p<0.05, p<0.01, p<0.001 and p<0.0001, respectively while (ns): Not significant



