- *Corresponding Author:
- Hayun Hayun
Faculty of Pharmacy, University of Indonesia, Depok 16424, West Java, Indonesia
E-mail: hayun@farmasi.ui.ac.id
| Date of Received | 10 November 2019 |
| Date of Revision | 28 January 2021 |
| Date of Acceptance | 28 October 2021 |
| Indian J Pharm Sci 2021;83(5):1074-1080 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The present study was to evaluate the two aminomethyl derivatives of dehydrozingerone: 5-[(4-methylpiperazin-1-yl)methyl]dehydrozingerone and 5-(morpholin-4-ylmethyl)dehydrozingerone as lipoxygenase and xanthine oxidase inhibitors. Nordihydroguaiaretic acid, allopurinol and the parent compound dehydrozingerone were used as comparative compounds. Results indicated that 5-[(4-methylpiperazin-1-yl)methyl] dehydrozingerone and 5-(morpholin-4-ylmethyl) dehydrozingerone inhibited the lipoxygenase enzyme with half-maximal inhibitory concentration of 219.13 and 269.39 μM, respectively. Their activities were comparable to nordihydroguaiaretic acid with a half-maximal inhibitory concentration of 216.84 μM. The compounds were also found to inhibit the xanthine oxidase enzyme with half-maximal inhibitory concentration of 102.34 and 230.52 μM, respectively, but their activities were lower than allopurinol with half-maximal inhibitory concentration of 34.09 μM. Dehydrozingerone was found inactive both as a lipoxygenase inhibitor and xanthine oxidase inhibitor. Besides, in silico study was carried out to predict the binding interaction between the enzymes and the compounds compared to each of positive controls. In conclusion, 5-[(4-methylpiperazin-1-yl)methyl] dehydrozingerone has lipoxygenase inhibitory activity comparable to nordihydroguaiaretic acid, but they have lower xanthine oxidase inhibitory activity than allopurinol.
Keywords
Lipoxygenase, xanthine oxidase inhibition, aminomethyl, dehydrozingerone
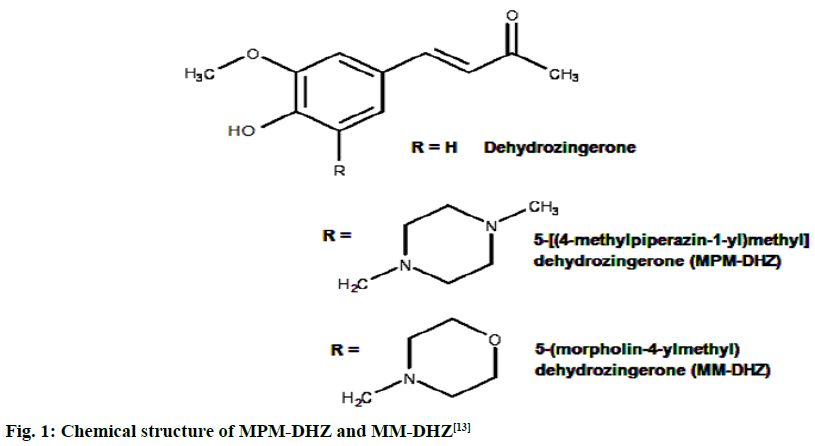
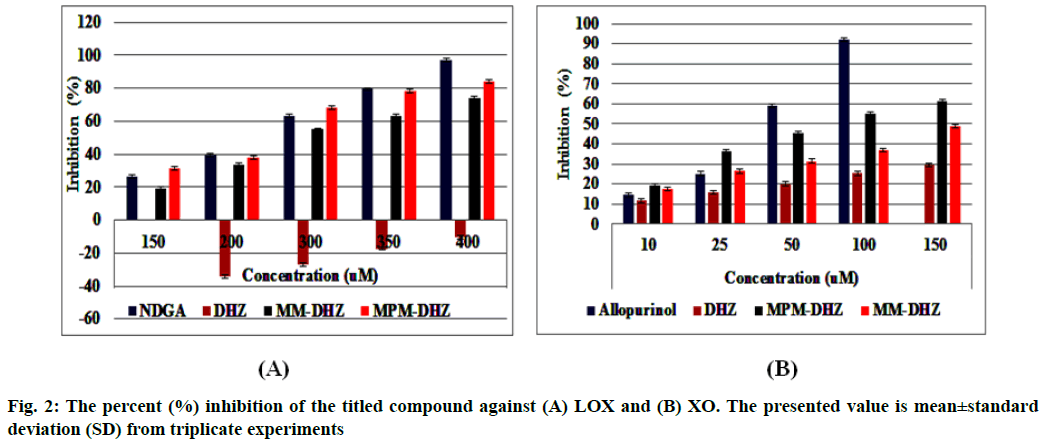
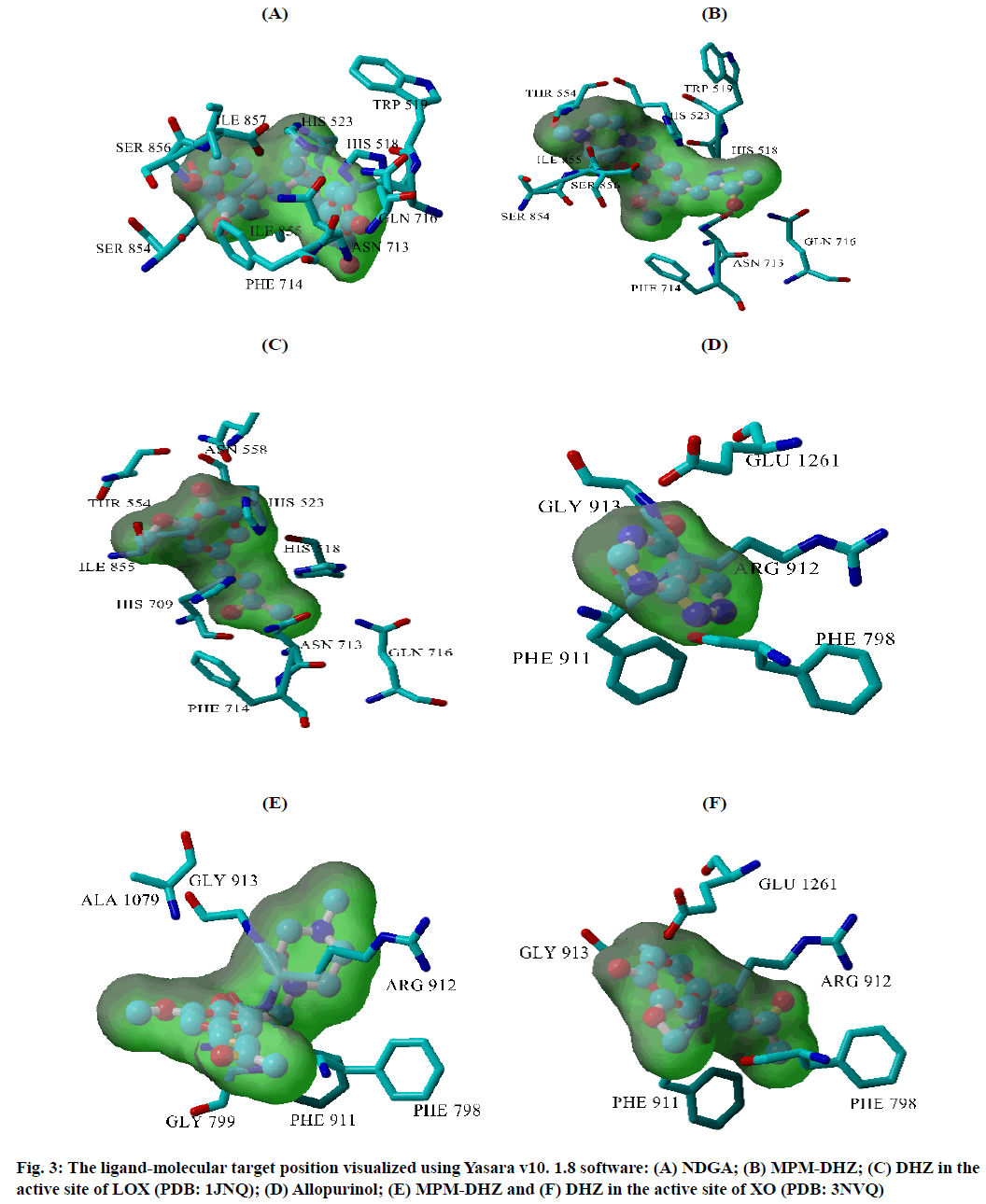
The prevention of potent inflammatory mediators, Leukotrienes (LTs) production by Lipoxygenase (LOX) inhibitors may be more useful for inflammation therapy than the prevention of prostanoids by cyclooxygenase inhibitors[1]. LOX converted arachidonic acid into LTs (LT-A4, LT-B4, LT-C4, LT-D4 and LT-E4) known to be involved in the formation of various diseases associated with inflammation and allergies, such as gastric ulcer, atherosclerosis and asthma[2-4]. Intensive research in this area has been conducted and some drugs were found, such as zafirlukast, montelukast, zileuton and Nordihydroguaiaretic acid (NDGA). The first three compounds have been approved for prophylaxis and chronic treatment of asthma[5]. NDGA has been shown to have potential medical uses in treating various diseases such as cardiovascular disease, neurological disorders and cancer[6]. However, there is no report about its clinical applications. Likewise, the inhibition of uric acid overproduction by Xanthine Oxidase (XO) inhibitor has been useful for gout arthritis therapy. XO catalyzes the hydroxylation of hypoxanthine to xanthine and then of xanthine to uric acid. Its overproduction causes hyperuricemia and deposition of the compound in the tissues around the joints, ultimately leading to chronic inflammation with severe pain, namely gout diseases[7]. Allopurinol is the only currently available XO competitive inhibitor for use in the clinic. However, the treatment often causes side effects such as allergic reactions and nephropathy[8]. Therefore, the discovery of new LOX and XO inhibitory agents is still needed. Some studies demonstrated that the Dehydrozingerone (DHZ) derivatives showed higher activity as antiinflammation[ 9], antimicrobial[10] and cytotoxic[11,12]. Our research group recently reported that some DHZ derivatives substituted an aminomethyl (Mannich base) in the aromatic ring exhibited anti-inflammatory activity by inhibition against protein denaturation[13]. However, there are no reports of the inhibitory activity mechanisms against the pro-inflammatory enzymes that play a role in the inflammation process. The present study aimed to evaluate two synthesized Mannich base derivatives of DHZ: 5-[(4-methylpiperazin-1-yl) methyl]dehydrozingerone (MPM-DHZ) and 5-(morpholin-4-ylmethyl)dehydrozingerone (MMDHZ) (fig. 1) as LOX and XO inhibitor. In addition, the compounds molecular docking studies were performed to estimate the binding interactions of the LOX and XO/tested compounds complex. The reagents and solvents used were analytical grades. Soybean LOX enzyme(enzyme commission number (EC number) 1.13.11.12) type I-B, linoleic acid, NDGA, XO microbial (EC number 1.17.3.2), xanthine and allopurinol were purchased from Sigma Aldrich, USA. Other reagents were purchased from Merck, Germany. Distilled water purchased from IKA Pharmindo, Indonesia; MPM-DHZ, MM-DHZ and DHZ, were obtained via synthesis as reported previously[13]. The LOX inhibition assay was performed spectrometrically according to the method previously reported[14] with little modifications. To the mixture of solvent or various concentrations of NDGA/test compound solutions and a borate buffer (0.2 M, pH 8.5, 1600 μl), a LOX enzyme solution (300 μl of 5000 units/ml) was added and then incubated at 25° for 10 min. After that, the reaction was initiated by the addition of 1000 μl of linoleic acid solution (substrate, 150 μM). The mixture was reincubated at 25° for 10 min and methanol (1000 μl) was added to stop the enzyme reaction. The absorbance values of the product, (9Z, 11E)-(13S)-13- hydroperoxyoctadeca-9,11-dienoate, were measured spectrophotometrically at 234 nm using a similar solution without solvent, test compounds and the enzyme as a blank solution for 100 % initial activity (IA) and using a similar solution without the enzyme as a blank solution for standard/test compounds inhibition. The percentage of the inhibition was calculated using the formula: Percentage (%) inhibition=(1–As/Aia) ×100 %, where Aia is the absorbance of initial activity and As is the absorbance of standard or test compound inhibition, respectively. The XO inhibition assay was performed spectrometrically according to the method previously reported[8,14,15] with little modifications. To the mixture of 1000 μl of solvent or allopurinol/test compound with various concentrations solutions and 1000 μl of phosphate buffer solution (0.05 M, pH 7.5), 1000 μl of XO enzyme solution was added and then incubated at 25° for 10 min. After that, the reaction was initiated by the addition of 2000 μl of xanthine solution (substrate). The mixtures were re-incubated at 25° for 10 min and 1000 μl of stop solution (HCl 1 N) was added to stop the reaction. The enzymatic conversion of xanthine to form uric acid was measured by observing the changes in absorbance at 295 nm spectrophotometrically using a similar solution without solvent, test compounds and the enzyme as a blank solution for 100 % IA and using a similar solution without the enzyme as a blank solution for standard/test compounds inhibition. The percentage of the inhibition was calculated using the formula: % inhibition=(1–As/ Aia)×100 %, where Aia is the absorbance of initial activity and As is the absorbance of standard or test compound inhibition, respectively. In silico study was done utilizing the Protein-Ligand ANT System (PLANTS v1.1) docking software (http://www.tcd.unikonstanz. de/research/plants.php) according to a previous procedure[16,17]. The crystal structure of soybean LOX-3 in complex with epigallocatechin (PDB code: 1JNQ, http://www.pdb.org/)[18] and bovine XO in complex with quercetin (Protein Data Bank (PDB) code: 3NVY, http://www.pdb.org/)[19], were used as the protein target in this study. The PDB format of the enzymes was converted into SYBYL mol2 format using Yet Another Scientific Artificial Reality Application (Yasara) software (http://www.yasara.org/) [20]. The compounds structures were prepared for docking as a combination of 10 conformations structure in SYBYL mol2 format using Chemaxon’s Marvin software (http://www.chemaxon.com)[21]. Yasara v10.1.8 software was utilized to visualize molecular docking results and analyze binding interaction in the protein/ligand complex. To validate the docking protocol, 1JNQ protein-bound ligand (epigallocatechin) and 3NVY protein-bound ligand (quercetin) were redocked into their binding pocket to obtain the docked poses and the Root Mean Square Deviations (RMSD) values of all atoms of the docked compounds and their references. The results of titled compounds in vitro evaluation as LOX and XO inhibitor agents were presented in fig. 2. All the compounds showed LOX and XO inhibitory activities in a concentrationdependent way. The Half-Maximal Inhibitory Concentration (IC50) values extrapolated from the linear regression curve obtained between percentage (%) inhibition and log10 concentration of the test solution using Probit analysis[22]. The IC50 values obtained were presented in Table 1. Among the compounds evaluated, MPM-DHZ showed the highest ability to inhibit LOX activity with IC50 219.13 μM. Its action is almost the same as NDGA, which is used as a standard (IC50 216.84 μM). The results for NDGA are not too different from those earlier reported[14]. NDGA (2,3-dimethyl-l, 4-bis (3,4-dihydroxyphenyl) butane) is a plant lignan derived from creosote bush leaves and twigs, Larrea tridentata[23]. The compound also has antioxidant and free radical scavenging properties[24]. The LOX inhibitory activity of MPM-DHZ, MM-DHZ and DHZ indicated to correlate their protein denaturation inhibitory activity reported earlier[13]. DHZ did not show denaturation activity in the previous report; the compound showed no activity to inhibit LOX activity. While the two aminomethyl derivatives exhibited denaturation activity in the previous report, the compounds showed the LOX inhibitory activity. The XO inhibitory activity of the titled compounds was found lower than allopurinol used as a positive standard. Among the compounds evaluated, MPM-DHZ exhibited the highest ability to inhibit XO with IC50 of 102.34 μM. However, their action is only about 33 % compared to allopurinol activity with IC50 of 34.09 μM. The inhibitory activity of allopurinol obtained is almost the same as the results previously studied[25]. The results indicated that DHZ did not inhibit XO. This result is similar to the results previously reported[26]. The XO inhibitory activity of the two aminomethyl derivatives of DHZ was higher than DHZ. It is consistent with aminomethyl derivatives in general, which show higher biological activity compared to the parent compound[27,28]. In order to support the inhibitory activity resulted from the in vitro experiment, the binding interaction between LOX and XO with tested compounds was predicted by molecular docking using PLANTS software. The results of the docking protocol validation showed that the values of the RMSD for the docked compounds (epigallocatechin and quercetin) to their reference ligands at the crystal structures considering all heavy atoms were 2.7284 Å (between 2.0 and 3.0 Å) and 0.8778 Å (less than 2.0 Å), indicating that the protocol found the acceptable and good binding mode of the ligands[29-31]. The protein-ligand interaction scores (total PLANTSCHEMPLP score) obtained during docking for 1JNQ and MPM-DHZ, MM-DHZ, DHZ, NDGA and epigallocatechin were -56.3845, -63.2693, -64.2494, -82.6026 and -50.3060, respectively. While the scores for 3NVY and MPM-DHZ, MM-DHZ, DHZ, allopurinol and quercetin were -82.4776, -62.3261, -78.0477, -68.3600 and -82.4984. The most negative of the score indicates a better binding affinity of a ligand with a molecular target. Nevertheless, the in silico study results indicated that the binding scores did not show a good correlation with the in vitro experiment obtained. The comparative study found that the binding affinity’s predictive power is relatively low than a structurebased feature[32]. Fig. 3 showed the protein-ligand complex obtained from the molecular docking study. Analysis of binding prediction between NDGA and LOX was found that two hydrogen bonds are formed: between the phenolic groups of the compound with amino acid residues N-Ser856 (0.917 Å) and HNAsn713 (2.657 Å). The distance between OH phenolic groups of NDGA and His-518 and Gln716 was 3418 and 3317 Å, respectively. Analysis of binding prediction between MPM-DHZ and LOX was found that two hydrogen bonds formed between N-4 from piperazine moiety of the compound with amino acid residue HOThr554 (1.057 Å) and the carbonyl group of alkyl chain with HN-Gln716 (2.375 Å), while DHZ and LOX without hydrogen bonding formation. The amino acid residues for hydrogen bonding interaction between epigallocatechin with LOX were HN-Gln716 and HNHis518[ 18]. Analysis of binding prediction between allopurinol and XO was found that five hydrogen bonds are formed: between HN-pyrazole moiety of the compound with amino acid residue O=C-Phe798 (2.335 Å), N-pyrazole moiety with HN-Arg912 (1.186 Å), HN-pyrimidine moiety with O-Glu1261 (2.618 Å), N-pyrimidine with HN-Phe911 (2.689 Å) and carbonyl of pyrimidine moiety with HN-Gly913 (1.754 Å). Analysis of binding prediction between MPM-DHZ and XO was found that three hydrogen bonds are formed between N-1 from piperazine moiety of the compound with amino acid residue HN-Arg912 (2.157 Å), the phenolic group with O-Phe911 (1.392 Å) and the carbonyl group of alkyl chain with HN-Gln799 (2.659 Å), while DHZ and XO by one hydrogen bond formed between O-methoxy group of the compound with amino acid residue HN-Phe911 (2.663 Å). The amino acid residues for hydrogen bonding interaction between quercetin with XO were Thr-1010, Glu802 and Arg880[19]. The results of the molecular docking study found by the indication that there was a correlation between the IC50 values experimentally obtained and the number of predicted hydrogen bonding formed in the protein-ligand complex. In conclusion, the compound MPM-DHZ was found to have LOX inhibitory activity comparable to NDGA but has lower XO inhibitory activity than allopurinol. The activities correlate with its binding prediction of the enzymeligand complex. Further study should be performed to evaluate the compound’s in vivo activity and toxicity as LOX inhibitors.
| Compound | IC50±SD (μM) | |
|---|---|---|
| LOX | XO | |
| MPM-DHZ | 219.13 | 102.34 |
| MM-DHZ | 269.39 | 230.52 |
| DHZ | - | 1540.73 |
| NDGA | 216.84 | - |
| Allopurinol | - | 34.09 |
Table 1: The IC50 Values of Inhibitory Activity of the Title Compounds Against Lox and Xo Enzymes
Fig. 1: Chemical structure of MPM-DHZ and MM-DHZ[13]
Acknowledgements:
Thanks to University of Indonesia, Depok, Indonesia, for the financial support of this study (PITTA Research Grant, 2019).
Conflicts of interest:
The authors have declared no conflict of interest
References
- Schneider I, Bucar F. Lipoxygenase inhibitors from natural plant sources. Part 1: Medicinal plants with inhibitory activity on arachidonate 5‐lipoxygenase and 5‐lipoxygenase [sol] cyclooxygenase. Phytother Res 2005;19(2):81-102.
- Nguyen MD, Nguyen DH, Yoo JM, Myung PK, Kim MR, Sok DE. Effect of endocannabinoids on soybean lipoxygenase-1 activity. Bioorg Chem 2013;49:24-32.
- Pontiki E, Hadjipavlou‐Litina D. Lipoxygenase inhibitors: A comparative QSAR study review and evaluation of new QSARs. Med Res Rev 2008;28(1):39-117.
- Radmark O, Werz O, Steinhilber D, Samuelsson B. 5-Lipoxygenase, a key enzyme for leukotriene biosynthesis in health and disease. Biochim Biophys Acta 2015;1851(4):331-9.
- Borne R, Levi M, Wilson N. Nonsteroidal Anti-Inflammatory Drugs. In: Lemke TL, Williams DA, Roche VF, Zito SW, editors. Foye’s Principles of Medicinal Chemistry. 7th ed. Baltimore: Lippincott Wiliams and Wilkins; 2013. p. 987-1042.
- Lu JM, Nurko J, Weakley SM, Jiang J, Kougias P, Lin PH, et al. Molecular mechanisms and clinical applications of nordihydroguaiaretic acid (NDGA) and its derivatives: an update. Med Sci Monit 2010;16(5):93-100.
- Kim Y, Oh HC, Park JW, Kim IS, Kim JY, Kim KC, et al. Diagnosis and treatment of inflammatory joint disease. Hip Pelvis 2017;29(4):211-22.
- Bhat MY, Gul MZ, Husain MK, Ghazi IA. In vitro evaluation of antiproliferative, lipoxygenase and xanthine oxidase inhibitory activities of Artemisia nilagirica (CB Clarke) Pamp. Leaf Extracts. Indian J Pharm Sci 2019;81(2):389-95.
- Jayasekhar P, Rao SB, Santhakumari G. Synthesis and pharmacological activity of some Mannich bases of dehydrozingerone. Indian J Pharm Sci 1998;60(4):191-5.
- Kubra IR, Bettadaiah BK, Murthy PS, Rao LJ. Structure-function activity of dehydrozingerone and its derivatives as antioxidant and antimicrobial compounds. J Food Sci Technol 2014;51(2):245-55.
- Burmudzija AZ, Muskinja JM, Kosanic MM, Rankovic BR, Novakovic SB, Dordevic SB, et al. Cytotoxic and antimicrobial activity of dehydrozingerone based cyclopropyl derivatives. Chem Biodivers 2017;14(8):e1700077.
- Pedotti S, Patti A, Dedola S, Barberis A, Fabbri D, Dettori MA, et al. Synthesis of new ferrocenyl dehydrozingerone derivatives and their effects on viability of PC12 cells. Polyhedron 2016;117:80-9.
- Hayun H, Arrahman A, Purwati EM, Yanuar A, Fortunata F, Suhargo F, et al. Synthesis, anti-inflammatory, and antioxidant activity of Mannich bases of dehydrozingerone derivatives. J Young Pharm 2018;10(2):S6.
- Fariza I, Fadzureena J, Zunoliza A, Chuah AL, Pin KY, Adawiah I. Anti-inflammatory activity of the major compound from methanol extract of Phaleria macrocarpa leaves. J Appl Sci 2012;12(11):1195-8.
- Thiombiano AM, Adama H, Jean BM, Bayala B, Nabere O, Samson G, et al. In vitro antioxidant, lipoxygenase and xanthine oxidase inhibitory activity of fractions and macerate from Pandiaka angustifolia (Vahl) Hepper. J App Pharm Sci 2014;4(1):9-13.
- Korb O, Stutzle T, Exner TE. Empirical scoring functions for advanced protein-ligand docking with PLANTS. J Chem Inf Model 2009;49(1):84-96.
- Hayun AY, Hanafi M, Pws SH. Virtual screening of 2, 3-disubstituted-4 (3H)-quinazolinones possessing benzenesulfonamide moiety for COX-2 inhibitor. Bioinformation 2011;7(5):246-50.
- Skrzypczak-Jankun E, Zhou K, Jankun J. Inhibition of lipoxygenase by (-)-epigallocatechin gallate: X-ray analysis at 2.1 Å reveals degradation of EGCG and shows soybean LOX-3 complex with EGC instead. Int J Mol Med 2003;12(4):415-20.
- Cao H, Pauff JM, Hille R. X-ray crystal structure of a xanthine oxidase complex with the flavonoid inhibitor quercetin. J Nat Prod 2014;77(7):1693-9.
- Krieger E, Koraimann G, Vriend G. Increasing the precision of comparative models with YASARA NOVA-a self‐parameterizing force field. Proteins 2002;47(3):393-402.
- Software solutions and services for chemistry & biology. Chemaxon; 2019.
- Finney DJ. Probit analysis: a statistical treatment of the sigmoid response curve. 2nd ed. Cambridge university press: Cambridge; 1952.
- Rahman S, Ansari RA, Rehman H, Parvez S, Raisuddin S. Nordihydroguaiaretic acid from creosote bush (Larrea tridentata) mitigates 12-O-tetradecanoylphorbol-13-acetate-induced inflammatory and oxidative stress responses of tumor promotion cascade in mouse skin. Evid Based Complement Alternat Med 2011;2011.
- Angerhofer CK, Maes D, Giacomoni PU. The Use of Natural Compounds and Botanicals in the Development of Anti-Aging Skin Care Products. In: Dayan N, editor. Skin aging handbook, Personal Care & Cosmetic Technology. Norwich: William Andrew Publising; 2009. p. 205-63.
- Hendriani R, Nursamsiar AT. In vitro and in silico evaluation of xanthine oxidase inhibitory activity of quercetin contained in Sonchus arvensis leaf extract. Asian J Pharm Clin Res 2017:50-3.
- Rajakumar DV, Rao MN. Dehydrozingerone and isoeugenol as inhibitors of lipid peroxidation and as free radical scavengers. Biochem Pharmacol 1993;46(11):2067-72.
- Wiji Prasetyaningrum P, Bahtiar A, Hayun H. Synthesis and cytotoxicity evaluation of novel asymmetrical mono-carbonyl analogs of curcumin (AMACs) against Vero, HeLa, and MCF7 Cell Lines. Sci Pharm 2018;86(2):25.
- Putri TN, Bachtiar A. Synthesis, antioxidant, and anti-inflammatory activity of morpholine Mannich base of AMACs ((2E, 6E)-2-((4-hydroxy-3-[morpholin-4-yl-) methyl] phenyl) methylidene)-6-(phenylmethylidene) cyclohexan-1-one) and its analogs. J App Pharm Sci 2018;8(5):19-25.
- Ramírez D, Caballero J. Is it reliable to take the molecular docking top scoring position as the best solution without considering available structural data? Molecules 2018;23(5):1038.
- Mena-Ulecia K, Tiznado W, Caballero J. Study of the differential activity of thrombin inhibitors using docking, QSAR, molecular dynamics and MM-GBSA. PLoS One 2015;10(11):e0142774.
- Jain AN, Nicholls A. Recommendations for evaluation of computational methods. J Comput Aided Mol Des 2008;22(3):133-9.
- Schneider M, Pons JL, Bourguet W, Labesse G. Towards accurate high-throughput ligand affinity prediction by exploiting structural ensembles, docking metrics and ligand similarity. Bioinformatics 2020;36(1):160-8.