- *Corresponding Author:
- B. Xia
Department of Hematology and Oncology, The Fourth Hospital of Changsha, Changsha, Hunan 410013, China
E-mail: xiabing1987@csu.edu.cn
| This article was originally published in a special issue, “Modern Applications in Biomedical Research and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2022:84(3) Spl Issue “172-178” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
This study aimed to investigate the expression and clinical significance of micro ribonucleic acid-132 and sex-determining region Y-box 4 gene in colon cancer. Luciferase reporter system was used to validate the target gene directly regulated by micro ribonucleic acid-132. Transwell invasion assay was used to test the effect of micro ribonucleic acid-132 on invasiveness of SW620 cells. Micro ribonucleic acid-132 and sex-determining region Y-box 4 expression levels in cancerous and peri-cancerous tissues were measured using in situ hybridization and EleVisionTM immunohistochemistry staining. Micro ribonucleic acid-132 downregulated sex-determining region Y-box 4 protein expression and inhibited the invasiveness of colon cancer cells. In the 72 cancerous tissue samples of colon cancer, micro ribonucleic acid-132 positive rate was 18.1 % (13/72) and sex-determining region Y-box 4 protein positive rate was 73.6 % (53/72), both expressions showed significant differences as compared to peri-cancerous tissues (p<0.01). Micro ribonucleic acid-132 expression was significantly lower in colon cancer tissues with lymph node metastasis than without lymph node metastasis (p<0.01). Sex-determining region Y-box 4 expression was elevated along with Dukes stage increase and higher expression was accompanied by more tumor invasion depth (both p<0.05). Sex-determining region Y-box 4 expression was significantly higher in colon cancer tissues with lymph node metastasis than without lymph node metastasis (p<0.01). Correlation analysis displayed significant negative correlation of micro ribonucleic acid-132 expression with sex-determining region Y-box 4 expression (r=-0.594, p=0.013). Micro ribonucleic acid-132 inhibits invasion and metastasis of colon cancer cell line SW620 via targeting sex-determining region Y-box 4. Low expression of micro ribonucleic acid-132 and high expression of sex-determining region Y-box 4 protein could be important biomarkers for malignant transformation of colonic mucosa and for invasion and metastasis of colon cancer. Measuring expression levels of both molecules is valuable for predicting invasion and metastasis of colon cancer.
Keywords
Colon cancer, micro ribonucleic acid-132, sex-determining region Y-box 4, luciferase reporter vector, in situ hybridization, immunohistochemistry staining
Micro Ribonucleic Acid (miRNA or miR) is an 18-25 nucleotides long non-coding RNA, which can incompletely or completely pair with bases on the 3’ Untranslated Region (UTR) of target genes and subsequently inhibit protein translation to exert a negative regulatory effect on target gene expression[1,2]. miRNAs participate in numerous cellular responses such as cell differentiation[3,4], metabolism[5], proliferation, apoptosis[6], etc. Previous studies revealed down-regulation of miR-132 (Homo sapiens) expressions in tissues or cells of glioma[7,8], Non-Small Cell Lung Cancer (NSCLC)[9,10], breast cancer[11] and colorectal cancer[12]. Sex-Determining Region Y (SRY)-Box 4 (SOX4) gene is a common oncogene, which encodes a protein of around 47 kD, with a Deoxyribonucleic Acid (DNA) binding domain at the N-terminal and a transcriptional activator structure at the C-terminal. The N-terminal of SOX4 protein binds to the minor groove of DNA helix, which contains the sequence of 5 (A/T)(A/T)CAA(A/T) G-3, to induce DNA conformational change and thereby facilitate binding of other transcription factors to DNA[13]. A meta-analysis revealed SOX4 as one of the 64 human tumor markers that plays an important role in tumor initiation and progression[14]. High expression of SOX4 was detected in colon cancer tissues, while downregulation of SOX4 could remarkably inhibit growth and metastasis of colon cancer[15]. However, the precise underlying mechanism of SOX4-mediated gene expression regulation remains unknown in colon cancer. By predictive analysis of the designated gene of SOX4 using prediction bioinformatics softwares TargetScanHuman 6.2 and mircoRNA.org, our study uncovered SOX4 as the potential target gene of miR-132. The biological activities of miR-132 in colon cancer cells and the differential expression between miR-132 and SOX4 in colon cancer tissues were subsequently explored using luciferase reporter vector, in situ hybridization and immunohistochemistry staining. This study aimed to uncover the association of both molecules with clinicopathological parameters in colon cancer and thus provide a theoretical basis for further elucidation of the molecular mechanisms of colon cancer initiation, progression, invasion and metastasis.
Materials and Methods
Materials:
Excised colon cancer specimens from 72 patients, who had undergone surgery at the General surgery department of Second Xiangya hospital between 2016 and 2017 (Table 1) and not received chemotherapy or radiotherapy before surgery, were collected. According to the World Health Organization (WHO) standard for histological classification and grading of colon cancer, 31 cases had lymph node metastasis, while 41 cases did not. Among the 72 patients, 30 were male and 42 were female, with a mean age of 48±11 y. Pathological diagnosis was confirmed by ≥2 pathology professors.
| Clinicopathological parameters | Case number | SOX4 expression | χ2 value | p value | miR-132 expression | χ2 value | p value |
|---|---|---|---|---|---|---|---|
| Gender | 0.408 | 0.523 | 0.007 | 0.933 | |||
| Male | 30 | 25 (83.3) | 5 (16.7) | ||||
| Female | 42 | 28 (66.7) | 8 (19.0) | ||||
| Age (years) | 0. 076 | 0.782 | 0.481 | 0.694 | |||
| <50 | 29 | 22 (75.9) | 4 (13.8) | ||||
| ≥50 | 43 | 31 (72.1) | 9 (20.9) | ||||
| Gross classification | 0. 195 | 0. 907 | 0.147 | 0.923 | |||
| Protruding type | 14 | 10 (71.4) | 2 (14.3) | ||||
| Ulcerative type | 23 | 16 (69.6) | 4 (17.4) | ||||
| Invasive type | 35 | 27 (77.1) | 7 (20.0) | ||||
| Differentiation degree | 2.427 | 0.297 | 0.123 | 0.941 | |||
| Highly differentiated | 18 | 13 (72.2) | 3 (16.7) | ||||
| Moderately differentiated | 25 | 18 (72.0) | 5 (20.0) | ||||
| Poorly differentiated | 29 | 22 (75.9) | 5 (17.2) | ||||
| Invasion depth | 13.1 | 0.0003 | 15.77 | <0.0001 | |||
| Serosal noninvolvement | 22 | 11 (50.0) | 12 (54.5) | ||||
| Serosal involvement | 50 | 42 (84.0) | 1 (2.0) | ||||
| Lymph node metastasis | 8.516 | 0.004 | 8.305 | 0.004 | |||
| Negative | 41 | 22 (53.7) | 11 (26.8) | ||||
| Positive | 31 | 31 (100) | 2 (5.2) | ||||
| Dukes staging | 20.51 | 0.0001 | 27.09 | <0.0001 | |||
| A stage | 12 | 5 (41.7) | 6 (50.0) | ||||
| B stage | 20 | 14 (70.0) | 4 (20.0) | ||||
| C stage | 19 | 15 (78.9) | 2 (10.5) | ||||
| D stage | 21 | 19 (90.5) | 1 (4.8) |
Table 1: Association of miR-132 and SOX4 Expression with Clinicopathological Parameters in Colon Cancer Tissues (Number, Positive Rate %).
Reagents:
SOX4 (ab56780) monoclonal antibody was purchased from Abcam. Digoxigenin-labelled in situ hybridization probe for miR-132 was purchased from Exiqon, while EleVisionTM plus two-step immunohistochemistry kit and enhanced 3,3’-Diaminobenzidine (DAB) chromogenic reagent kit were obtained from Beijing Zhongshan Golden Bridge Biotechnology Co., Ltd. (Beijing, China).
Cell lines and cell culture:
Human colon cancer cell line (SW620) was purchased from Shanghai Cell Bank of Chinese Academy of Sciences and cultured under recommended conditions.
Transient transfection of miR-132:
Cells were cultured at a density of 1×105 cells/well in a 6-well plate until 30 % confluency. Then 5 μl liposome was mixed with 200 μl serum-free culture media and kept for 15 min. 100 pmol miR-132 mimics or inhibitor was diluted in 200 μl serum-free culture media. The liposome and DNA dilution solutions were then mixed and kept for 30 min at room temperature.
Cells in the 6-well plate were sequentially rinsed with Hanks buffer and serum-free culture media, followed by addition of the liposome-DNA mixture and 1600 μl serum-free culture media. The plate was incubated for 6 h in a 37° Carbon dioxide (CO2) incubator. The culture supernatant was then discarded and complete culture medium was added to cells for overnight culture.
Validation of SOX4 as the target gene:
Target genes regulated by miR-132 were predicted with online software TargetScan 5.2. SOX4 3’ UTR sequences (synthesized by Invitrogen) with miR-132 binding sites were chosen for constructing a luciferase reporter vector (upstream primer: 5’-AGCTT-AT GTTGCCAAATTCAATGTAGAAAGAATGT GACAA-CACACCTTGGGTAGTTCTGA-3’; downstream primer: 5’-AGCTTCAGAACTACCCAAGGTGTGTTGTCACAT TCTTTC-TACATTGAATTTGGCAACATA-3’). SOX4 3’ UTR mutated sequences without miR-132 binding site (synthesized by Invitrogen) were designed for constructing another luciferase reporter vector (upstream primer: 5’-AGCTTATGTTGCCAAATTCAATGTAGAAAGGA CAACAC-ACCTTGGGTAGTTCTGA-3 ’ ; downstream primer: 5’-AGCTTCAGAACTACCCAAGGTGTGTTGTCCTTTC TACATT-GAATTTGGCAACATA-3’). miR-132 mimics and pmiR-Report/SOX4 UTR vector were cotransfected into SW620 cells, followed by luciferase activity measurement. SW620 cells were transfected with miR-132 mimics or the negative control, followed by extraction of cellular proteins for measurement of SOX4 protein levels by Western blotting.
Transwell invasion assay:
Matrigel was diluted in pre-chilled serum-free Roswell Park Memorial Institute (RPMI) 1640 culture media prior to coating the upper chamber, which had 8 μm polycarbonate filter on the bottom. 100 μl transfected SW620 cells (5×105/ml) in serum-free culture media were then seeded on the upper chamber, while the lower chamber was filled with 600 μl of 10 % serum RPMI 1640 culture media. The system was incubated for 24 h at 37° in a 5 % CO2 incubator. The upper chamber was then removed and fixed in 4 % formaldehyde, followed by crystal violet staining. The chamber was inverted, observed and photographed under an optical microscope. Four low magnification fields (10×) were chosen for cell counting and mean cell number was calculated. The experiment was repeated three times.
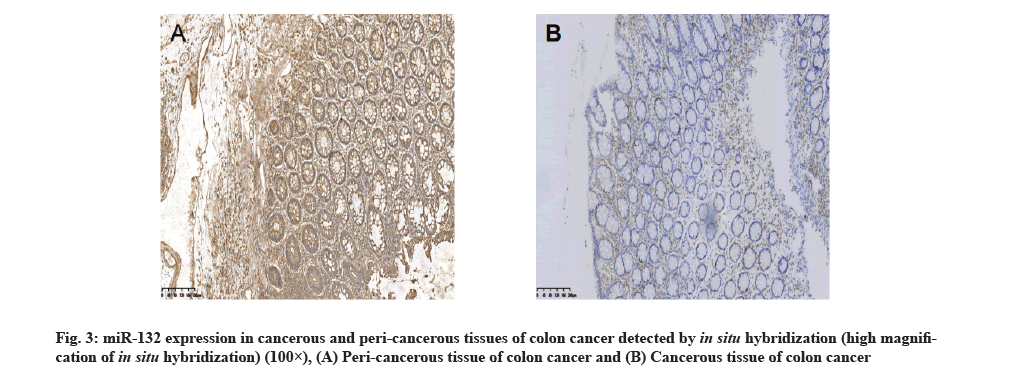
miR-132 expression in cancerous and peri-cancerous tissues of colon cancer detected by in situ hybridization:
The slides were dewaxed and hydrated, followed by soaking in 3 % Hydrogen peroxide (H2O2) for 10 min at room temperature and rinsing with enzyme-free water three times, prior to 3 % pepsin digestion for 15-20 min at room temperature and three rinses with Phosphate Buffered Saline (PBS) for 5 min and one rinse with enzyme-free water. The slides were then soaked in 1 % paraformaldehyde for 10 min at room temperature, followed by three rinses with enzyme-free water. The appropriate volume of prehybridization solution was added to the slides and incubated for 4 h at 55°, followed by addition of miR-132 probe according to the instruction, mixing and incubation overnight at 55°. The slides were rinsed twice with 37° Saline Sodium Citrate (SSC) buffer for 5 min each time, followed by sequential rinsing with 0.5×SSC and 0.2×SSC, 15 min each time. Blocking solution was then added to the slide and incubated for 30 min at room temperature, followed by addition of biotinylated mouse antidigoxin, incubation for 2 h at room temperature, rinsing with PBS four times, and 5 min each time, prior to color development with DAB reagents. The slides were restained with hematoxylin, washed and mounted with neutral gum.
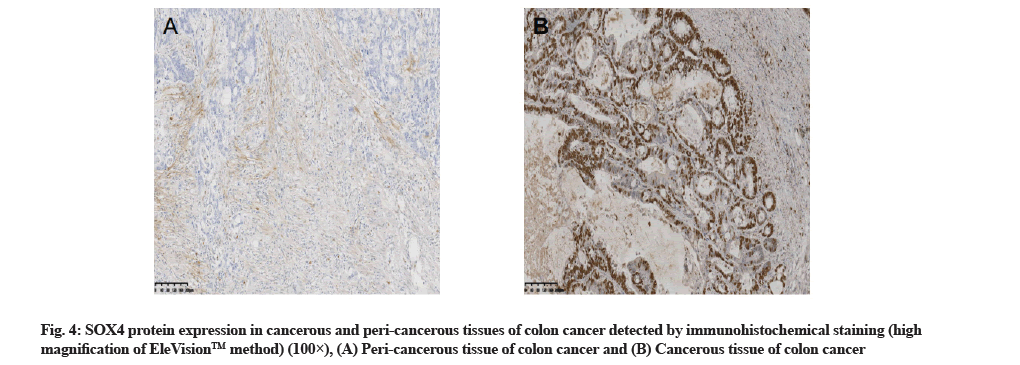
SOX4 expression in cancerous and pericancerous tissues of colon cancer detected by immunohistochemistry staining:
Slides were dewaxed and hydrated prior to staining according to the instruction of EleVisionTM plus twostep kit.
Statistical analysis:
Statistical analysis was conducted using Statistical Package for the Social Sciences (SPSS) 19.0 software. Measurement data were analyzed by t-test, enumeration data by χ2 test and correction by Fisher exact probability method. Spearman rank correlation analysis was performed, p<0.05 was defined as statistical significance.
Results and Discussion
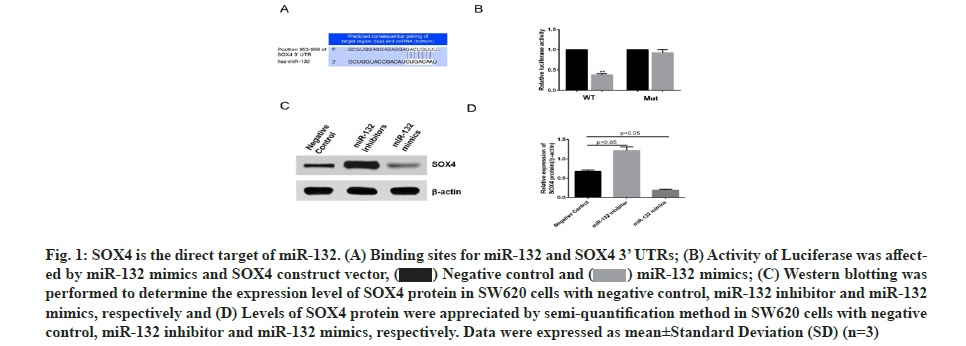
SOX4 was found to be one of the target genes regulated by miR-132 via prediction using online software, http://www.targetscan.org/ (fig.1A). SOX4 3’ UTR (wild type and mutant) luciferase reporter vectors were constructed and respectively co-transfected into colon cancer cell line, SW620, along with miR-132 mimics. Subsequent luciferase activity measurement revealed that miR-132 inhibited the luciferase activity of wild type SOX4 3’ UTR, without influencing the mutant SOX4 3’ UTR (fig. 1B) expression, while miR-132 inhibitor promoted SOX4 protein expression (fig. 1C and fig. 1D). These data indicated that SOX4 is the direct target gene of miR-132.
Fig. 1: SOX4 is the direct target of miR-132. (A) Binding sites for miR-132 and SOX4 3’ UTRs; (B) Activity of Luciferase was affected by miR-132 mimics and SOX4 construct vector, (C) Western blotting was performed to determine the expression level of SOX4 protein in SW620 cells with negative control, miR-132 inhibitor and miR-132 mimics, respectively and (D) Levels of SOX4 protein were appreciated by semi-quantification method in SW620 cells with negative control, miR-132 inhibitor and miR-132 mimics, respectively. Data were expressed as mean±Standard Deviation (SD) (n=3).
(C) Western blotting was performed to determine the expression level of SOX4 protein in SW620 cells with negative control, miR-132 inhibitor and miR-132 mimics, respectively and (D) Levels of SOX4 protein were appreciated by semi-quantification method in SW620 cells with negative control, miR-132 inhibitor and miR-132 mimics, respectively. Data were expressed as mean±Standard Deviation (SD) (n=3).
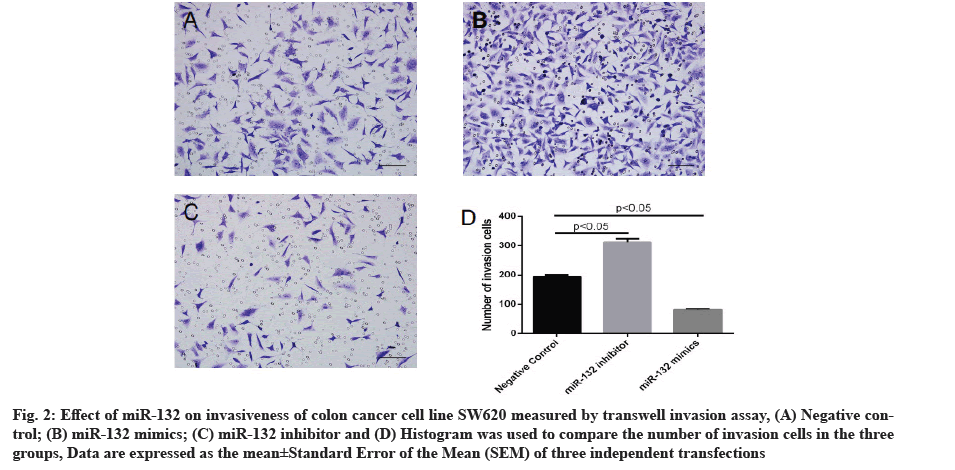
Effect of miR-132 on invasiveness of colon cancer cells measured by transwell invasion assay is explained clearly. In order to further probe the effect of miR-132 on colon cancer cell phenotype, SW620 cells were transfected with miR-132 mimics or inhibitor and cell invasiveness was measured 48 h post-transfection by transwell assay. As compared to the control group, transfection of miR-132 mimics led to an evident reduction in cells penetrating out of the upper chamber, whereas miR-132 inhibitor transfection caused an evident increase in cells penetrating out of the upper chamber (p<0.05 for both, fig. 2).
Fig. 2: Effect of miR-132 on invasiveness of colon cancer cell line SW620 measured by transwell invasion assay, (A) Negative control; (B) miR-132 mimics; (C) miR-132 inhibitor and (D) Histogram was used to compare the number of invasion cells in the three groups, Data are expressed as the mean±Standard Error of the Mean (SEM) of three independent transfections.
The hybridization data revealed that out of the 72 cancerous tissue samples, 13 showed positive miR-132 expression, with the positive rate of 18.1 %. Out of the 72 peri-cancerous tissue samples, 68 showed positive miR-132 expression, with the positive rate of 94.4 %, with a significant difference between the two types of tissues (p<0.01,Table 1 and fig. 3). Immunohistochemistry staining showed that out of 72 cancerous tissue samples, 53 had positive SOX4 expression, with the positive rate of 73.6 %. Out of 72 peri-cancerous tissue samples, 11 had positive SOX4 expression, with the positive rate of 15.3 %, with a significant difference between the two types of tissues (p<0.01, Table 1 and fig. 4).
No significant association of miR-132 and SOX4 protein expressions with patient’s gender, age, tumor gross classification, or tumor differentiation degree was detected (p>0.05), miR-132 expression was significantly reduced along with Dukes stage increase (p<0.05). MiR-132 expression was significantly lower in colon cancer tissues with lymph node metastasis than without lymph node metastasis (p<0.01). In contrast, SOX4 expression was elevated along with Dukes stage increase and higher expression was accompanied by more tumor invasion depth (both p<0.05). SOX4 expression was significantly higher in colon cancer tissues with lymph node metastasis than without lymph node metastasis (p<0.01, Table 1).
Spearman rank correlation analysis displayed significant negative correlation of miR-132 expression with SOX4 expression in colon cancer tissues (r=-0.594, p=0.013), whereby increase in miR-132 expression led to decrease in SOX4 expression (Table 2).
| SOX4 | miR-132 | Total | ||
|---|---|---|---|---|
| + | - | |||
| + | 10 | 43 | 53 | |
| - | 3 | 16 | 19 | |
| Total | 13 | 59 | 72 | |
Table 2: Association of miR-132 Expression with SOX4 Protein Expression in Colon Cancer Tissues.
Colon cancer is a common gastrointestinal neoplasm. With changes in lifestyle and dietary composition, the incidence of colon cancer is steadily increasing, especially in large and medium cities. An epidemiological survey revealed that morbidity and mortality of colon cancer in China was 1 720 000 and 990 000, respectively, in 2005. The key for cure and prevention of colon cancer lies in the identification of its etiology and pathogenesis[16].
As important post-transcriptional regulatory factors, miRNAs are widely involved in tumor initiation and development. Dysregulation of miRNA expression has been detected in various tumors and some miRNA families have similar functions to oncogenes or tumor suppressor genes[17-19]. miRNAs exhibit specific expression patterns in malignant neoplasms and their expressions change along with clinicopathological progression, thus enabling comprehensive tumor diagnosis and precise tracking of tumor changes[20]. Down-regulation of miR-132 expression was detected in tissues of multiple tumors[7-12]. SOX4 was found to be a target gene regulated by miR-132 via prediction using online software, http://www.targetscan.org/ and expression of SOX4 gene, located on chromosome 6p22.3, was overexpressed in many types of human cancers, including leukemias[21], glioblastomas[22] and cancers of the liver[23], lung[24], urinary bladder[25] and prostate[26]. High miR-132 expression could specifically inhibit SOX4 expression and is thus involved in tumor initiation and development.
In order to examine changes in miR-132 and SOX4 expressions in colon cancer and explore their association, luciferase reporter vector and Western blotting were used in this study to identify whether miR-132 specifically inhibited SOX4 expression in colon cancer cell line SW620 and miR-132 inhibited the invasiveness of colon cancer cells. Moreover, in situ hybridization and immunohistochemistry staining detected 18.1 % miR-132 positive rate in cancerous tissues of 72 colon cancer cases and 73.6 % SOX4 protein positive rate, with statistical difference as compared to control peri-cancerous tissues. Correlation analysis revealed significant negative correlation of miR-132 expression with SOX4 expression. miR-132 expression was decreased along with clinicopathological progression and increasing depth of invasion. miR-132 expression was significantly lower in tissues of patients with lymph node metastasis than those without lymph node metastasis. SOX4 expression was upregulated along with clinicopathological progression and increasing depth of invasion. SOX4 expression was significantly higher in tissues of patients with lymph node metastasis than those without lymph node metastasis. These data indicated that low expression of miR-132 and high expression of SOX4 in cancerous tissues of colon cancer could be important biomarkers for malignant transformation of colonic mucosa, and invasion and metastasis of colon cancer. Detection of miR-132 and SOX4 expression levels could help to develop personalized therapeutic strategy by clinicians and also facilitate early diagnosis of colon cancer in patients without conspicuous symptoms. This would enable early discovery and treatment, and does not miss the optimal treatment timing.
In summary, identification of novel susceptible genes or miRNA of colon cancer, followed by exploration of their functions, in order to identify key genes involved in colon cancer initiation and development, or specific molecular markers that reflect the processes of carcinogenesis and progression was accomplished in this study. Thus, genetically susceptible factors of colon cancer could be defined and the molecular mechanism of pathogenesis could be delineated. These findings are of great significance in the diagnosis and prognosis prediction for colon cancer, as well as in novel antitumor research and discovery.
Conflict of interests:
The authors report no conflicts of interest in this work.
References
- Yu X, Zheng H, Chan MT, Wu WK. MicroRNAs: New players in cataract. Am J Transl Res 2017;9(9):3896-903.
[Google Scholar] [PubMed]
- Zhang M, Lygrisse K, Wang J. Role of MicroRNA in osteoarthritis. J Arthritis 2017;6(2):239.
[Crossref] [Google Scholar] [PubMed]
- Yan Z, Guo Y, Wang Y, Li Y, Wang J. MicroRNA profiles of BMSCs induced into osteoblasts with osteoinductive medium. Exp Ther Med 2018;15(3):2589-96.
[Crossref] [Google Scholar] [PubMed]
- Soltanzadeh-Yamchi M, Shahbazi M, Aslani S, Mohammadnia-Afrouzi M. MicroRNA signature of regulatory T cells in health and autoimmunity. Biomed Pharmacother 2018;100:316-23.
[Crossref] [Google Scholar] [PubMed]
- Mirra P, Nigro C, Prevenzano I, Leone A, Raciti GA, Formisano P, et al. The destiny of glucose from a microRNA perspective. Front Endocrinol 2018;9:1-15.
[Crossref] [Google Scholar] [PubMed]
- Liu C, Jian M, Qi H, Mao WZ. MicroRNA 495 inhibits proliferation and metastasis and promotes apoptosis by targeting twist1 in gastric cancer cells. Oncol Res 2019;27(3):389-97.
[Crossref] [Google Scholar] [PubMed]
- Wang YZ, Han JJ, Fan SQ, Yang W, Zhang YB, Xu TJ, et al. miR-132 weakens proliferation and invasion of glioma cells via the inhibition of Gli1. Eur Rev Med Pharmacol Sci 2018;22(7):1971-8.
[Crossref] [Google Scholar] [PubMed]
- Wang H, Li XT, Wu C, Wu ZW, Li YY, Yang TQ, et al. miR-132 can inhibit glioma cells invasion and migration by target MMP16 in vitro. Onco Targets Ther 2015;8:3211-8.
[Crossref] [Google Scholar] [PubMed]
- Liu X, Yan S, Pei C, Cui Y. Decreased microRNA-132 and its function in human non-small cell lung cancer. Mol Med Rep 2015;11(5):3601-8.
[Crossref] [Google Scholar] [PubMed]
- You J, Li Y, Fang N, Liu B, Zu L, Chang R, et al. MiR-132 suppresses the migration and invasion of lung cancer cells via targeting the EMT regulator ZEB2. PLoS One 2014;9(3):e91827.
- Zhang ZG, Chen WX, Wu YH, Liang HF, Zhang BX. MiR-132 prohibits proliferation, invasion, migration and metastasis in breast cancer by targeting HN1. Biochem Biophys Res Commun 2014;454(1):109-14.
[Crossref] [Google Scholar] [PubMed]
- Zheng YB, Luo HP, Shi Q, Hao ZN, Ding Y, Wang QS, et al. miR-132 inhibits colorectal cancer invasion and metastasis via directly targeting ZEB2. World J Gastroenterol 2014;20(21):6515-22.
[Crossref] [Google Scholar] [PubMed]
- Xia X, Wan R, Huo W, Zhang L, Xia X, Chang Z. Molecular cloning and mRNA expression pattern of Sox4 in Paramisgurnus dabryanus. Gene Expr Patterns 2017;25:109-17.
[Crossref] [Google Scholar] [PubMed]
- Chen J, Ju HL, Yuan XY, Wang TJ, Lai BQ. SOX4 is a potential prognostic factor in human cancers: A systematic review and meta-analysis. Clin Transl Oncol 2016;18(1):65-72.
[Crossref] [Google Scholar] [PubMed]
- Lin CM, Fang CL, Hseu YC, Chen CL, Wang JW, Hsu SL, et al. Clinical and prognostic implications of transcription factor SOX4 in patients with colon cancer. PLoS One 2013;8(6):e67128.
[Crossref] [Google Scholar] [PubMed]
- Sengupta N, Gill KA, MacFie TS, Lai CS, Suraweera N, Mcdonald S, et al. Management of colorectal cancer: A role for genetics in prevention and treatment? Pathol Res Pract 2008;204(7):469-77.
[Crossref] [Google Scholar] [PubMed]
- Rohan TE, Wang T, Weinmann S, Wang Y, Lin J, Ginsberg M, et al. A miRNA expression signature in breast tumor tissue is associated with risk of distant metastasis. Cancer Res 2019;79(7):1705-13.
[Crossref] [Google Scholar] [PubMed]
- Si W, Shen J, Zheng H, Fan W. The role and mechanisms of action of microRNAs in cancer drug resistance. Clin Epigenetics 2019;11(1):1-24.
[Crossref] [Google Scholar] [PubMed]
- Bhandari V, Hoey C, Liu LY, Lalonde E, Ray J, Livingstone J, et al. Molecular landmarks of tumor hypoxia across cancer types. Nat Genet 2019;51(2):308-18.
[Crossref] [Google Scholar] [PubMed]
- Harrandah AM, Mora RA, Chan EK. Emerging microRNAs in cancer diagnosis, progression and immune surveillance. Cancer Lett 2018;438:126-32.
[Crossref] [Google Scholar] [PubMed]
- Andersson A, Ritz C, Lindgren D, Edén P, Lassen C, Heldrup J, et al. Microarray-based classification of a consecutive series of 121 childhood acute leukemias: Prediction of leukemic and genetic subtype as well as of minimal residual disease status. Leukemia 2007;21(6):1198-203.
[Crossref] [Google Scholar] [PubMed]
- Lin B, Madan A, Yoon JG, Fang X, Yan X, Kim TK, et al. Massively parallel signature sequencing and bioinformatics analysis identifies up-regulation of TGFBI and SOX4 in human glioblastoma. PLoS One 2010;5(4):e10210.
[Crossref] [Google Scholar] [PubMed]
- Liao YL, Sun YM, Chau GY, Chau YP, Lai TC, Wang JL, et al. Identification of SOX4 target genes using phylogenetic footprinting-based prediction from expression microarrays suggests that overexpression of SOX4 potentiates metastasis in hepatocellular carcinoma. Oncogene 2008;27(42):5578-89.
[Crossref] [Google Scholar] [PubMed]
- Medina PP, Castillo SD, Blanco S, Sanz-Garcia M, Largo C, Alvarez S, et al. The SRY-HMG box gene, SOX4, is a target of gene amplification at chromosome 6p in lung cancer. Hum Mol Genet 2009;18(7):1343-52.
[Crossref] [Google Scholar] [PubMed]
- Aaboe M, Birkenkamp-Demtroder K, Wiuf C, Sørensen FB, Thykjaer T, Sauter G, et al. SOX4 expression in bladder carcinoma: Clinical aspects and in vitro functional characterization. Cancer Res 2006;66(7):3434-42.
[Crossref] [Google Scholar] [PubMed]
- Liu P, Ramachandran S, Seyed MA, Scharer CD, Laycock N, Dalton WB, et al. Sex-determining region Y box 4 is a transforming oncogene in human prostate cancer cells. Cancer Res 2006;66(8):4011-9.
[Crossref] [Google Scholar] [PubMed]