- *Corresponding Author:
- Xiaoyue ChangRespiratory Department, Baotou Central Hospital, Baotou 014000
China
E-mail: changxiaoyue1986@sina.com
| This article was originally published in a special issue, “Therapeutic Perspectives in Biomedical Research and Pharmaceutical Sciences and their Nursing Methods” |
| Indian J Pharm Sci 2021:83(4)Spl issue “165-172” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
To investigate the expression of elicitin response protein and alpha-chemokines in lung cancer, inflammatory tissues and normal tissues and to observe the relationship between the expression of age, sex, tumor type, stage, differentiation and vascular endothelial growth factor. To detect the expression of chemokines in squamous cell carcinoma, adenocarcinoma and small cell lung cancer and analyze the relationship between the expression of age, gender, pathological grade and stage. The high expression rate of elicitin response protein and alpha-chemokines in lung cancer was higher than that in normal tissue and the difference was statistically significant (p<0.05). The expression of elicitin response protein and alpha-chemokines in different pathological types was different (p<0.001). The expression of elicitin response protein and alphachemokines in lung cancer was independent of age and gender. C-X-C motif chemokine ligand 3, C-X-C motif chemokine ligand 5 and C-X-C motif chemokine ligand 7, as elicitin response protein members, may play a more important role in lung cancer formation than other C-X-C motif chemokine ligand members. There was correlation between the expression of alpha-chemokine and vascular endothelial growth factor. Elicitin response protein and alpha-chemokines may play important role in development of lung cancer, especially the C-X-C motif chemokine ligand 3, C-X-C motif chemokine ligand 5 and C-X-C motif chemokine ligand 7 may become new potential diagnostic and therapeutic targets.
Keywords
Chemokine, lung cancer, REL proto-oncogene, elicitin response
Lung cancer has the highest morbidity and mortality at present. It has been found that the occurrence and development of lung cancer involve a lot of changes in gene structure and expression regulation[1,2]. Chemokine is a small protein (8-14 kDa), which plays an important role in the immune response process by binding with the receptor on the surface of host cells[3] and plays an important role in the defense mechanism of human body[4]. They are involved in tissue damage repair[3], inflammatory response[5], physiological or pathological angiogenesis[6-8], cell proliferation and apoptosis[9]. Elicitin response protein (ELR) and α-chemokine (CXC) binds to C-X-C Motif Chemokine Receptor 2 (CXCR2), promotes angiogenesis and inhibiting CXCR2 can inhibit the growth and metastasis of human lung cancer cells[10]. However, the role of various chemokines in lung cancer is not necessarily the same. In this experiment, by comparing the expression of various types of chemokines in lung cancer, we can find more accurate targets.
Inflammatory chemokine is a kind of small molecule bioactive peptide which is produced by inflammatory cells and tissue cells with various biological functions. The combination of chemokine and homologous G-protein coupled receptor can cause a series of intracellular reactions, which can promote the migration of related cells. Chemokine can be divided into four subfamilies according to the position of N-terminal residues, i.e. XC family, CC family, CXC family and CX3C family. Chemokine of CXC family can be further divided into two groups according to whether there is glutamate-leucine-arginine (ELR) motif: ELR+CXC chemokine and ELR-CXC chemokine. ELR+CXC chemokine, such as C-X-C motif chemokine ligand 1 (CXCL1), C-X-C motif chemokine ligand 2 (CXCL2), C-X-C motif chemokine ligand 3 (CXCL3), C-X-C motif chemokine ligand 5 (CXCL5), C-X-C motif chemokine ligand 7 (CXCL7), C-X-C motif chemokine ligand 8 (CXCL8), can bind to the C-X-C motif chemokine receptor 1 (CXCR1) or CXCR2 of neutrophils, chemotactic neutrophils or endothelial cells, thus promoting the synthesis and storage of factors to advance vascular production and angiogenesis.
In recent years, chemokine has been found to play a crucial role in the development of various cancer diseases. CXCL1 is highly expressed in melanoma, breast cancer[11,12], bladder cancer, colorectal cancer, epithelial ovarian cancer and gastric cancer[13]. The continuous expression of CXCL1 chemokine can stimulate the growth of microvasculature to tumor through paracrine and promote the malignant transformation and progression of melanoma in an autocrine manner; over expression of CXCL1 in ovarian epithelial cancer cells can promote cell proliferation; the positive expression rate of CXCL1 in prostate cancer is significantly increased and it is found that the positive expression intensity is closely related to the grading and staging of prostate cancer, which provide reference value for the diagnosis and prognosis of prostate cancer[13]. CXCL1 can activate protein kinase B (Akt)/nuclear factor-kappa B (NF-κB) signal transduction pathway and promote the progression of breast cancer by binding to its receptor CXCR2[14]. CXCL2 is highly expressed in oral squamous cell carcinoma cells. In the blood of esophageal cancer patients, serum CXCL2 is significantly over expressed and plays a vital part in the formation and progression of esophageal cancer tumors, proliferation and metastasis of tumor cells. Serum CXCL2 can be used as a prognostic indicator[15]. CXCL2 may be a marker of adverse prognosis in gastrointestinal stromal cell tumors[16]. CXCL2 expression is significantly increased in colon cancer and even in precancerous adenoma[17]. CXCL3 is highly expressed in breast cancer and participates in the metastasis of breast cancer, which can be the therapeutic target of breast cancer[18]. CXCL3 and its receptor CXCR2 are over expressed in prostate cancer cells, prostate epithelial cells and prostate cancer tissues, which may play multiple roles in the progression and metastasis of prostate cancer[19,20]. The expression of CXCL5 in breast cancer is significantly only be used as a preliminary indicator to assess the severity of the disease, but also as an independent predictor of postoperative recurrence[21]; In cervical cancer, the expression of CXCL5 is up-regulated, which promotes the proliferation and migration of cervical cancer cells[22]. Currently, CXCL7 can modify breast cancer cells and the surrounding extracellular matrix to promote cancer cell metastasis. CXCL8 is widely involved in systemic inflammatory response and can stimulate the proliferation of various human melanocytes in a dose dependent manner, suggesting that it can be an autocrine growth factor, stimulate human epithelial cells and participate in the formation of metastasis.
Materials and Methods
Research object:
Lung cancer tissue chip (60 cases of squamous cell carcinoma, 60 cases of adenocarcinoma, 8 cases of small cell lung cancer, 10 cases of normal lung tissue, 15 cases of paracancerous tissue).
Immunohistochemical antibody:
Immunohistochemical polyclonal antibody against CXCL1 (Immuoway YT2074), CXCL2 polyclonal antibody (cloud-clone PAB603Hu02), CXCL3 polyclonal antibody (Immuoway YT2075), CXCL5 polyclonal antibody (Immuoway YT5961), CXCL7 polyclonal antibody (Immuoway YT5963), CXCL8 polyclonal antibody (Immuoway YT2342), vascular endothelial growth factor (VEGF) monoclonal antibody (Immuoway YM3681), immunohistochemical second antibody, goat anti rabbit immunoglobulin G (IgG) purchased from Beijing Zhongshan Jinqiao Company.
Experimental steps of immunohistochemical elivision:
Each piece of lung cancer tissue chip costs 30 min; by performing regular dewaxing hydration to these chips; blocking inactivated endogenous peroxidase in these lung cancer tissue chips: this is done by first using 3 % hydrogen peroxide (H2O2), with citric acid buffer (pH 6.0) and 2.5 min high pressure to repair antigen in these tissues, until the temperature of the tissues cool to room temperature and then washing with phosphate buffer solution (PBS) for 5 min for three times; washing them with H2O2 3 % methanol to close endogenous peroxidase for 10 min at room temperature and then washing with PBS for 3 min for three times; adding primary antibody (CXCL1 1:100; CXCL2 1:75; CXCL3 1:100; CXCL5 1:100; CXCL7 1:100; CXCL8 1:100) to these tissues and putting them into 4° refrigerator overnight; washing with 0.1 % tween-20 PBS for 5 min for three times; adding amplifying agent to these tissues and incubating them at room temperature for 15 min; washing with 0.1 % tween-20 PBS for 5 min for three times; adding polymerase conjugate drop to these tissues and incubating them for 15 min at room temperature; washing with 0.1 % tween-20 PBS for 5 min for three times; conduct 3,3′-Diaminobenzidine (DAB) color development to these tissues, DAB color development time was controlled under microscope and the color development was terminated by distilled water washing; hematoxylin in the cells of these tissues was re-dyed, washed and fully washed after differentiation; after normal dehydration, these tissues should be transparent and using neutral gum to seal the tablets of these tissues.
Determination of immunohistochemical results:
The negative control was stained with PBS and the results showed that the cancer tissues were not stained. The positive tissue was determined by the staining intensity score and the percentage score of positive cells. Percentage score of positive cells: 0 points when the positive rate ≤5 %, 1 point when the positive rate ≤5 %, 2 points when the positive rate ≤ 50 %, 3 points when the positive rate ≤100 %. Color intensity score: colorless for 0, light yellow for 1, brown for 2 and brown for 3. Take the product of the two for grading: 0 is negative; Record 1-3 points (+), record 4-6 points (++); record 7-9 points (+++); 9-12 points recorded (++++). In this study, 7-12 scores were defined as high expression of chemokines.
Statistical methods:
Statistical package for the social sciences (SPSS) 22.0 software was used for statistical analysis of the data and the measurement data were expressed. For comparison of measurement data between the two groups, the t-test was used in the case of normal distribution, while Wilcoxon symbol rank sum test was used in the case of non-normal distribution. Counting data were expressed by frequency or rate, Pearson chi square test was used for comparison, Spearman correlation was used in correlation analysis when the data was in order grade and Pearson correlation was used in dichotomous data. The p<0.05 was considered to be statistically significant and the pairwise comparison was corrected by Bonferroni method on the basis of 0.05.
Results and Discussion
The expression of ELR+CXC factors in different tissue types is shown in Table 1. No high expression of ELR+CXC was detected in normal tissues and the high expression rate of ELR+CXC factors in cancer tissues was higher than that in normal tissues, with statistically significant differences (p<0.05) (Table 1).
| ELR+CXC correlation factor |
Organization type | Statistics | Parameter | ||
|---|---|---|---|---|---|
| Normal tissue | Cancer tissue | ||||
| CXCL1 | Low expression | 25 (100.00 %) | 109 (83.85 %) | χ2=4.671 | p=0.031 |
| High expression | 0 (0.00 %) | 21 (16.15 %) | |||
| CXCL2 | Low expression | 25 (100.00 %) | 103 (79.23 %) | χ2=4.927 | p=0.026 |
| High expression | 0 (0.00 %) | 27 (20.77 %) | |||
| CXCL3 | Low expression | 25 (100.00 %) | 59 (45.38 %) | χ2=25.195 | p<0.001 |
| High expression | 0 (0.00 %) | 71 (54.62 %) | |||
| CXCL5 | Low expression | 25 (100.00 %) | 69 (53.08 %) | χ2=19.343 | p<0.001 |
| High expression | 0 (0.00 %) | 61 (46.92 %) | |||
| CXCL7 | Low expression | 25 (100.00 %) | 68 (52.31 %) | χ2=19.872 | p<0.001 |
| High expression | 0 (0.00 %) | 62 (47.69 %) | |||
| CXCL8 | Low expression | 25 (100.00 %) | 100 (76.92 %) | χ2=5.752 | p=0.016 |
| High expression | 0 (0.00 %) | 30 (23.08 %) | |||
Table 1: Comparison of Expression of ELR+CXC Related Factors in Different Tissue Types
The expression of ELR+CXC factor in squamous cell carcinoma is shown in Table 2. Comparison showed that the expression of ELR+CXC factor in squamous cell carcinoma was statistically significant (χ2=24.000, p<0.001). The highest expression rate was CXCL3 factor. Pair comparison showed that the high expression rate of CXCL3 and CXCL5 factors was higher than that of CXCL1 factor, with statistically significant difference (p<0.003). The high expression rate of CXCL7 factors was higher than that of CXCL2 factor, with statistically significant difference (p<0.003) and the high expression rate of other factors was not statistically significant after comparison (p>0.0033).
| ELR+CXC correlation factor | Expression situation | Statistics | Parameter | |
|---|---|---|---|---|
| Low expression | High expression | |||
| CXCL1 | 60 (100.00 %) | 0 (0.00 %) | χ2=24.000 | p<0.001 |
| CXCL2 | 57 (95.00 %) | 3 (5.00 %) | ||
| CXCL3*# | 45 (75.00 %) | 15 (25.00 %) | ||
| CXCL5* | 48 (80.00 %) | 12 (20.00 %) | ||
| CXCL7 | 51 (85.00 %) | 9 (15.00 %) | ||
| CXCL8 | 54 (90.00 %) | 6 (10.00 %) | ||
Note: * is p<0.003 compared with CXCL1; # is p<0.003 compared with CXCL2
Table 2: Expression of ELR+CXC Factors In Squamous Cell Carcinoma
The expression of ELR+CXC factors in adenocarcinoma is shown in Table 3. There was significant difference in the expression of ELR+CXC in adenocarcinoma (χ2=108.592, p<0.001). The comparison between the two groups showed that the high expression rate of CXCL3, CXCL5 and CXCL7 factors was higher than that of CXCL1, CXCL2 and CXCL8 factors (p<0.003).
| ELR+CXC correlation factor |
Expression situation | Statistics | Parameter | |
|---|---|---|---|---|
| Low expression | High expression | |||
| CXCL1 | 48 (80.00 %) | 12 (20.00 %) | χ2=108.592 | p<0.001 |
| CXCL2 | 45 (75.00 %) | 15 (25.00 %) | ||
| CXCL3*#▽ | 12 (20.00 %) | 48 (80.00 %) | ||
| CXCL5*#▽ | 20 (33.33 %) | 40 (66.67 %) | ||
| CXCL7*#▽ | 10 (16.67 %) | 50 (83.33 %) | ||
| CXCL8 | 46 (76.67 %) | 14 (23.33 %) | ||
Note: * refers to p<0.003 compared with CXCL1; # refers to p<0.003 compared with CXCL2; ▽ refers to p<0.003 compared with CXCL8
Table 3: Expression of ELR+CXC Factors In Adenocarcinoma
The age of patients with different expression of ELR+CXC factors is shown in Table 4. There was no significant difference in the age of patients with different expression of each factor (p>0.05).
| ELR+CXC correlation factor | Expression situation | Statistics | Parameter | |
|---|---|---|---|---|
| Low expression | High expression | |||
| CXCL1 (109:21) | 55.30±8.70 | 55.81±8.88 | t=0.244 | p=0.808 |
| CXCL2 (103:27) | 55.20±8.77 | 56.07±8.57 | t=0.461 | p=0.645 |
| CXCL3 (59:71) | 56.17±8.45 | 54.73±8.91 | t=0.937 | p=0.350 |
| CXCL5 (69:61) | 56.51±8.61 | 54.11±8.69 | t=1.574 | p=0.118 |
| CXCL7 (68:62) | 55.87±8.83 | 54.85±8.59 | t=0.662 | p=0.509 |
| CXCL8 (100:30) | 55.40±8.81 | 55.33±8.48 | t=0.037 | p=0.970 |
Table 4: Correlation between Expression of ELR+CXC Factors and Age of Patients
The expression of ELR+CXC factors in patients of different genders is shown in Table 5. There was no significant difference in the expression of ELR+CXC between different genders (p>0.05).
| ELR+CXC correlation factor | Gender | Statistics | Parameter | ||
|---|---|---|---|---|---|
| Male | Female | ||||
| CXCL1 | Low expression | 78 (85.71 %) | 31 (79.49 %) | χ2=0.782 | p=0.377 |
| High expression | 13 (14.29 %) | 8 (20.51 %) | |||
| CXCL2 | Low expression | 74 (81.32 %) | 29 (74.36 %) | χ2=0.804 | p=0.370 |
| High expression | 17 (18.68 %) | 10 (25.64 %) | |||
| CXCL3 | Low expression | 46 (50.55 %) | 13 (44.83 %) | χ2=3.264 | p=0.071 |
| High expression | 45 (49.45 %) | 26 (55.17 %) | |||
| CXCL5 | Low expression | 53 (58.24 %) | 16 (55.17 %) | χ2=3.249 | p=0.071 |
| High expression | 38 (41.76 %) | 23 (44.83 %) | |||
| CXCL7 | Low expression | 50 (54.95 %) | 18 (62.07 %) | χ2=0.846 | p=0.358 |
| High expression | 41 (45.05 %) | 21 (37.93 %) | |||
| CXCL8 | Low expression | 71 (78.02 %) | 29 (74.36 %) | χ2=0.206 | p=0.50 |
| High expression | 20 (21.98 %) | 10 (25.64 %) | |||
Table 5: Correlation between the Expression of ELR+CXC Factors and Patients Gender
The expression of ELR+CXC factors in different TNM stages is shown in Table 6. The high expression rate of CXCL7 in tumor, nodes and metastases (TNM) stage I-II was 54.26 % and that in TNM stage III-IV was 30.56 %. The comparison showed that the high expression rate of CXCL7 in TNM stage I-II was higher than that in TNM stage III-IV, the difference was statistically significant (χ2=5.861, p=0.015). There was no statistical difference in the expression of other ELR+CXC factors in different TNM stages. Significance is (p>0.05).
| ELR+CXC correlation factor | TNM stages | Statistics | Parameter | ||
|---|---|---|---|---|---|
| Stage I~II | Stage III~IV | ||||
| CXCL1 | Low Expression | 81 (86.17 %) | 28 (77.78 %) | χ2=1.354 | p=0.245 |
| High expression | 13 (13.83 %) | 8 (22.22 %) | |||
| CXCL2 | Low expression | 78 (82.98 %) | 25 (69.44 %) | χ2=2.898 | p=0.089 |
| High expression | 16 (17.02 %) | 11 (30.56 %) | |||
| CXCL3 | Low expression | 41 (43.62 %) | 18 (50.00 %) | χ2=0.428 | p=0.513 |
| High expression | 53 (56.38 %) | 18 (50.00 %) | |||
| CXCL5 | Low expression | 46 (48.94 %) | 23 (63.89 %) | χ2=2.337 | p=0.126 |
| High expression | 48 (51.06 %) | 13 (36.11 %) | |||
| CXCL7 | Low expression | 43 (45.74 %) | 25 (69.44 %) | χ2=5.861 | p=0.015 |
| High expression | 51 (54.26 %) | 11 (30.56 %) | |||
| CXCL8 | Low expression | 70 (74.47 %) | 30 (83.33 %) | χ2=1.152 | p=0.283 |
| High expression | 24 (25.53 %) | 6 (16.67 %) | |||
Table 6: Comparison of Expression of ELR+CXC Factors In Different TNM Stages
The expression of ELR+CXC factors in different tissue grades is shown in Table 7. There was no statistical significance in the comparison of different tissue grades (χ2=5.920, p=0.052) and there was no statistical significance in the correlation analysis. The expression of ELR+CXC factors in lung cancer tissues is shown in fig. 1.The expression of ELR+CXC factors in lung cancer tissues is significantly higher than that in normal lung tissues.
| ELR+CXC correlation factor | Tissue category | Statistics | Parameter | |||
|---|---|---|---|---|---|---|
| Category 1 | Category 2 | Category 3 | ||||
| CXCL1 | Low expression | 13 (86.67 %) | 75 (89.29 %) | 9 (100.00 %) | χ2=1.209 | p=0.546 |
| High expression | 2 (13.33 %) | 9 (10.71 %) | 0 (0.00 %) | |||
| CXCL2 | Low expression | 12 (80.00 %) | 75 (89.29 %) | 6 (66.67 %) | χ2=4.021 | p=0.134 |
| High expression | 3 (20.00 %) | 9 (10.71 %) | 3 (33.33 %) | |||
| CXCL3 | Low expression | 4 (26.67 %) | 40 (47.62 %) | 7 (77.78 %) | χ2=5.920 | p=0.052 |
| High expression | 11 (73.33 %) | 44 (52.38 %) | 2 (22.22 %) | |||
| CXCL5 | Low expression | 9 (60.00 %) | 43 (51.19 %) | 5 (55.56 %) | χ2=0.427 | p=0.808 |
| High expression | 6 (40.00 %) | 41 (48.81 %) | 4 (44.44 %) | |||
| CXCL7 | Low expression | 6 (40.00 %) | 44 (52.38 %) | 3 (33.33 %) | χ2=1.754 | p=0.416 |
| High expression | 9 (60.00 %) | 40 (47.62 %) | 6 (66.67 %) | |||
| CXCL8 | Low expression | 15 (100.00 %) | 67 (79.76 %) | 8 (88.89 %) | χ2=3.971 | p=0.137 |
| High expression | 0 (0.00 %) | 17 (20.24 %) | 1 (11.11 %) | |||
Table 7: Comparison of Expression of ELR+CXC Factors In Different Tissue Grades
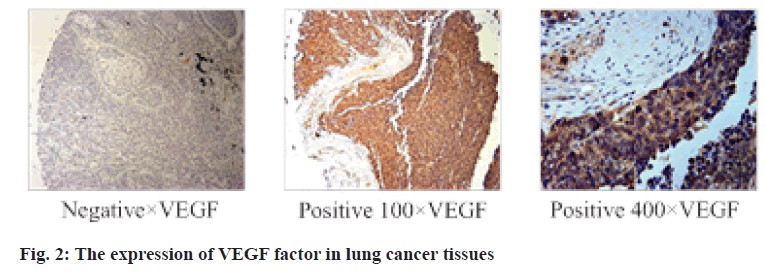
The expression of VEGF in 130 cancer tissues was detected. The correlation between the expression of ELR+CXC and VEGF was shown in Table 8. Correlation analysis showed that the expression of ELR+CXC factor and VEGF factor had correlation and had statistical significance (p<0.05). The expression of VEGF factor in lung cancer tissues is showed in fig. 2. The expression of VEGF factor in lung cancer tissues is significantly higher than that in normal lung tissues.
| ELR+CXC correlation factor | VEGF Factor | Correlation analysis | Contingency coefficient* | ||
|---|---|---|---|---|---|
| Low expression | High expression | ||||
| CXCL1 | Low expression | 56 | 53 | χ2=18.954, p<0.001 | r=0.382, p<0.001 |
| High expression | 0 | 21 | |||
| CXCL2 | Low expression | 49 | 54 | χ2=4.088, p=0.043 | r=0.177, p=0.044 |
| High expression | 7 | 20 | |||
| CXCL3 | Low expression | 40 | 19 | χ2=26.921, p<0.001 | r=0.455, p<0.001 |
| High expression | 16 | 55 | |||
| CXCL5 | Low expression | 46 | 23 | χ2=33.372, p<0.001 | r=0.507, p<0.001 |
| High expression | 10 | 51 | |||
| CXCL7 | Low expression | 46 | 22 | χ2=35.103, p<0.001 | r=0.520, p<0.001 |
| High expression | 10 | 52 | |||
| CXCL8 | Low expression | 53 | 47 | χ2=17.401, p<0.001 | r=0.366, p<0.001 |
| High expression | 3 | 27 | |||
Note: * is Pearson coefficient
Table 8: Correlation between ELR+CXC Factor Expression and VEGF Factor Expression
The above studies show that ELR+CXC are widely involved in the formation and progression of tumor cells. In non-small cell lung cancer, the binding of ELR+CXC chemokine and CXCR2 receptor can promote angiogenesis, while inhibition of CXCR2 can obstruct the growth and metastasis of human lung cancer cells, suggesting that ELR+CXC is involved in the process of lung cancer development, but the role of each factor in this process is not clear. In this experiment, the expression of each factor in tumor cells was preliminarily detected and the expressions of CXCL3, CXCL5 and CXCL7 were found significantly higher in tumor cells than in others, suggesting that it may play a more important role in tumor and may become a new target for the treatment of lung cancer. The expression of ELR+CXC is correlated to VEGF. Considering the synergistic effect of ELR+CXC and VEGF in the process of tumor formation, the possibility of common pathway is not excluded. At present, there are relevant researches showing that CXCL5 and CXCL7 are highly expressed in the serum of patients with non-small cell lung cancer, but there is no relevant research on the expression of CXCL3 in lung cancer. The expression of ELR+CXC in small cell lung cancer is higher than that of squamous cell carcinoma and adenocarcinoma, considering that ELR+CXC have a strong effect on the formation of small cell lung cancer. However, since the number of small cell lung cancer specimens in this experiment is relatively small, it is impossible to carry out systematic statistics and currently there is no relevant research. Therefore, the role of ELR+CXC in small cell lung cancer needs further experimental study.
The high expression rate of ELR+CXC in lung cancer was significantly higher than that in normal tissues (p<0.05). The expression of ELR+CXC factors in different pathological types was different (p<0.001). The expression of ELR+CXC in lung cancer was unrelated to age and gender. CXCL3, CXCL5 and CXCL7 and REL proto-oncogene (REL) members may play a more vital role in lung cancer formation than other ELR+CXC members. There was correlation between the expressions of ELR+CXC and VEGF factors.
Acknowledgement:
The research is supported by Scientific and Technological Innovation Capacity Building Project (No. 2018C2007-2-14).
Conflicts of interest:
The authors declared no conflict of interest.
References
- Zlotnik A, Yoshie O. Chemokines: a new classification system and their role in immunity. Immunity 2000;12(2):121-7.
- Clarke CN, Kuboki S, Tevar A, Lentsch AB, Edwards M. CXC chemokines play a critical role in liver injury, recovery and regeneration. Am J Surg 2009;198(3):415-9.
- Wang B, Hendricks DT, Wamunyokoli F, Parker MI. A growth-related oncogene/CXC chemokine receptor 2 autocrine loop contributes to cellular proliferation in esophageal cancer. Cancer res 2006;66(6):3071-7.
- Strieter RM, Burdick MD, Gomperts BN, Belperio JA, Keane MP. CXC chemokines in angiogenesis. Cytokine Growth Factor Rev 2005;16(6):593-609.
- Charo IF, Ransohoff RM. The many roles of chemokines and chemokine receptors in inflammation. N Engl J Med 2006;354(6):610-21.
- Rossi D, Zlotnik A. The biology of chemokines and their receptors. Annu Rev Immunol 2000;18(1):217-42.
- Thorburn E, Kolesar L, Brabcova E, Petrickova K, Petricek M, Jaresova M, et al. CXC and CC chemokines induced in human renal epithelial cells by inflammatory cytokines. APMIS 2009;117(7):477-87.
- Doll D, Keller L, Maak M, Boulesteix AL, Siewert JR, Holzmann B, et al. Differential expression of the chemokines GRO-2, GRO-3 and interleukin-8 in colon cancer and their impact on metastatic disease and survival. Int J Colorectal Dis 2010;25(5):573-81.
- Oue E, Lee JW, Sakamoto K, Iimura T, Aoki K, Kayamori K, et al. CXCL2 synthesized by oral squamous cell carcinoma is involved in cancer-associated bone destruction. Biochem Biophys Res Commun 2012;424(3):456-61.
- Li Y, Wang Y, Zhang P. Clinical significance of serum expression of GROβ in hepatocellular carcinoma. Tumor Biol 2015;36(8):6445-9.
- Lu Y, Li S, Ma L, Li Y, Zhang X, Peng Q, et al. Type conversion of secretomes in a 3D TAM2 and HCC cell co-culture system and functional importance of CXCL2 in HCC. Sci Rep 2016;6(1):1-3.
- Ye Q, Zhai X, Wang W, Zhang S, Zhu H, Wang D, et al. Overexpression of growth-related oncogene-β is associated with tumorigenesis, metastasis and poor prognosis in ovarian cancer. Dis Markers 2015;2015.
- Yang TG, Hu ZH. The Expression and significance of SPARC and CXCL1 in prostate cancer. Pract Clin Med 2017;18(10):21-3.
- Dong QM, Zhang JQ, Li Q, Bracher JC, Hendricks DT, Zhao XH. Clinical significance of serum expression of GROβ in esophageal squamous cell carcinoma. World J Gastroenterol 2011;17(21):2658-62.
- Zhao H, Zhu H, Jin Q, Zhang S, Wang W, Wang D, et al. Association of high expression of Groβ with clinical and pathological characteristics of unfavorable prognosis in gastrointestinal stromal tumors. Dis Markers 2015;2015.
- Zhang H, Ye YL, Li MX, Ye SB, Huang WR, Cai TT, et al. CXCL2/MIF-CXCR2 signaling promotes the recruitment of myeloid-derived suppressor cells and is correlated with prognosis in bladder cancer. Oncogene 2017;36(15):2095-104.
- Lay Pin See A, Kuan Chong P, Lu SY, Pin Lim Y. CXCL3 is a potential target for breast cancer metastasis. Curr Cancer Drug Targets 2014;14(3):294-309.
- Gui SL, Teng LC, Wang SQ, Liu S, Lin YL, Zhao XL, et al. Overexpression of CXCL3 can enhance the oncogenic potential of prostate cancer. Int Urol Nephrol 2016;48(5):701-9.
- An XQ, Lv JD, Su F, Zhao J. The sensitivity and specificity of G2 serum SDF-1 combined with CXCL5 and CEA in the diagnosis of breast cancer. Chin J Lab Diagn 2018;22(1):10-3.
- Ni F, Du YH, Zheng L, Hui P, Hu X, Zhang Z, et al. Effect of serum CXCL5 on recurrence of bladder cancer patients after surgical treatment. Pract J Cancer 2017;32(9):1444-6.
- Feng X, Zhang D, Li X, Ma S, Zhang C, Wang J, et al. CXCL5, the upregulated chemokine in patients with uterine cervix cancer, in vivo and in vitro contributes to oncogenic potential of Hela uterine cervix cancer cells. Biomed Pharmacother 2018;107:1496-504.
- Wu YC, Tang SJ, Sun GH, Sun KH. CXCR7 mediates TGFβ1-promoted EMT and tumor-initiating features in lung cancer. Oncogene 2016;35(16):2123-32.