- *Corresponding Author:
- Bin Liu
Department of Otorhinolaryngology Head and Neck Surgery, Xiangtan Municipal Central Hospital, Xiangtan, Hunan 411000, China
E-mail: dr455412@163.com
| This article was originally published in a special issue, “Role of Biomedicine in Pharmaceutical Sciences” |
| Indian J Pharm Sci 2023:85(2) Spl Issue “74-79” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
To clarify the expression level and methylation level of esophageal cancer related gene 4 in laryngeal malignant tumor and tumor adjacent tissues, and to explore its potential correlation with the clinicopathological characteristics of patients with laryngeal cancer is the objective of the study. The clinical samples were collected from 20 patients with primary laryngeal cancer in Xiangtan Central Hospital from December 2017 to December 2018. The level of expression and methylation of esophageal cancer related gene 4 in the samples were detected by the next-generation sequencing technology. In 15 cases, the messenger ribonucleic acid expression of esophageal cancer related gene 4 of tumor was significantly lower than that in adjacent tissues (p<0.05). In 18 cases, the methylation level of esophageal cancer related gene 4 in tissue of laryngeal squamous carcinoma was 46.7 %, which was significantly higher than that in adjacent tissues (26.2 %, p<0.01). Combined with clinical data analysis, it was found that, in laryngeal high and middle differentiated squamous cell carcinoma, the expression level of esophageal cancer related gene 4 was low and the methylation level was significantly increased. However, the expression of esophageal cancer related gene 4 in small round cell carcinoma was high, while the methylation level was significantly reduced. Smoking and drinking habits are the related factors of laryngeal high and middle differentiated squamous cell carcinoma. These results indicated significant difference of the esophageal cancer related gene 4 expression and methylation level between laryngeal malignant tumor and tumor adjacent tissues. Furthermore, the correlation between esophageal cancer related gene 4 expression and methylation level in laryngeal malignant cancer was found. Esophageal cancer related gene 4 could be regarded as a potential therapeutic target gene for laryngeal malignant cancer.
Keywords
Laryngeal malignant tumor, esophageal cancer related gene 4, methylation, tumor suppressor gene
Laryngeal carcinoma is a common primary epithelial cancer of the head and neck, accounting for 5.7 %-7.6 % of the total malignant tumors. Its main pathological type is laryngeal squamous cell carcinoma. In recent years, the incidence rate of laryngeal cancer has increased significantly. The causes of laryngeal cancer are complicated, including smoking, drinking, human papillomavirus infection, laryngopharyngeal reflux and gastroesophageal reflux[1]. At present, the main treatment methods are surgery combined with radiotherapy, drug therapy and gene therapy. Although the treatment effect of early laryngeal cancer is better, the effect and prognosis of late laryngeal cancer are still unsatisfactory. Therefore, it is of great significance for the prevention and treatment of laryngeal cancer to find an effective diagnosis and treatment target.
Esophageal Cancer Related Gene 4 (ECRG4) is a differentially expressed gene isolated from normal esophageal tissues and high cancer families by differential display Polymerase Chain Reaction (PCR) in Institute of oncology, Chinese Academy of Medical Sciences. The gene is located in chromosome 2ql4.1~2ql4.3 and composed of about 12 500 bases. The encoding open reading frame is composed of 444 bases and contains 4 exons. It encodes a peptide composed of 148 amino acids[2]. By Northern blot analysis, Steck et al.[3] showed that ECRG4 is widely expressed in esophageal carcinoma, breast cancer, Esophageal Squamous Cell Carcinoma (ESCC), prostate cancer, colorectal carcinoma and glioma[4]. In addition, the research of tumor shows that ECRG4 show low expression or lack of expression in some tumors[5]. It can be inferred that the expression of ECRG4 has a certain correlation with the occurrence and development of tumor. Therefore, ECRG4 has been recognized as a candidate tumor suppressor gene.
The reasons of downregulation and low expression of ECRG4, besides gene mutation and gene deletion, are epigenetic modification which is more prevalent. Epigenetic modification refers to the heritable change of gene function and the change of phenotype when the Deoxyribonucleic Acid (DNA) sequence of gene does not change. At present, DNA methylation, histone modification, chromosome configuration and noncoding Ribonucleic Acid (RNA) regulation are the main epigenetic mechanisms. It is found that hypermethylation is one of the early molecular abnormalities in carcinogenesis. The methylation frequency of some genes in the diseased cells was significantly higher than that in the adjacent tissues, which belonged to the high tendency methylation genes[6]. The methylation level in the pathological cells is significantly higher than that in the adjacent tissues, which is of great significance to the early detection, treatment and prognosis of tumors. In this study, the expression and methylation level of ECRG4 in laryngeal malignant cancer tissues were studied by the next-generation DNA sequencing technology and the correlation analysis was made with clinical data.
Materials and Methods
Clinical samples:
The tumor tissue samples and tumor adjacent tissue samples (1-2 cm from the edge of the tumor) were collected from 20 patients (19 males, 1 female, 52-73 y old, without preoperative radiotherapy and chemotherapy) with primary laryngeal malignant cancer. The 20 patients were diagnosed and treated in Xiangtan Central Hospital from December 2017 to December 2018.
After washing with normal saline, part of each sample was cut into 2-5 mm slices and immediately put into RNA, then fully infiltrated overnight at 4°, cooled down at -20° and finally transferred to -80° refrigerator for cryopreservation. According to the standard operating procedures of the hospital, pathological analysis was carried out on the remaining tissues.
This study was approved by the Xiangtan Central Hospital. Written informed consent was obtained from all the participants.
Total RNA extraction:
The tumor tissue and its adjacent tissue were taken out from -80° refrigerator and melted on ice. 25 mg tissues were cut with scissors and put into 1.5 ml DNA LoBind® (Eppendorf, Germany) centrifuge tube. Add a proper amount of liquid nitrogen. Use the high-speed tissue grinder kit (OSE-Y30, TGrinder, Tiangen Biotech Beijing Co., Ltd.) with a disposable plastic grinding pestle to carry out tissue grinding on ice. After grinding, RNA was extracted with MagJET™ viral RNA purification kit (Thermo Fisher Scientific, USA). In order to avoid DNA pollution, Deoxyribonuclease I (DNase I) (Thermo Fisher Scientific, USA) was added into the samples for digestion in the extraction process. The extracted RNA was quantified by NanoDrop™ 2000c ultra micro spectrophotometer (Thermo Fisher Scientific, United States of America (USA)).
Tissue DNA extraction and bisulfite transformation:
25 mg of tumor tissue and 25 mg of tumor adjacent tissue were taken respectively, and DNA was extracted from two groups of samples using QIAamp DNA Mini Kit (QIAGEN, Germany). The time for tissue lysis was prolonged to 1 h because of the tissue hardening due to long preservation time in RNA. The extracted DNA was quantified by Qubit 3.0 fluorescence quantitative instrument (Thermo Fisher Scientific, USA) and its purity was determined by NanoDrop™ 2000c ultra micro spectrophotometer (Thermo Fisher Scientific, USA).
The rapid sulfite transformation of the two groups of samples was carried out by using EpiTect Fast DNA bisulfite kit (50) 59824 (QIAGEN, Germany). 1 μg of DNA was used for transformation and the transformation time at 60° was extended from 10 min to 15 min. Other steps were carried out according to the instructions of the kit. The transformed DNA was quantified by NanoDrop™ 2000c ultra micro spectrophotometer (Thermo Fisher Scientific, USA).
Quantitative real-time PCR:
HiFiScript genomic DNA (gDNA) removal complementary DNA (cDNA) synthesis kit (CWBIO, China) was used for RNA reverse transcription. According to the operation instructions, the gDNA was digested (42°, 2 min) and then it was incubated at 42° for 15 min to reverse and convert into cDNA, and incubated at 85° for 5 min to terminate the reaction.
Beta (β)-Actin (ACTB) was selected as the internal reference gene and the relative expression of ECRG4 was detected by QuantStudio™ Dx real-time fluorescence quantitative PCR (Applied Biosystems, USA). Fluorescence quantitative detection was carried on by using TB Green™ Premix DimerEraser (Perfect Real Time) (Takara, Japan). For the reaction system, refer to the operation instructions. Take 1.5 μl of the reverse transcripts as the reaction template. PCR primers are synthesized from Sangon Biotech Co., Ltd. in Wuhan (Table 1). The amplification program was 98° for 2 min, (98° for 15 s, 60° for 30 s, 40 cycles). ACTB and ECRG4 were detected three times and quantitative analysis was carried out by 2-ΔΔCt method.
| Primer | Sequence (5'-3') |
|---|---|
| ECRG4-Exp-forward | GTTCTCCCTCGCAGCACC |
| ECRG4-Exp-reverse | TTCCACTTATGCCACCTGGG |
| ACTB-Exp-forward | CTCGCCTTTGCCGATCC |
| ACTB-Exp-reverse | GGGGTACTTCAGGGTGAGGA |
| ECRG4-MSP-forward | AGTGGGGGAGTTAAGGAGATATTTT |
| ECRG4-MSP-reverse | CTAAACTCCAAAACCAAAAATACTTAA |
Note: ECRG4-Exp and ACTB-Exp are primers for detecting mRNA expression; Exp: Expression; ECRG4-MSP is Methylation Specific Primer (MSP).
Table 1: Sequence of Primers.
Amplification and sequencing of core region:
Using TaKaRa Taq™ HS perfect mix (Takara, Japan) amplified the core region of ECRG4 promoter (Chr2: 106065304-106065523) and the length of amplified product was 220 base pair (bp), including 4 homeopathic elements and 16 Cytosines followed by Guanine residues (CpG) sites. The amplification system is configured according to the operation instructions, DNA template 50-100 ng and the amplification primer sequence is shown in Table 1. The amplification program was 98° for 2 min, (98° for 10 s, 60° for 20 s, 72° for 30 s, 30 cycles), 72° for 5 min. The amplified products were detected by 2 % agarose gel electrophoresis and the PCR products were recovered by AMPure XP Beads (Beckman, USA).
50 ng of the amplified products were used to construct the library by using Ion Plus Fragment Library Kit (Thermo Fisher Scientific, USA). Use Agilent 2100 chip (Agilent, China) for library quality inspection and use KAPA KK4827-Library Quantification Kit (KAPA Biosystems, USA) to accurately quantify the library. Dilute 20 pairs of sample libraries to 100 pM and mix equally, and 10 μl was taken to prepare the template with Ion PI™ Hi-Q™ OT2 200 kit (Thermo Fisher Scientific, USA). Ion PI™ Hi-Q™ sequencing 200 kit (Thermo Fisher Scientific, USA) and Ion PI™ V3 chips were used for sequencing in Ion Torrent Proton gene analyzer.
Results and Discussion
RNA extraction and DNA recovery was shown here.The concentration of RNA extracted from tissue and DNA was 25-500 ng/μl and 28-609 ng/μl respectively. The values of A260/A280 were higher than 1.8, which indicated that the purity of extraction was higher and the recovery of DNA was 50 %-70 %.
Difference of relative expression level of ECRG4 was explained here. In 20 pairs of matched tissue samples, at least one sample in the four groups of A/J/U/V failed to form valid experimental repetition, so these four pairs of samples were not included in the statistical analysis in this study. As shown in Table 2, by calculating the relative expression of ECRG4 in tumor tissue and tumor adjacent tissue, it was found that the messenger RNA (mRNA) expression level in 15 tumor tissues was significantly lower than that in tumor adjacent tissue (p<0.05). It is worth noting that, for samples in group H, the expression of ECRG4 gene in the tumor tissue is upregulated compared with that in the adjacent tissues. And the pathological results showed that the tumor was small round cell type malignant tumor. The results of immunohistochemistry indicate that the tumor cells express B lymphocyte and plasmocytic cell related antibodies, but not T lymphocyte related antibodies. The tumor cells are monoclonal and show proliferation and some tumor cells are binucleate and multinucleated. The concurrent proliferation index is high (Ki-67 is about 40 % positive (+)) and some of them are of plasmocytic type.
| Groups | Relative expression level | ECRG4 methylation level | ||||
|---|---|---|---|---|---|---|
| DDCt | Difference of expression (%) | Changes in expression | Tumor adjacent tissue | Tumor tissue | ||
| A | ** | ** | ** | 7.98 | 28.5 | |
| B | 4.52 | -95 | ---- | 44.9 | 29 | |
| C | 2.96 | -87 | --- | 34.3 | 24.5 | |
| D | 1.62 | -67 | --- | 51.9 | 39 | |
| E | 1.71 | -69 | --- | 30.4 | 24.4 | |
| F | 0.37 | -22 | - | 69.4 | 20.8 | |
| G | 1.1 | -53 | -- | 46 | 15 | |
| H | -7.9 | 25200 | ++++ | 36.8 | 21.7 | |
| I | 2.43 | -81 | --- | 64 | 19 | |
| J | ** | ** | ++ | 64.3 | 19.4 | |
| K | 0.11 | -7 | - | 56.4 | 30.6 | |
| L | 3.67 | -92 | ---- | 63.1 | 26.9 | |
| M | 1.78 | -70 | --- | 51.4 | 31 | |
| N | 8.73 | -99 | ---- | 57.8 | 35.7 | |
| O | 5.68 | -98 | ---- | 1.94 | 28.9 | |
| P | 1.6 | -67 | --- | 49.9 | 29.5 | |
| Q | 0.87 | -45 | -- | 49.9 | 29.3 | |
| R | 1.39 | -62 | -- | 59.3 | 18.2 | |
| p=0.038 | p=5.7e-3 | |||||
Note: **The data is not available, (+) Upregulation and (-) Downregulation. The data of expression and methylation level in samples of group S and group T are not available and are not included in the statistics and samples in U and V group could not provide available data.
Table 2: Expression level and Methylation level of ECRG4.
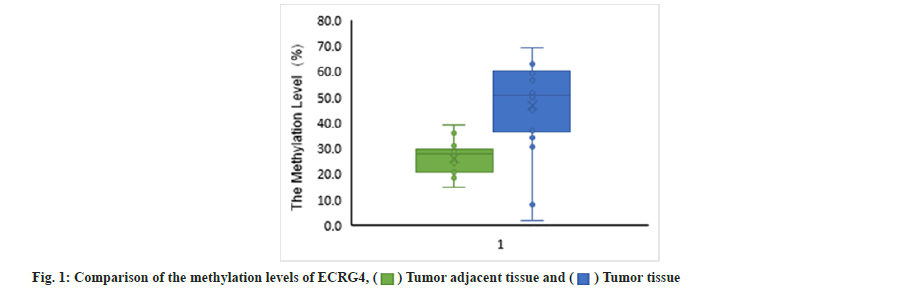
Difference of methylation level in core region of ECRG4 promoter was shown here. For analysis of ECRG4 methylation level, samples in U and V group could not provide available data, so they are not included in the analysis. As shown in fig. 1, the overall methylation level of ECRG4 gene in cancer tissues (46.7 %) was significantly higher than that in adjacent tissues (26.2 %) (p<0.01). As shown in Table 2, comparing the expression level and methylation level of ECRG4 gene in the two groups of samples, it could be found that the downregulation of ECRG4 gene mRNA expression level was related to the upregulation of ECRG4 gene methylation level.
Statistical results of clinical data are shown here. A total of 18 pairs of samples, of which, laryngeal squamous carcinoma accounts for a high proportion of laryngeal malignant cancer and the patients are mostly male, aged 52-73 y. Among them, 17 cases were smokers, 13 cases were drinkers and 13 cases were both smokers and drinkers. 17 cases werelaryngeal high and middle differentiated squamous carcinoma. 1 case was small round cell malignant tumor. Among them, ECRG4 gene expression was low in 16 cases of laryngeal high and middle differentiated squamous carcinoma and methylation level was significantly increased in 17 cases. However, the expression level of ECRG4 gene in small round cell malignant carcinoma was high and the methylation level was significantly reduced.
ECRG4 has been proved to be related to epithelial squamous cell carcinoma for many times since it was found. It has been proved to be a potential tumor suppressor gene in esophageal cancer, breast cancer, prostate cancer, nasopharyngeal cancer, laryngeal squamous cell carcinoma and so on. In these cancers, ECRG4 is significantly down regulated[7-11]. ECRG4 gene is composed of four exons, which is highly conserved among species. Gene sequencing showed that CpG island existed in the first exon and part of the first intron of the ECRG4 promoter region[12]. ECRG4 encodes a 17 kDa protein, which contains a leading peptide at residues 1-30, a furan like cleavage site at residues 68-71 and a thrombin like cleavage site at residues 130-134[13,14]. Unlike p53 and Retinoblastoma 1 (Rb1), which act by encoding intracellular proteins, the suppressor gene ECRG4 acts in a secretory manner like a cytokine or a growth factor which is similar to a chemical kinase [15,16]. The protein encoded by ECRG4 is an auxiliary molecule, secreted by endocrine tissue in vivo and is involved in the regulation of fluid dynamic balance of cerebrospinal fluid, the stimulation of precursor nerve cells and the induction of cell induction in central nervous system after brain injury. Although the exact function of ECRG4 has not been determined yet, it is regarded as a potential tumor suppressor gene and plays an important role in vivo.
By calculating the relative expression of ECRG4 in tumor tissue and tumor adjacent tissue, the results of this study showed that the expression of ECRG4 mRNA in tumor tissue was significantly lower than that in tumor adjacent tissue, but ECRG4 gene is highly expressed in small round cell tumors. According to the results of immunohistochemistry, it suggested that the expression mechanism of ECRG4 gene in different pathological types of laryngeal cancer might be different. Previous studies have shown that the expression level of ECRG4 in different organs of squamous carcinoma is significantly lower than that in adjacent tissues. The downregulation of ECRG4 expression is significantly related to the prognosis of patients. It can be used as an independent prognostic indicator in clinical practice.
This study also explored the methylation level of ECRG4 gene. It was found that the methylation level of tumor tissue was significantly higher than that of tumor adjacent tissue. Moreover, the downregulation of ECRG4 gene mRNA expression level was related to the upregulation of ECRG4 gene methylation level. Previous studies have shown that the expression of ECRG4 is related to hypermethylation of CpG island[7-11]. Yue et al.[12] first found that the downregulation of ECRG4 expression in ESCC was related to the methylation of CpG island in the core promoter region of the gene by Reverse Transcription-Polymerase Chain Reaction (RT-PCR) and denaturing high performance liquid chromatography, and proposed that the methylation of ECRG4 might be an early event in the development of ESCC. The CpG island is a key factor in the regulation of ECRG4 gene expression [17]. The methylation of ECRG4 promoter can be detected in peripheral blood of patients with Nasopharyngeal Carcinoma (NPC). Abnormal methylation of ECRG4 promoter can be used for early cancer monitoring and pathological staging prediction[18].
In this study, through different methods, it was reconfirmed that the ECRG4 gene expression in human laryngeal high and middle differentiated squamous carcinoma was downregulated and related to hypermethylation. Smoking and drinking habits are related to the low expression of ECRG4 gene, which may be related to the rapid downregulation of ECRG4 expression in the normal and still Middle Ear (ME) mucosa after infection, as well as similar reactions in acute skin injury and other chronic inflammation[19,20].
Different from squamous cell carcinoma, laryngeal cancer with pathological differentiation of other types have the opposite phenomenon. This is related to the different progression between malignant tumor with other pathological types and squamous cell carcinoma. Further research is needed.
In conclusion, with the explosive development of research for ECRG4, the mechanism of ECRG4 in the development of injury, inflammation, infection, tumor and other diseases is becoming more and more clear and the properties of ECRG4 are being elucidated. As a potential biomarker in precision medicine, it may play an important role in the future treatment of laryngeal cancer. In this study, we found that the expression level and methylation level of ECRG4 gene in laryngeal malignant tumor tissues and adjacent tissues were significantly different. The expression of ECRG4 gene was low and the methylation level was significantly increased in laryngeal high and middle differentiated squamous carcinoma, while the expression of ECRG4 gene was high and the methylation level was significantly decreased in small round cell carcinoma. For older men, smoking and drinking habits may be related to the development of laryngeal high and middle differentiated squamous carcinoma. As a biomarker of tumor diagnosis and prediction of prognosis, ECRG4 has potential value.
Author’s contributions:
Zhengjia Peng and Bin Liu contributed equally to this work.
Conflict of interests:
The authors declared no conflict of interest.
References
- Huang ZG. The status and prospects of the basic research in larynx carcinoma. Chin Arch Otolaryngol Head Neck Surg 2010;14(2):57-8.
- Li LW, Li YY, Li XY, Zhang CP, Zhou Y, Lu SH. A novel tumor suppressor gene ECRG4 interacts directly with TMPRSS11A (ECRG1) to inhibit cancer cell growth in esophageal carcinoma. BMC Cancer 2011;11(1):1-7.
[Crossref] [Google scholar] [PubMed]
- Steck E, Breit S, Breusch SJ, Axt M, Richter W. Enhanced expression of the human chitinase 3-like 2 gene (YKL-39) but not chitinase 3-like 1 gene (YKL-40) in osteoarthritic cartilage. Biochem Biophys Res Commun 2002;299(1):109-15.
[Crossref] [Google scholar] [PubMed]
- Gilmore TD. Multiple mutations contribute to the oncogenicity of the retroviral oncoprotein v-Rel. Oncogene 1999;18(49):6925-37.
[Crossref] [Google scholar] [PubMed]
- Matsuzaki J, Torigoe T, Hirohashi Y, Tamura Y, Asanuma H, Nakazawa E, et al. Expression of ECRG 4 is associated with lower proliferative potential of esophageal cancer cells. Pathol Int 2013;63(8):391-7.
[Crossref] [Google scholar] [PubMed]
- Götze S, Feldhaus V, Traska T, Wolter M, Reifenberger G, Tannapfel A, et al. ECRG4 is a candidate tumor suppressor gene frequently hypermethylated in colorectal carcinoma and glioma. BMC Cancer 2009;9(1):1-11.
[Crossref] [Google scholar] [PubMed]
- Xiang TX, Yuan Y, Li LL, Wang ZH, Dan LY, Chen Y, et al. Aberrant promoter CpG methylation and its translational applications in breast cancer. Chin J Cancer 2013;32(1):12-20.
[Crossref] [Google scholar] [PubMed]
- Sabatier R, Finetti P, Adelaide J, Guille A, Borg JP, Chaffanet M, et al. Down-regulation of ECRG4, a candidate tumor suppressor gene, in human breast cancer. PLoS One 2011;6(11):e27656.
[Crossref] [Google scholar] [PubMed]
- Li LW, Yu XY, Yang Y, Zhang CP, Guo LP, Lu SH. Expression of esophageal cancer related gene 4 (ECRG4), a novel tumor suppressor gene, in esophageal cancer and its inhibitory effect on the tumor growth in vitro and in vivo. Int J Cancer 2009;125(7):1505-13.
[Crossref] [Google scholar] [PubMed]
- Li W, Liu X, Zhang B, Qi D, Zhang L, Jin Y, et al. Overexpression of candidate tumor suppressor ECRG4 inhibits glioma proliferation and invasion. J Exp Clin Cancer Res 2010;29(1):1-7.
[Crossref] [Google scholar] [PubMed]
- Jia J, Dai S, Sun X, Sang Y, Xu Z, Zhang J, et al. A preliminary study of the effect of ECRG4 overexpression on the proliferation and apoptosis of human laryngeal cancer cells and the underlying mechanisms. Mol Med Rep 2015;12(4):5058-64.
[Crossref] [Google scholar] [PubMed]
- Yue CM, Deng DJ, Bi MX, Guo LP, Lu SH. Expression of ECRG4, a novel esophageal cancer-related gene, downregulated by CpG island hypermethylation in human esophageal squamous cell carcinoma. World J Gastroenterol 2003;9(6):1174-8.
[Crossref] [Google scholar] [PubMed]
- Dang X, Podvin S, Coimbra R, Eliceiri B, Baird A. Cell-specific processing and release of the hormone-like precursor and candidate tumor suppressor gene product, Ecrg4. Cell Tissue Res 2012;348(3):505-14.
[Crossref] [Google scholar] [PubMed]
- Gonzalez AM, Podvin S, Lin SY, Miller MC, Botfield H, Leadbeater WE, et al. Ecrg4 expression and its product augurin in the choroid plexus: Impact on fetal brain development, cerebrospinal fluid homeostasis and neuroprogenitor cell response to CNS injury. Fluids Barriers CNS 2011;8(1):1-8.
[Crossref] [Google scholar] [PubMed]
- Carrasco‐Garcia E, Moreno M, Moreno‐Cugnon L, Matheu A. Increased Arf/p53 activity in stem cells, aging and cancer. Aging Cell 2017;16(2):219-25.
[Crossref] [Google scholar] [PubMed]
- Dyson NJ. RB1: A prototype tumor suppressor and an enigma. Genes Dev 2016;30(13):1492-502.
[Crossref] [Google scholar] [PubMed]
- Kao S, Shaterian A, Cauvi DM, Dang X, Chun HB, de Maio A, et al. Pulmonary preconditioning, injury and inflammation modulate expression of the candidate tumor suppressor gene ECRG4 in lung. Exp Lung Res 2015;41(3):162-72.
[Crossref] [Google scholar] [PubMed]
- Wang YB, Ba CF. Promoter methylation of esophageal cancer-related gene 4 in gastric cancer tissue and its clinical significance. Hepatogastroenterology 2012;59(118):1696-8.
[Crossref] [Google scholar] [PubMed]
- Kurabi A, Pak K, Dang X, Coimbra R, Eliceiri BP, Ryan AF, et al. Ecrg4 attenuates the inflammatory proliferative response of mucosal epithelial cells to infection. PLoS One 2013;8(4):e61394.
[Crossref] [Google scholar] [PubMed]
- Shaterian A, Kao S, Chen L, DiPietro LA, Coimbra R, Eliceiri BP, et al. The candidate tumor suppressor gene ECRG4 as a wound terminating factor in cutaneous injury. Arch Dermatol Res 2013;305:141-9.
[Crossref] [Google scholar] [PubMed]