- *Corresponding Author:
- M. Tang
Medical College, Hunan Polytechnic of Environment and Biology, Hengyang City, 421005, China
E-mail: tangmin5463@126.com
| This article was originally published in a special issue: Special issue on “Drug Development and Human Health in China” |
| Indian J Pharm Sci 2020:82(1)spl issue2;31-35 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
In order to study the application of genomic DNA methylation in non-small cell lung cancer in early screening of tumour, one human normal bronchial epithelial cell line and two non-small cell lung cancer cell lines, 95C and 95D were selected as experimental subjects. The cell lines were cultured, RNA was extracted and DNA methylation analysis was performed using the real-time quantitative polymerase chain reaction and plasmid DNA extraction. It was found that in the analysis of RNF111 mRNA expression, the mRNA expression levels of RNF111 gene in the 3 cell lines were significantly different and the difference between 95C and 95D was more prominent (p<0.01). Analysis of DNA methylation in the proximal promoter region of RNF111 gene indicated that the frequency of Cp G site methylation in the positions of -309, -109 and +3 in 95C and 95D cell lines had significant difference (p<0.01). These results indicated that the methylation status of this site of RNF111 gene can be specifically targeted or changed, which is beneficial to the development of drugs for lung cancer and to stop lung cancer metastasis process. Although there were some shortcomings in the experimental process, it still provided a reference for the clinical treatment of non-small cell lung cancer.
Keywords
NSCLC, DNA methylation, tumour, RT-PCR
At present, the incidence of lung cancer is increasing and it is already one of the common malignant tumours. Among those, non-small cell lung cancer (NSCLC) is more than 80 %, including adenocarcinoma (AC), squamous cell carcinoma (SCC), adenosquamous carcinoma (ASC), and large cell lung cancer (LCLC)[1]. Its morbidity and mortality are at the forefront of the world and it is still dominated by surgery[1,2]. Studies in China and overseas are focussing more and more on this subject. Traditionally, the occurrence of NSCLC is caused only by classical genetic factors and DNA changes and now as the new force of genetics, epigenetics in non-small cells, the role of lung cancer in the occurrence and development has been paid more and more attention too[3,4].
NSCLC includes AC, SCC, ASC and LCLC[5]. Compared to other types of lung cancer, NSCLCs grow relatively slowly with relatively late metastasis[6]. For patients with early stage NSCLC, the clinical symptoms are not obvious, so most patients miss the best treatment time at the time of diagnosis or need to undergo greater surgical trauma or pain. Although the current treatment of lung cancer is becoming more and more abundant and the cure rate has increased, the five-year survival rate of patients with NSCLC is still less than 15 % and majority of patients die due to recurrence or metastasis[7,8]. DNA methylation, an epigenetic modification method, is mediated by the DNA methyltransferase family. DNA methylation changes include whole genome hypomethylation and hypermethylation of the tumour suppressor gene promoter region[9-11]. Epigenetic treatment is gradually becoming a potential treatment for solid tumors. In recent years, there have been more and more studies on DNA methylation of epigenetic therapeutics and the role of apparent treatment in lung cancer has received much attention.
To sum up, in order to study the application of genomic DNA methylation in NSCLC in early screening of tumour, real-time quantitative PCR and plasmid DNA extraction were used to analyse DNA methylation in order to provide experimental evidence for the clinical treatment of NSCLC by selecting cell line HBE and NSCLC lines 95C and 95D.
Human normal bronchial epithelial cell line (HBE) and 2 types of NSCLC cell lines 95C (low metastatic lung giant cell carcinoma cell line) and 95D (high metastatic lung giant cell carcinoma cell line) were used in this study. The HBE cell line, 95C and 95D cell lines were purchased from Beijing Crisprbio Biotechnology Co., Ltd., China.
HBE, 95C and 95D cell lines were cultured in RPMI- 1640 medium containing 10 % fetal bovine serum in a cell incubator at 37°, 5 % CO2. Every 3-4 d, when the cells are in the logarithmic growth phase, they were digested with 0.25 % trypsin and passaged. All cell lines would adhere to the wall under normal conditions. The specific steps were as follows, the temperature of the water bath to adjusted to 37-40°, on an ultraclean workbench about 8 ml of complete medium was drawn into a 10 ml centrifuge tube. The frozen cell lines in liquid nitrogen were taken out, the upper end of the cryotube was placed quickly in a 37° water bath and gently shaken to uniformly heat and completely thaw. The cells were aspirated, placed in the prepared 10 ml centrifuge tube, centrifuged at 800 to 1000 rpm for 5 min and the supernatant was removed. The cells were suspended in 6-8 ml of medium, transferred to a culture flask, observed under a microscope, after which incubated in a CO2 incubator at 37° with medium change after 24 h and subcultured until the number of cells grows to 80 %.
After the cells were grown to a certain density, the culture solution was drained and washed twice with 5 ml phosphate buffered saline (PBS). One millilitre of 0.25 % trypsin was added to cover the culture surface of the flask, gently shaken and allowed to stand for a while. When the adherent cells became spherical and some cells were detached, 3 ml of the culture medium was added to terminate the digestion reaction. Blow the four corners of the culture flask until the surface of the culture flask becomes transparent and 2 ml of liquid was dispensed into a new culture flask. Six millilitres of medium was added to each flask, capped loosely and placed in an incubator for cultivation.
After trypsinization and washing with PBS, the cells were collected into a 1.5 ml centrifuge tube and 1 ml of Trizol was added and allowed to stand at room temperature for 15 min. Two hundred microliters of chloroform was added and shaken vigorously for 30 s, kept aside for 5 min, thoroughly mixed and centrifuged at 12 000 rpm for 15 min at 4°. The supernatant was transferred to a new 1.5 ml centrifuge tube, 500 μl of isopropanol was added, mixed, allowed to stand at room temperature for 10 min and centrifuged at 12 000 rpm for 20 min at 4°. The supernatant was discarded, 1 ml of pre-cooled 75 % ethanol was added and the pellet was gently pipetted out.
The total RNA template was diluted to 200 ng/l with diethyl pyrocarbonated (DEPC) water. Two microliters random primer, 1 μl deoxynucleoside triphosphate, 1 μl RNA, and 9 μl DEPC water were sequentially added to a 20 μl Eppendorf tube to form a 20 μl reaction system. After incubating for 5 min at 65°, it was placed on ice for cooling and briefly centrifuged. Added 4 μl 5×buffer and 2 μl 0.1 MTT to a 200 μl EP tube. Mixed well and placed at 37° for 2 min. At room temperature, 1 μl Moloney murine leukaemia virus was added and mixed by pipetting, incubated at 25° for 10 min, at 37° for 50 min, at 70° for 15 min and then the reaction was completed.
The lung cancer cell line in the logarithmic growth phase was taken, and the total RNA was extracted using Trizol method. The reverse transcription reaction system was prepared according to the Bestar qPCR RT Kit specification with 2 μg total RNA as a template and the total system was 20 μl to synthesize the first strand of cDNA. The reaction conditions of PCR (Nanjing Shengxing Biotechnology Co., Ltd., China) were, 94° for 2 min, 94° for 20 s, 58° for 20 s, 72° for 20 s, 40 cycles. Melting curve analysis was at temperature 62°- 95°. Three experiments were repeated for each set of samples. Fluorescence quantitative PCR experiments were performed using an Agilent Stratagene PCR instrument Mx3000P. The sequence of the target gene is as follows, gene length of RNF11: 186 bp; primer sequence: GAGGCAGAATGCAGCAGAAGTTGT (upstream); ACTGTGGAAGGTTGGCTACTGGTG (downstream). Gene length of β-actin: 216 bp; primer sequence: GGCATCCTCACCCTGAAGT (upstream); GGGATAGCACAGCCTGGAT (downstream). Fifty microliters of PCR amplification reaction system was prepared according to Table 1. The reaction procedure was as follows, 94°, 5 min; 94°, 30 s, 52°, 30 s, 72°, 30 s, 45 cycles; 72°, 10 min; 4°, store and amplify target gene. After the reaction was completed, 5 μl of the amplified product was subjected to 2 % agarose gel electrophoresis analysis.
Table 1: PCR amplification reaction system
| Components | Volume |
|---|---|
| 2×PCR Taq mix | 25 µl |
| Primers (10 µM) | 2.0 µl (F/R) |
| DNA | 3 µl |
| H2O | 18 µl |
| Total | 50 µl |
After rinsing the cell culture flask with PBS, the cells were digested with 1 ml trypsin, transferred to a 2 ml EP tube, centrifuged at 12 000 g, for 5 min at room temperature and the supernatant was discarded, the pellet was transferred into a mortar, a small amount of liquid nitrogen was added and the pellet was ground to a powder. Then, 1.5 ml TE, 40 μl 20 % SDS, and 15 μl proteinase K (20 mg/ml) were added while stirring in the mortar and finally transfer all the liquid to a 1.5 ml centrifuge tube. Incubate at 50° for 12-18 h, fully lyse the cells and digest the protein with gently mixing the contents. The same volume of phenol:chloroform:isoamyl alcohol (25:24:1) was added to the centrifuge tube and mixed thoroughly for 10 min. The supernatant was transferred to a new centrifuge tube, centrifuged at 13 000 g at 4° for 15 min. An equal volume of chloroform:isoamyl alcohol (24:1) was added to the centrifuge tube and mixed by inversion for 10 min. It was ultra-centrifuged at low temperature for 15 min, and the supernatant was taken to a new 1.5 ml centrifuge tube. A small volume of NaAC, pH 4.8 was added along with 2 volumes of pre-cooled ethanol, mixed well, kept at -20° for 2 h, centrifuged at 13 000 g at 4° for 15 min, the supernatant was discarded and 200 μl of 70 % pre-cooled ethanol was added to rinse the DNA. The DNA was collected by centrifugation at 13 000 g 4° for 15 min and allowed to air dry at room temperature. The DNA was dissolved in 20 μl of sterile water and stored at -20°. The purity and concentration of genomic DNA were determined on an UV spectrophotometer.
The obtained genomic DNA was amplified. The reaction parameters of PCR were, pre-denaturation at 98° for 2 min, denaturation at 98° for 10 s, extension at 68° for 1 min, for a total of 35 cycles. After completion, the PCR product was separated by agarose gel electrophoresis and finally stained with ethidium bromide and observed under an ultraviolet lamp. In order to know the methylation status of the Cp G site of a specific fragment of a gene, the specific fragment was usually ligated to the T vector by TA cloning, then transfected into Escherichia coli and finally the monoclonal for sequencing was selected by blue-white spot screening. The fragment of the gene was identified by agarose gel electrophoresis, and the target fragment of the recovered gene was purified using Biomiga Gel/PCR extraction kit. One microliter of pMD19-T vector, 3 μl of ddH2O, 15 μl of solution and 1 μl of the purified DNA fragment were placed in a 200 μl centrifuge tube to construct a 10 μl reaction system. The reaction was allowed to stand at 16° for 30 min. Ten microliters of the ligation product was added to 10 μl of competent cells for 30 min in an ice bath. Immediately after the completion of the ice bath, transfer to a 42° metal bath and put it on ice for 2 min after heat shock for 90 s (be careful not to shake the tube), then 250 μl of LB liquid medium (without Amp) was added to the centrifuge tube, gently mixed with a pipette tip, placed in a 37° incubator and centrifuged at 150 rpm for 40 min. Forty microliters of X-gal and Isopropyl β-d-1-thiogalactopyranoside were uniformly coated on an LB plate containing Amp (the LB plate was preheated at 37°), then 200 μl of the transformant was applied to the LB plate in the above step, cultured in a 37° incubator for 40 min and then inverted and cultured overnight. After the formation of a single colony, the white spot was picked and placed in a liquid LB medium (containing Amp), and shaken overnight in an incubator.
One and a half millilitres of the overnight culture solution was pipetted out into a new centrifuge tube, centrifuged at 13 000 pm for 1 min, the supernatant was discarded then 250 μl of buffer S1 was added to the centrifuge tube to uniformly suspend the bacterial body precipitate, and no small bacteria remained. Added 250 μl of buffer S2 to the centrifuge tube, and thoroughly and gently flip the tube for 4-6 times to mix well, so that the cells are fully lysed until the solution becomes translucent. Then 350 μl of buffer S3 was added to the centrifuge tube and the tube was thoroughly and gently inverted 6 to 8 times to mix well, centrifuged at 13 000 rpm for 10 min. The supernatant was transferred to a preparation tube, centrifuged at 13 000 rpm for 1 min and the filtrate was discarded. The preparation tube was placed back into the centrifuge tube, and buffer W1 500 μl was added, centrifuged at 13 000 rpm for 1 min and the filtrate was discarded. The preparation tube was returned to the centrifuge tube, buffer W2 700 μl was added, centrifuged at 13 000 rpm for 1 min, and the filtrate was discarded. The previous step was repeated and the preparation tube was returned to a 2 ml centrifuge tube, centrifuged at 13 000 rpm for 1 min. The preparation tube was transferred to a new 1.5 ml centrifuge tube, 60-80 μl eluent or deionized water was added to the center of the preparation tube membrane, allowed to stand at room temperature for 1 min and centrifuged at 13 000 rpm for 1 min. Finally, the concentration was measured using a micro spectrophotometer and the purity was observed by electrophoresis.
All data of the experimental results were expressed as mean±standard deviation and statistical methods used were one-way ANOVA and t-test for data processing. All data were analysed using SPSS software.
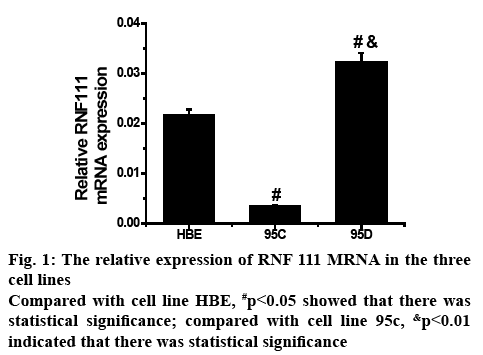
To determine the expression level of RNF111 gene mRNA in HBE, 95C and 95D cell lines, the histogram of the post-test statistical analysis using real-time fluorescent quantitative PCR is shown in figure 1. In the 3 cell lines, the mRNA expression of RNF111 gene was significantly different between the two, and the difference between 95C and 95D was more prominent. Therefore, in the metastatic tumour cells, the upregulation of mRNA expression of RNF111 gene might be closely related to the degree of malignancy of tumour metastasis.
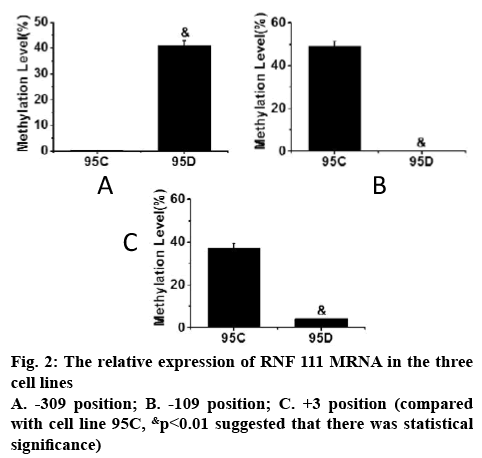
The methylation status of DNA in the promoter region of RNF111 gene in 95C and 95D cell lines was determined and the methylation status of each Cp G site in both cell lines was analysed. The frequency of Cp G site methylation at the 3 positions of the promoter regions of RNF111 gene -309, -109 and +3 is shown in figure 2. As can be seen, the frequency of Cp G site methylation in the three positions of -309, -109 and +3 in 95C and 95D cell lines was significantly different (p<0.01).
In order to study the application of genomic DNA methylation in NSCLC in early screening of tumour, one human normal bronchial epithelial cell line HBE and two NSCLC cell lines 95C and 95D were selected as test system. The cell lines were cultured, the RNA in the cells was extracted, and DNA methylation analysis was performed by real-time quantitative PCR and plasmid DNA extraction. It was found that in the analysis of RNF111 mRNA expression, the mRNA expression levels of RNF111 gene in the 3 cell lines were significantly different, and the difference between 95C and 95D was more prominent (p<0.01). In the analysis of DNA methylation in the proximal promoter region of RNF111 gene, it was found that there was a significant difference in the frequency of Cp G site methylation occurred in the three positions of -309, -109 and +3 in 95C and 95D cell lines (p<0.01).
In summary, through the application of genomic DNA methylation of NSCLC in early screening of tumour, it was found that the methylation status of this site of RNF111 gene can be targeted or changed, which was conducive to the development of lung cancer targeted drugs to delay the lung cancer metastasis process and could provide evidence for the clinical treatment of NSCLC. However, there were still some shortcomings in this investigation. For example, only the effects on cell line were evaluated but further research is needed to assess clinical applicability.
References
- Huang Q, Ma XC, Yang X. Expression of IL-17A, E, and F and their receptors in non-small-cell lung cancer. J Biol Regul Homeost Agents 2018;32(5):1105-16.
- Zeng Y, Zhu J, Qin H. Methylated+ 322-327 CpG site decreases hOGG1 mRNA expression in non-small cell lung cancer. Oncol Rep 2017;38(1):529-37.
- O'Leary K, Shia A, Schmid P. Epigenetic regulation of EMT in non-small cell lung cancer. Curr Cancer Drug Targets 2018;18(1):89-96.
- Wu Z, Qiu M, Guo Y. OTU deubiquitinase 4 is silenced and radiosensitizes non-small cell lung cancer cells via inhibiting DNA repair. Cancer Cell Int 2019;19(1):99.
- Liu L, Yuan P, Dogan S. The Impact of Post-Translational Regulation of Histone on Cancer Metastasis and Cancer Chemoresistance. Curr Pharmacol Rep 2017;3(5):253-67.
- Yan X, Xiong X, Chen YG. Feedback regulation of TGF-β signaling. Acta Biochim Biophys Sin 2017;50(1):37-50.
- Wright JD, Mace PD, Day CL. Secondary ubiquitin-RING docking enhances Arkadia and Ark2C E3 ligase activity. Nat Struct Mol Biol 2016;23(1):45.
- Holcomb N, Goswami M, Han SG. Exposure of human lung cells to tobacco smoke condensate inhibits the nucleotide excision repair pathway. PloS One 2016;11(7):e0158858.
- Agarwal P, Miller KM. The nucleosome: orchestrating DNA damage signaling and repair within chromatin. Biochem Cell Biol 2016;94(5):381-95.
- Bhargava A, Bunkar N, Aglawe A. Epigenetic biomarkers for risk assessment of particulate matter associated lung cancer. Curr Drug Targets 2018;19(10):1127-47.
- Xia Y, Xiong N, Huang Y. Relationship between methylation status of RASSF2A gene promoter and endometriosis-associated ovarian cancer. J Biol Regul Homeost Agents 2018;32(1):21-28.