- *Corresponding Author:
- Daikai Lu
Department of Otolaryngology, Hwamei Hospital, University of Chinese Academy of Sciences, Ningbo, Zhejiang 315000, China
E-mail: nbludakai@163.com
| This article was originally published in a special issue, “Recent Developments in Biomedical Research and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2022:84(4) Spl Issue “77-83” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Objective in order to discover correlated gene modules and hub genes for recurrent laryngeal squamous cell carcinoma through weighted gene co-expression network analysis method. The microarray dataset of recurrent laryngeal cancer, namely GSE27020, were obtained from the gene expression Omnibus database. Weighted gene co-expression network analysis was introduced to establish a gene co-expression network, mining key clinical trait correlated hub genes. Gene ontology enrichment analyses were performed for the genes in modules related to recurrent laryngeal squamous cell carcinoma. Then, we build up a proteinprotein interaction network with the genes in interest gene module and identified hub genes through analyze such protein-protein interaction network. The hub genes were mined using cytohubba plus-in. Finally, we analyzed these hub genes overall survival using gene expression profiling interactive analysis database. Forty four gene co-expression gene modules were achieved via weighted gene co-expression network analysis analysis. We found that the orangered4 module was the most correlated module with recurrence in laryngeal squamous cell carcinoma patients. Genes in the orangered4 module were related to organelle organization, response to chemical, regulation of catalytic activity and regulation of cell differentiation. Two genes were discovered as hub genes which were related to poor prognosis, namely annexin A2 and S100 calcium binding protein A10. Here, we found several hub genes that played important roles in recurrent laryngeal squamous cell carcinoma, which may improve our understanding of the mechanisms underlying recurrence laryngeal squamous cell carcinoma.
Keywords
Laryngeal squamous cell carcinoma, gene ontology, bioinformatics, laryngectomy, tumor
Laryngeal Squamous Cell Carcinoma (LSCC), nearly 20 % of all head and neck malignant tumors, is a common malignant tumor of upper respiratory tract[1]. The 5 y survival rate of LSCC (about 60 %) has changed little in the past decade[2]. Recurrence is the crucial factor of failure of anti-tumor therapy in LSCC patients[3]. A better understanding of the mechanism of recurrence of LSCC will help to inhibit tumor progression and improve the survival rate and effect of treatment. It is very important to identify hub biomarkers and discover potential mechanism for recurrence LSCC.
With the tremendous development of Ribonucleic Acid (RNA) microarray, the study of co-expression genes related to clinical trait has improved our understanding of the mechanism of recurrence LSCC[4]. Weighted Gene Co-Expression Network Analysis (WGCNA) is an influential tool which was used to analyze gene expression datasets and to discovery gene modules which are highly correlated to clinical trait. Here, we constructed a WGCNA network and identified hub genes associated with recurrence LSCC.
In this study, the modules which were most related to the progression of clinical staging and recurrence was obtained. The Gene Ontology (GO) analysis and functional annotation showed that genes in orangered4 module which were mostly correlated to recurrence LSCC were enriched in organelle organization, response to chemical, regulation of catalytic activity and regulation of cell differentiation in LSCC patients. Finally, we discovered two hub genes (Annexin A2 (ANXA2) and S100 Calcium Binding Protein A10 (S100A10)) that could surely predict recurrence of LSCC. Also, we combined Gene Expression Profiling Interactive Analysis (GEPIA) databases to verify if these two hub genes be able to predict the progression and prognosis of LSCC.
Materials and Methods
Data processing:
GSE27020, obtained from Gene Expression Omnibus (GEO) database[5] was introduced to build up coexpression networks and mine hub genes which were correlated to recurrence LSCC. The dataset GSE27020 provided gene expression profile from 34 recurrence LSCC patients and 75 non-recurrence LSCC patients. After log2 conversion and quantile normalization, the GSE27020 data set is normalized using Robust Multiarray Average (RMA)[6].
Construction of WGCNA co-expression network:
The co-expression network[7] was constructed with GSE27020 dataset using the WGCNA package in R-project. The soft-thresholding power of the coexpression network we set was 6 and 0.9 was set as the correlation coefficient threshold. The minimum number of genes in the module was set to 30. In order to merge possible similar modules, we define 0.2 as the threshold of cutting height.
Identification of significant modules correlated to radioresistence:
Eigengene and gene significance[8] were introduced to discover gene modules correlated to recurrence of LSCC. The association between module eigengenes and clinical trait was used to identify the significant clinical module. The gene significance was a mediated p-value of each gene in the linear regression between expression and clinical traits. And the module significance was the average the gene significance of all genes associated with the module. The average absolute gene significance was defined as module significance.
Functional enrichment analysis:
In order to achieve deeper understanding of the biological function of the genes in the interested module related to recurrence LSCC, we introduced the GO analysis[9] using the powerful online bioinformatics tool Database for Annotation, Visualization, and Integrated Discovery (DAVID) database[10] (https://david.ncifcrf. gov/home.jsp/) and p<0.05 was set as the cut-off.
Hub genes identification:
Gene module which had the highest connectivity to recurrence LSCC was identified by the WGCNA algorithm. The Protein-Protein Interaction (PPI) network[11] was established with the genes in such gene module using Cytoscape v3.7.0[12]. The network is analyzed by Molecular Complex Detection (MCODE), and the hub genes were discovered using the cytoHubba in Cytoscape with Matthews Correlation Coefficient (MCC) algorithms[13].
Overall survival of these hub genes:
GEPIA is a web-based powerful tool for cancer bioinformatics study which was based on The Cancer Genome Atlas (TCGA) and Genotype-Tissue Expression (GTEx) data[14]. GEPIA offers several key functions including patient survival analysis. Since there is no record of LSCC in TCGA database, we analyzed the Head and Neck Squamous Cell Carcinoma (HNSC) data set in TCGA database as a substitute. In this study, we performed survival analysis with GEPIA to investigate the relationship between hub genes expression level and HNSC patient’s prognosis.
Results and Discussion
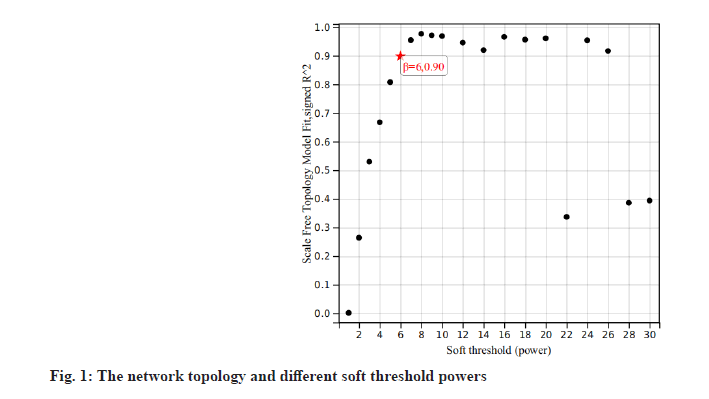
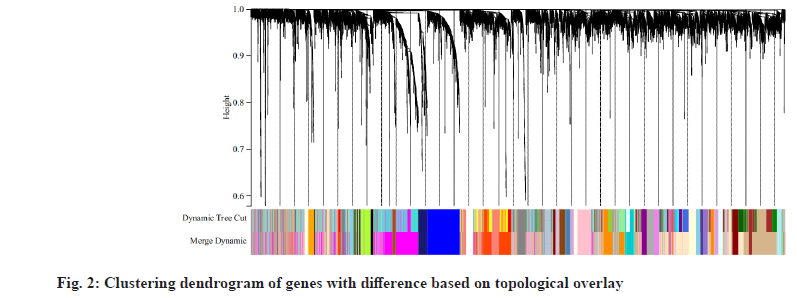
We downloaded the dataset GSE27020 of LSCC patients together and its clinical trait data from the GEO database. There were 34 recurrence LSCC patients and 75 non-recurrence patients in GSE27020. The raw microarray data of GSE27020 were normalized using the limma package in R. When the soft thresholding power beta (β) was set at 6, the scale independence reached 0.90 (fig. 1). We used the one-step network to construct the functional to identification module using WGCNA in R and 45 gene co-expression modules were finally obtained as shown in fig. 2.
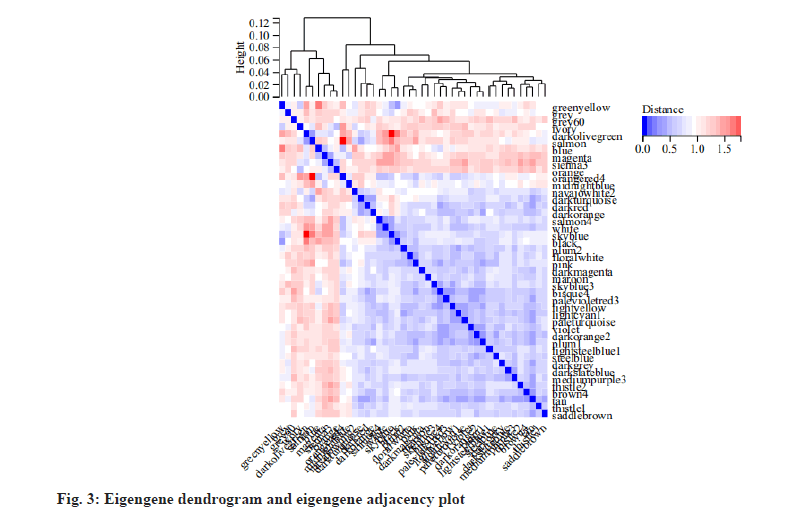
To obtain the information about the relationship between the gene co-expression modules, we analyzed the correlation of eigengenes. We found that the eigengenes were clustered into several modules. As the results, 45 modules could be divided into two clusters (fig. 3) and a few modules had a high degree of interaction connectivity.
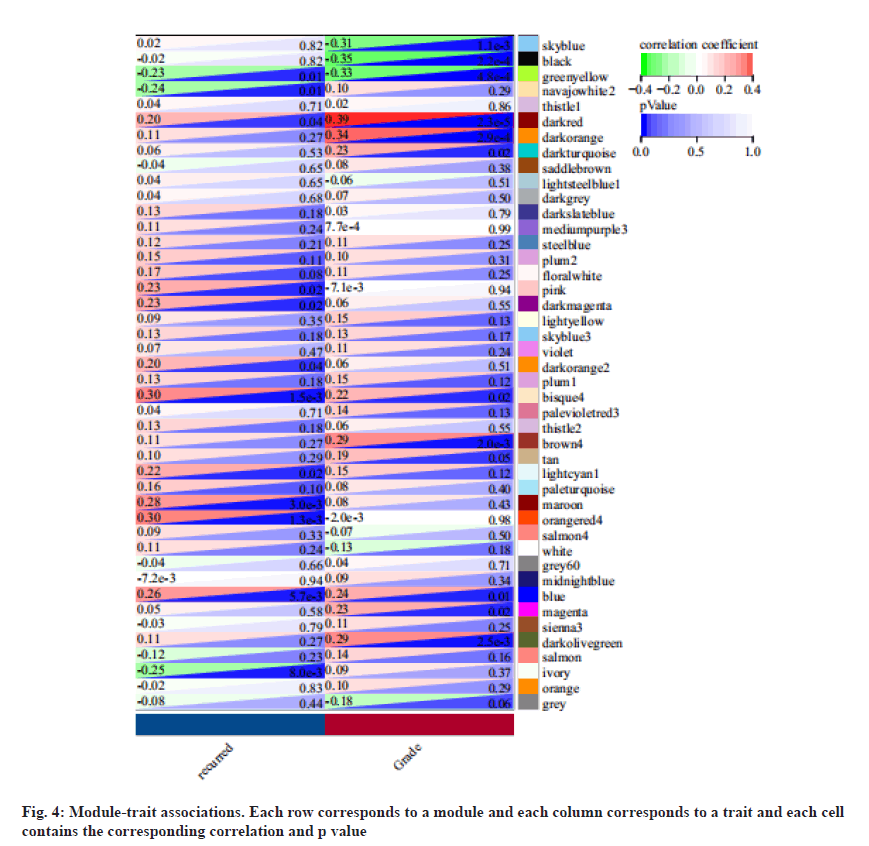
We associate the gene module with the recurrence and identify the most significant correlation. As the result, module orangered4 was mainly related to recurrence LSCC as shown in fig. 4.
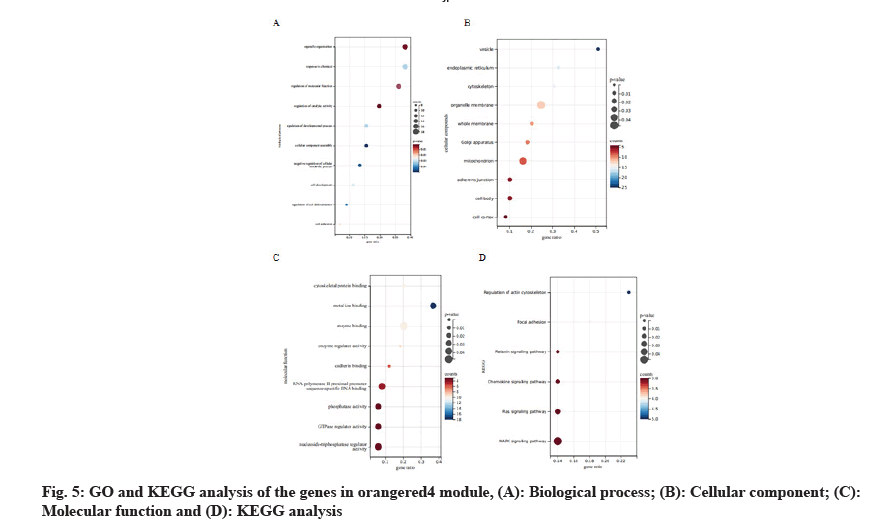
In this study, we introduced GO analysis for the genes in the module orangered4 to identify potential molecular mechanism. The results showed that genes in orangered4 module were primarily enriched in organelle organization, response to chemical, regulation of catalytic activity and regulation of cell differentiation in biological process. As for the cellular component, these genes were mainly enriched in vesicle, endoplasmic reticulum, cytoskeleton, organelle membrane and mitochondrion. Regarding molecular function, these genes were enriched in cytoskeletal protein binding, metal ion binding, enzyme regulator activity and phosphatase activity. As for the Kyoto Encyclopedia of Genes and Genomes (KEGG) signal pathway analysis, these genes were enriched in Relaxin signaling pathway, Chemokine signaling pathway, Ras signaling pathway and Mitogen Activated Protein Kinase (MAPK) signaling pathway (fig. 5).
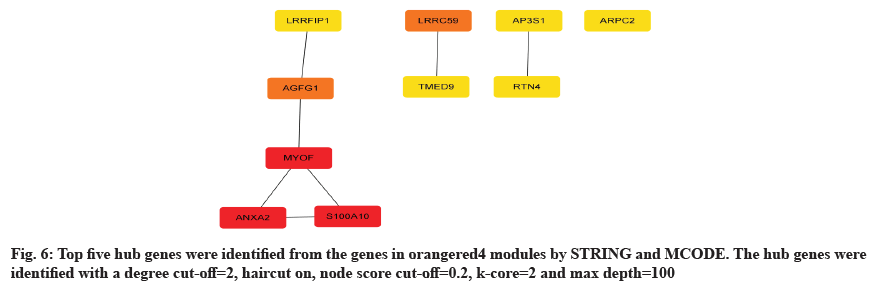
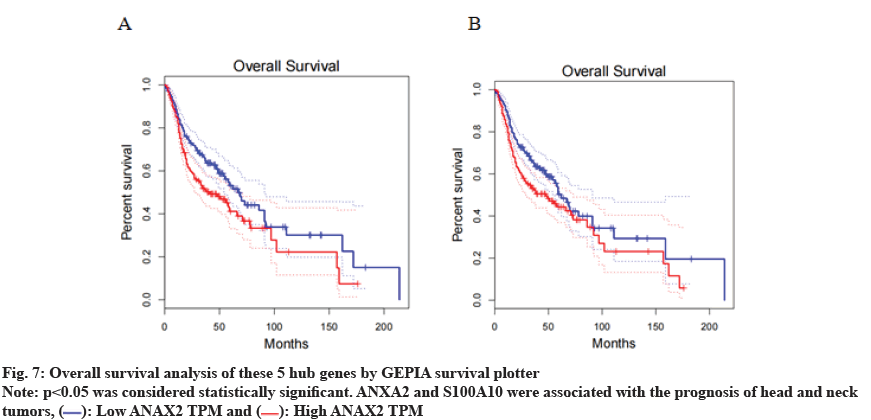
To achieve a deeper understanding of the associations between genes in the orangered4 gene modules, a PPI network was constructed with these genes. As the results shown, top 5 hub genes were discovered in the orangered4 module, including Leucine-Rich Repeat Binding FLII Interacting Protein 1 (LRRFIP1), ArfGAP with FG Repeats 1 (AGFG1), Myoferlin (MYOF), ANXA2 and S100A10 as shown in fig. 6. We introduced an overall survival analysis for these hub genes in HNSCA using GEPIA. The results showed that ANXA2 and S100A10 could cause poor prognosis in HNSC patients as shown in fig. 7.
In 2016, more than 13 000 new cases of LSCC were diagnosed, and nearly 3600 patients will die from LSCC[15]. About 60 % of patients were advanced LSCC at initial diagnosis[16-18]. LSCC is one of the oncologic diseases with low 5 y survival rate[15]. The pathogenesis of LSCC is related to many risk factors. The most important ones are tobacco and alcohol consumption[19-22]. Besides, exposure to other environmental factors, including asbestos, polycyclic aromatic hydrocarbons and textile dust, is firmly believed to increase the risk of LSCC[23,24].
Before the early 1990s, total laryngectomy was the standard treatment for advanced LSCC[25]; however, due to surgery related complications and the existence of anastomoses, the treatment has been changed to chemotherapy combined with radiotherapy[26]. Although the treatment of LSCC had improved, the overall 5 y survival rate has not increased[27], so it is necessary to find new and improved diagnosis, prognosis evaluation and treatment methods. Disclosuring the mechanism of recurrence in LSCC will help to inhibit tumor progression and improve quality of life of LSCC patients. Therefore, exploring susceptibility modules and genes for recurrence LSCC patients is important.
Here, we established the co-expression network by WGCNA using the published data of recurrence LSCC patients. All the genes in this dataset were included in the network. Genes with similar expression patterns were clustered into 45 modules. We identified the mostly correlated modules with recurrence LSCC patients. The genes in this key module were mainly enriched in organelle organization, response to chemical, regulation of catalytic activity and regulation of cell differentiation. As for the KEGG signal pathway analysis[28], the genes in key module were mainly enriched in relaxin signaling pathway, chemokine signaling pathway, Ras signaling pathway and MAPK signaling pathway. We identified two huh genes which were related to recurrence LSCC and prognosis, namely ANXA2 and S100A10.
WGCNA is widely used to analyze large-scale gene expression data sets and find gene modules highly related to clinical characteristics[29]. Through in-depth analysis of the GSE27020 data set, we determined that the orangered4 module was significantly associated with the recurrence of LSCC patients. GO analyses shows that organelle organization, response to chemical, regulation of catalytic activity and regulation of cell differentiation was activated during recurrence in LSCC patients. Moreover, we identified two hub genes which were highest correlated to recurrence LSCC and prognosis, including ANXA2 and S100A10. Studies have confirmed that high expression of ANXA2 promotes tumor progression by promoting the migration, invasion and metastasis in several types of tumors, including breast cancer[30], esophageal squamous cell carcinoma[31], glioblastoma[32] and human cervical cancer[33]. Besides, S100A10, mainly binding to annexin A2, mediates the conversion of plasminogen to plasmin[34]. Studies had shown that higher S100A10 expression linked to worse outcome and chemo resistance in a number of cancer types in lung, breast, ovary, pancreas, gall bladder and colorectal and leukemia[35]. And it plays a key role in cancer progression, prognostic and was a potential cancer therapy target. These researches are consistent with the results of this study. Totally, we discovered a gene module and two hub genes that acted as essential roles in recurrence LSCC, which may be novel therapeutic targets of LSCC.
Conflict of interests:
The authors declared no conflict of interest.
References
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68(6):394-424.
[Crossref] [Google Scholar] [Pub Med]
- da Silva GH, Miranda MG, Rodrigues DS, de Souza GR, Ribeiro CV. Epidemiological factors in patients with larynx cancer treated by surgery, radiotherapy or therapeutic associations. Arch Otolaryngol Rhinol 2019;5(1):43-9.
- Brandstorp-Boesen J, Sørum Falk R, Folkvard Evensen J, Boysen M, Brøndbo K. Risk of recurrence in laryngeal cancer. PLoS One 2016;11(10):e0164068.
[Crossref] [Google Scholar] [Pub Med]
- Li H, Sun Y, Zhan M. Exploring pathways from gene co-expression to network dynamics. Methods Mol Biol 2009;541:249-67.
[Crossref] [Google Scholar] [Pub Med]
- Barrett T, Edgar R. Gene expression omnibus: Microarray data storage, submission, retrieval and analysis. Methods Enzymol 2006;411:352-69.
[Crossref] [Google Scholar] [Pub Med]
- López-Romero P, González MA, Callejas S, Dopazo A, Irizarry RA. Processing of agilent microRNA array data. BMC Res Notes 2010;3(1):1-6.
- Langfelder P, Horvath S. WGCNA: An R package for weighted correlation network analysis. BMC Bioinform 2008;9(1):599.
[Crossref] [Google Scholar] [Pub Med]
- Langfelder P, Horvath S. Eigengene networks for studying the relationships between co-expression modules. BMC Syst Biol 2007;1(1):54.
[Crossref] [Google Scholar] [Pub Med]
- Gene Ontology Consortium. The gene ontology resource: 20 years and still going strong. Nucleic Acids Res 2019;47(D1):D330-8.
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for annotation, visualization and integrated discovery. Genome Biol 2003;4(5):P3.
[Crossref] [Google Scholar] [Pub Med]
- Hao T, Peng W, Wang Q, Wang B, Sun J. Reconstruction and application of protein–protein interaction network. Int J Mol Sci 2016;17(6):907.
[Crossref] [Google Scholar] [Pub Med]
- Thomas S, Bonchev D. A survey of current software for network analysis in molecular biology. Hum Genomics 2010;4(5):853-60.
[Crossref] [Google Scholar] [Pub Med]
- Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8(4):1-7.
[Crossref] [Google Scholar] [Pub Med]
- Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z. GEPIA: A web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 2017;45(W1):W98-102.
[Crossref] [Google Scholar] [Pub Med]
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA Cancer J Clin 2019;69(1):7-34.
[Crossref] [Google Scholar] [Pub Med]
- Goodwin WJ, Thomas GR, Parker DF, Joseph D, Levis S, Franzmann E, et al. Unequal burden of head and neck cancer in the United States. Head Neck 2008;30(3):358-71.
[Crossref] [Google Scholar] [Pub Med]
- Shin JY, Truong MT. Racial disparities in laryngeal cancer treatment and outcome: A population-based analysis of 24 069 patients. Laryngoscope 2015;125(7):1667-74.
[Crossref] [Google Scholar] [Pub Med]
- DeSantis C, Naishadham D, Jemal A. Cancer statistics for African Americans, 2013. CA Cancer J Clin 2013;63(3):151-66.
[Crossref] [Google Scholar] [Pub Med]
- Rothman KJ, Cann CI, Flanders D, Fried MP. Epidemiology of laryngeal cancer. Epidemiol Rev 1980;2(1):195-209.
- Kuper H, Boffetta P, Adami HO. Tobacco use and cancer causation: Association by tumour type. J Int Med 2002;252(3):206-24.
[Crossref] [Google Scholar] [Pub Med]
- Boffetta P, Hashibe M. Alcohol and cancer. Lancet Oncol 2006;7(2):149-56.
[Crossref] [Google Scholar] [Pub Med]
- Bosetti C, Gallus S, Franceschi S, Levi F, Bertuzzi M, Negri E, et al. Cancer of the larynx in non-smoking alcohol drinkers and in non-drinking tobacco smokers. Br J Cancer 2002;87(5):516-8.
[Crossref] [Google Scholar] [Pub Med]
- Stell PM, McGill T. Asbestos and laryngeal carcinoma. Lancet 1973;302(7826):416-7.
[Crossref] [Google Scholar] [Pub Med]
- Paget-Bailly S, Cyr D, Luce D. Occupational exposures and cancer of the larynx-systematic review and meta-analysis. J Occup Environ Med 2012;54(1):71-84.
[Crossref] [Google Scholar] [Pub Med]
- Department of veterans affairs laryngeal cancer study group. Induction chemotherapy plus radiation compared with surgery plus radiation in patients with advanced laryngeal cancer. N Engl J Med 1991;324(24):1685-90.
[Crossref] [Google Scholar] [Pub Med]
- Forastiere AA, Goepfert H, Maor M, Pajak TF, Weber R, Morrison W, et al. Concurrent chemotherapy and radiotherapy for organ preservation in advanced laryngeal cancer. N Engl J Med 2003;349(22):2091-8.
[Crossref] [Google Scholar] [Pub Med]
- Hoffman HT, Porter K, Karnell LH, Cooper JS, Weber RS, Langer CJ, et al. Laryngeal cancer in the United States: changes in demographics, patterns of care and survival. Laryngoscope 2006;116:1-3.
[Crossref] [Google Scholar] [Pub Med]
- Kanehisa M, Furumichi M, Tanabe M, Sato Y, Morishima K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 2017;45(D1):D353-61.
[Crossref] [Google Scholar] [Pub Med]
- Zhao W, Langfelder P, Fuller T, Dong J, Li A, Hovarth S. Weighted gene coexpression network analysis: State of the art. J Biopharm Stat 2010;20(2):281-300.
[Crossref] [Google Scholar] [Pub Med]
- Gibbs LD, Chaudhary P, Mansheim K, Hare RJ, Mantsch RA, Vishwanatha JK. ANXA2 expression in African American triple-negative breast cancer patients. Breast Cancer Res Treatment 2019;174(1):113-20.
[Crossref] [Google Scholar] [Pub Med]
- Ma S, Lu CC, Yang LY, Wang JJ, Wang BS, Cai HQ, et al. ANXA2 promotes esophageal cancer progression by activating MYC-HIF1A-VEGF axis. J Exp Clin Cancer Res 2018;37(1):183.
[Crossref] [Google Scholar] [Pub Med]
- Maule F, Bresolin S, Rampazzo E, Boso D, Della Puppa A, Esposito G, et al. Annexin 2A sustains glioblastoma cell dissemination and proliferation. Oncotarget 2016;7(34):54632-49.
[Crossref] [Google Scholar] [Pub Med]
- Buttarelli M, Babini G, Raspaglio G, Filippetti F, Battaglia A, Ciucci A, et al. A combined ANXA2-NDRG1-STAT1 gene signature predicts response to chemoradiotherapy in cervical cancer. J Exp Clin Cancer Res 2019;38(1):279.
[Crossref] [Google Scholar] [Pub Med]
- Kwon M, MacLeod TJ, Zhang Y, Waisman DM. S100A10, annexin A2, and annexin a2 heterotetramer as candidate plasminogen receptors. Front Biosci 2005;10(1):300-25.
[Crossref] [Google Scholar] [Pub Med]
- Saiki Y, Horii A. Multiple functions of S100A10, an important cancer promoter. Pathol Int 2019;69(11):629-36.
[Crossref] [Google Scholar] [Pub Med]
 : Low ANAX2 TPM and
: Low ANAX2 TPM and  : High ANAX2 TPM
: High ANAX2 TPM



