- *Corresponding Author:
- Yiming Wu
Clinical Research Center in Mental Health, Shanghai Yangpu District Mental Health Center, Shanghai University of Medicine and Health Sciences, Shanghai 200090, China
E-mail: dr_yimingwu@163.com
| This article was originally published in a special issue, “Current Trends in Pharmaceutical and Biomedical Sciences” |
| Indian J Pharm Sci 2022:84(5) Spl Issue “159-167” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Dementia is most frequently caused by Alzheimer’s disease. The objective of this study was to investigate the potential targets of Alzheimer’s disease and its associated biological processes. We obtained gene expression profiles of the GSE5281, GSE48350 and GSE11882 datasets from the gene expression Omnibus database. Among the total datasets, 263 were of Alzheimer’s disease tissues and 204 were of healthy tissues. We selected differentially expressed genes from Alzheimer’s disease tissues and healthy tissues using the gene expression Omnibus 2R tool and Venn diagram software. Next, we used database for annotation, visualization and integrated discovery database, gene ontology database and Kyoto encyclopedia of genes and genomes pathways for analysis. Then, by using Cytoscape’s search tool for the retrieval of interacting genes, it is possible to see how these differentially expressed genes and protein-protein interactions interact with one another. In total, there were 270 consistently expressed genes, containing 193 down-regulated genes and 77 up-regulated genes. Additionally, 270 consistently expressed genes were found by chip analysis as the test set and 242 acquisitions of causative differentially expressed genes were sought via gene list automatically derived for you as the training set. ToppGene was then utilized to optimize these genes. Of the protein-protein interaction network, analysed by the molecular complex detection plugin, 14 regulated genes were selected. In conclusion, using integrated bioinformatics techniques, we have attempted to discover the substantially altered expression genes associated with a poorer prognosis in Alzheimer’s disease and the investigations may benefit their predictive and prognostic value in Alzheimer’s disease.
Keywords
Alzheimer’s disease, bioinformatics analysis, microarray, differentially expressed genes
Alzheimer’s Disease (AD) is a prevalent neurodegenerative condition marked by progressive neuronal deterioration and unique neuropathological alterations. AD may lead to irreversible damage to the cerebrum with cognitive and functional impairments[1]. It was discovered that genetic variables play a significant influence in the vulnerability to AD[2,3]. Despite the fact that a number of putative biomarkers have been used, the specific pathogenic mechanism of AD has yet to be demonstrated. It is important to explore more reliable prognostic biomarkers to improve the treatment effect and to better understand the underlying mechanisms of AD.
In this study, we selected the Gene Expression Omnibus (GEO) datasets: GSE5281, GSE48350 and GSE11882. The GEO2R web tool and Venn diagram software were used to access the Differentially Expressed Genes (DEGs) in the three datasets mentioned above. From Gene List Automatically Derived for You (GLAD4U), we obtained genes with differential expression that are causative. To make these genes more effective, we used ToppGene. To examine the screened DEGs with Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis, we used the Database for Annotation, Visualization and Integrated Discovery (DAVID, http://david.abcc.ncifcrf.gov/). Then, we analyzed the DEGs using the Protein-Protein Interaction (PPI) network and also screened the PPI network modules in AD. We used Cytoscape Molecular Complex Detection (MCODE) to screen the important gene modules and co-expression networks in order to look for the relative relevance in the three aforementioned datasets. The underlying gene pathways and putative molecular processes were therefore discovered and the outcomes could help with AD therapy, diagnosis and prevention.
Materials and Methods
Microarray data information:
The National Center for Biotechnology Information (NCBI)-GEO, a well-known and open database of microarray/gene profiles, made the gene expression profiles for AD and healthy brain tissues in the GSE5281, GSE48350 and GSE11882 datasets public. Affymetrix HGU 133 plus 2.0 microarray data for GSE5281, GSE48350 and GSE11882 were all based on the GeneChip Human Genome U133 plus 2.0 (GPL570) platform (Affymetrix, Inc., Santa Clara, California (CA), United States of America (USA)). GSE48350 contains microarray data from healthy controls (aged 20-99 y) and AD cases from four different brain regions, the hippocampus, entorhinal cortex, superior frontal cortex and post-central gyrus. GSE5281 contained 151 samples from 84 AD patients and 67 healthy individuals. GSE48350 included 273 samples, including 80 control samples and 153 AD samples. GSE11882 comprises information from 26 Alzheimer’s patients (aged 74 to 95 y) and 57 neurologically and cognitively normal controls (aged 20 to 99 y).
Data processing of DEGs:
DEGs between AD tissues and healthy brain tissues were identified via GEO2R online tools[4] with adjusted p value<0.05 and |log Fold Change (FC)|>0.5. Online Venn diagram software was used to examine the common DEGs in the three datasets mentioned above by choosing the original findings. Genes with a logFC>0 were up-regulated, whereas genes with a logFC<0 were down-regulated, according to the logFC analysis of the DEGs.
Examination of the pathways and gene ontologies:
A technique to unifying biology known as GO (http://www.geneontology.org) analysis provides details on how genes work by utilizing ontologies to express biological knowledge[5]. A significant database that includes medications, genomes, illnesses, chemical substances, biological processes and more may be found in the KEGG[6]. Several genes or proteins functions may be checked online by using the bioinformatics program, DAVID[7]. GO categories and pathways were found by using DAVID (p<0.05).
Analysis of PPI’s network and modules:
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database’s data was used to build the PPI network[5]. To find the prognosis link among the aforementioned DEGs, the STRING database was then displayed using the Cytoscape software version, a well-known open-source software program from www.cytoscape.org[6], with a confidence score of 0.4. The modules of the PPI network were additionally investigated using Cytoscape’s MCODE software (node score cut off=0.2, maximum depth=100, k-core=2 and degree cut off=2).
Acquisition of causal DEGs:
The GLAD4U database is considered as an open public database of human disease genes[8]. The pathogenic gene of the disease was determined through literature in PubMed. We searched for the acquisition of causal DEGs with “Alzheimer’s Disease (AD)” as the key word in the GLAD4U database. The genes found in the GLAD4U database are all genes that have proven disease associations by relevant studies, but there are many genes related to AD that have not been studied and these genes cannot be searched in GLAD4U. The DEGs obtained by chip analysis are the genes that have changed expressions in the experimental group compared with the control group.
Optimization of DEGs:
ToppGene can optimize the candidate genes of diseases by comparing the functional similarity between two groups of genes and marking the candidate genes according to the similarity[6]. However, we cannot completely exclude sample pollution and some deviations in the data analysis process in the chip experimental process. Some of these DEGs obtained by chip analysis may not be reliable. We took the relatively reliable genes obtained in the GLAD4U database as the training set and the DEGs obtained through the chip analysis as the test set. We took advantage of the ToppGene database to enter the genes as follows, p<0.01 and score≥0.6. These genes were selected as reliable genes in the DEGs and those genes from the GLAD4U database were integrated into the pathogenic genes of AD.
Results and Discussion
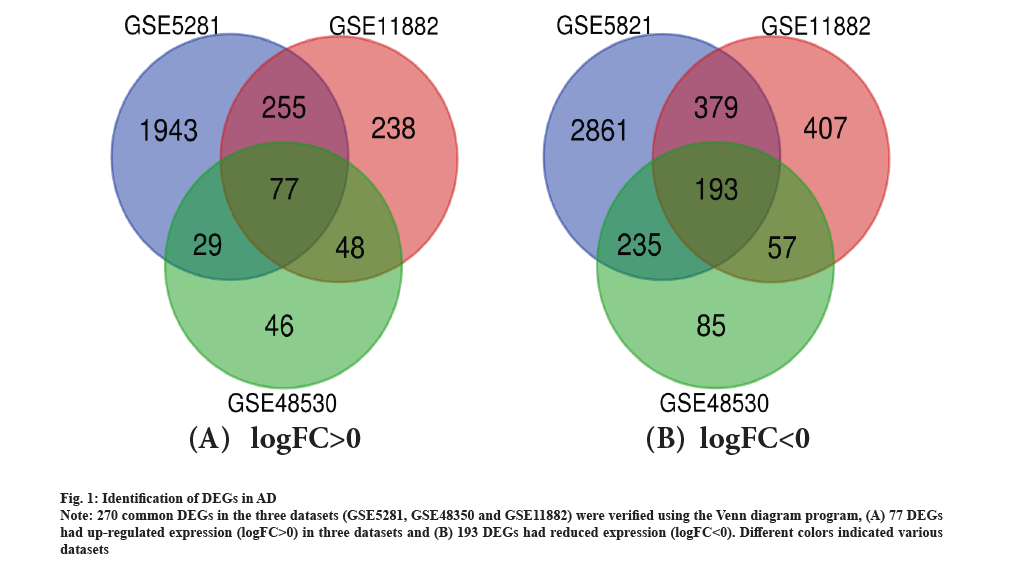
Identification of DEGs in AD was shown in fig. 1. There were 69 AD tissues and 26 healthy tissues in the current investigation. We were able to extract 2842, 261 and 866 DEGs from the three databases, GSE5281, GSE48350 and GSE11882, respectively, using GEO2R online tools. Utilizing Venn diagram software, the common DEGs between the three datasets were found. The findings indicated that 270 frequent DEGs in total were investigated, including 193 down-regulated genes and 77 up-regulated genes in the tissues of people with Alzheimer’s illness (Table 1 and fig. 1). As shown in fig. 1A and fig. 1B, 270 common DEGs in the three datasets (GSE5281, GSE48350 and GSE11882) were verified using the Venn diagram program, which is accessible at http://bioinformatics.psb.ugent.be/webtools/Venn/.
Fig. 1: Identification of DEGs in AD.
Note: 270 common DEGs in the three datasets (GSE5281, GSE48350 and GSE11882) were verified using the Venn diagram program, (A) 77 DEGs had up-regulated expression (logFC>0) in three datasets and (B) 193 DEGs had reduced expression (logFC<0). Different colors indicated various datasets.
| DEGs | Genes name |
|---|---|
| Up-regulated | CD44 CXCR4 PALLD EMX2 BDH2 GNG12 ZBTB20 AHNAK SLC39A12 MT1G PLSCR4 SOX9 SYTL4 IL13RA1 NOTCH2 MIR6883///PER1 DDIT4L RGS1 GALNT15 PTPRC VAC14-AS1 ACSS3 ZFP36L1 LOC101930416///LOC101929792///LOC100996724///PDE4DIP ITPKBGLIS3 EMP3 ERBIN VCAN MT1F LOC101929787 TGFBR3 CD84 DTNA KCNE4 RNASE4 CHST3 RNF135 HSPB8 ID3 GJA1 MAML2 HIF3A ABCA1 AQP4 CNN3 NUPR1 STON2 SLC16A9 IL17RB SRGN C4B_2///C4B///C4A ANGPT1 ANGPT2 SLC7A2 CD163 TBL1X LATS2 DDIT4 ID4 CLU AEBP1 GFAP MIR612///NEAT1 WWTR1 FAM107A TJP2 YAP1 SPATA13 BMPR1B RFX4 GEM SLC14A1 PARD3 NPL PHYHD1 GRAMD3 |
| Down-regulated | RIT2 PTPRR STYK1 PRKCB LINC00460 SLC8A1 ZNF385B FAXC RGS4 CACNB1 SEZ6 EPHA4 CDH8 KRT222 NMNAT2 LMO4 ABCC12 DLGAP1 LYPD8 EIF5A FGF12 SLC44A5 SNCA HN1 GRIN2A PCNX2 NRXN1 NR4A3 HRASLS SLIT1 UBE2QL1 PLPPR5 SYN2 CALM3///CALM2///CALM1 FAM19A2 MYT1L FAM71E1 TOMM70 CACNB2 PPP2CA KCNV1 NREP COL26A1 RIMKLA TMEM132D SLC30A3 HECW1 ARL6 ZNF385D CALB1 ARPP21 SYNGR3 STMN3 EPHA5 EMC3 OLFM1 EGR1 CNR1 DLG3 SLC32A1 MAPRE3 ARPP19 RHEBL1 SVOP SEZ6L2 PAK3 TMEM169 KCNK1 PCDHAC2 LRRC73 C1QTNF4 LINC01128 TMEM155 MFSD2B///FKBP1B STX1A LARGE1 FXYD7 CA10 LRRC7 CNTNAP2 ITFG1 CAP2 GLRB CAMKK2 HS6ST3 PPP1R14C SCN2B RASGRF2 KIF3C GAD1 C17orf51 ADD2 IGF1 PAK1 B9D1 SLC4A8 GOT1 LOC401442 ENTPD3 BBS7 TASP1 ACOT7 PGM2L1 YWHAZ GNAO1 MFSD4A PRMT8 THY1 FAM19A1 GAD2 SST KCNAB1 GRM1 GNG2 RIMS1 PHF24 SAR1A ASAH2B CNKSR2 UBE2T RPH3A ARHGAP20 SLC1A6 ACTR3B CRH GNG3 SRD5A1 TUSC3 GABRD KCNJ3 CAMK4 RBFOX2 GABRB3 GLS STS TMEM59L NRXN3 ARHGDIG NSG1 GABBR2 LY86-AS1 TOLLIP GRIA4 HTR2A OLFM3 LRTM2 LNX1 NXPH2 SLC6A17 LINC00643 WTH3DI///RAB6A KCNF1 NETO2 RAB3C FRRS1L CREG2 MAEL KIAA1549L GUCY1B3 RAB27B LMBRD2 KIAA1107 CADPS RGS7 RAB6B HS6ST2 MCHR1 C10orf35 PDE1A NWD2 PIAS2 PACSIN1 KCNAB2 NELL1 SCN2A ST8SIA3 ST6GAL2 GABRA4 7-Sep CLVS1 GNB5 ROBO2 KLC1 NCALD RNF144A-AS1 CAMKV PTPN3 SCN8A KCTD16 ATOH7 MAL2 NDRG3 |
Note: All 270 commonly DEGS were detected from three profile datasets, including 193 down-regulated genes and 77 up-regulated genes in the AD tissues and normal brain tissues.
Table 1: Identification of DEGs.
DEGs expression and activity in AD was explained here. The dendritic membrane, the synapse and the neuron projection membrane were all enriched in GO keywords in a number of up-regulated and down-regulated DEG categories. The activation of the translocation of the protein to membrane, ruffles by growth factors was the most important Biological Process (BP) of up-regulated and down-regulated DEGs. The up-regulated DEGs were considerably enriched in the BPs of cell adhesion, controlling synaptic stability in addition to the enrichment in translocation of the protein to membrane ruffles, whereas the down-regulated DEGs strongly enriched in the function of existing synapses which may be damaged.
The DEG optimization was explained here. We obtained 242 optimized DEGs, which were compared with the known genes in GLAD4U and serve a similar function to those from GLAD4U. The known genes in GLAD4U and the optimized DEGs make up the pathogenic genes of AD.
DEG enrichment analysis using GO and KEGG was described here. To study the biological categorization of DEGs by DAVID, we carried out functional and pathway enrichment studies. BP, Cellular Component (CC) and Molecular Function (MF) were the three categories used in the GO analysis. According to GO pathway enrichment analysis, the genes in the BP-associated category were mostly engaged in the mitotic cell cycle, complement activation, defense response, protein activation cascade and cell cycle processes (Table 2). Carbohydrate binding, oxidoreductase activity, mannose binding, scavenger receptor activity and monosaccharide binding were all substantially enriched in the MF of DEGs. Extracellular space, the membrane attack complex and the chromosomes alterations in the CCs of DEGs were most prevalent (Table 2).
| Expression | Category | Term | Count | Percentage (%) | p-value | False Discovery Rate (FDR) | |
|---|---|---|---|---|---|---|---|
| Up-regulated | GOTERM_BP_FAT | GO: 0040007~growth | 16 | 22.22 | 4.03E-06 | 0.007066 | |
| GOTERM_BP_FAT | GO: 0051240~positive regulation of multicellular organismal process | 18 | 25 | 4.52E-05 | 0.079287 | ||
| GOTERM_BP_FAT | GO: 0070887~cellular response to chemical stimulus | 25 | 34.72 | 7.19E-05 | 0.12607 | ||
| GOTERM_BP_FAT | GO: 0030855~epithelial cell differentiation | 11 | 15.28 | 8.98E-05 | 0.15733 | ||
| GOTERM_BP_FAT | GO: 0048585~negative regulation of response to stimulus | 17 | 23.61 | 9.70E-05 | 0.1699340.188256 | ||
| GOTERM_BP_FAT | GO: 0071310~cellular response to organic substance | 22 | 30.56 | 1.07E-04 | |||
| GOTERM_CC_FAT | GO: 0098552~side of membrane | 9 | 12.5 | 5.69E-04 | 0.713752 | ||
| GOTERM_CC_FAT | GO: 0098562~cytoplasmic side of membrane | 5 | 6.944 | 0.00686 | 8.305208 | ||
| GOTERM_CC_FAT | GO: 0030055~cell-substrate junction | 7 | 9.722 | 0.00696 | 8.414906 | ||
| GOTERM_CC_FAT | GO: 0005887~integral component of plasma membrane | 15 | 20.83 | 0.00792 | 9.534688 | ||
| GOTERM_CC_FAT | GO: 0031226~intrinsic component of plasma membrane | 15 | 20.83 | 0.01109 | 13.11106 | ||
| GOTERM_MF_FAT | GO: 0003714~transcription co-repressor activity | 7 | 9.722 | 3.65E-04 | 0.499886 | ||
| GOTERM_MF_FAT | GO: 0000989~transcription factor activity, transcription factor binding | 9 | 12.5 | 0.003724 | 4.991662 | ||
| GOTERM_MF_FAT | GO: 0000988~transcription factor activity, protein binding | 9 | 12.5 | 0.003953 | 5.290087 | ||
| GOTERM_MF_FAT | GO: 0030165~PDZ domain binding | 4 | 5.556 | 0.005645 | 7.473767 | ||
| GOTERM_MF_FAT | GO: 0004896~cytokine receptor activity | 4 | 5.556 | 0.006403 | 8.437672 | ||
| Down-regulated | GOTERM_BP_FAT | GO: 0099537~trans-synaptic signaling | 34 | 18.09 | 1.75E-16 | 4.00E-13 | |
| GOTERM_BP_FAT | GO: 0099536~synaptic signaling | 34 | 18.09 | 1.75E-16 | 4.00E-13 | ||
| GOTERM_BP_FAT | GO: 0098916~anterograde trans-synaptic signaling | 34 | 18.09 | 1.75E-16 | 4.00E-13 | ||
| GOTERM_BP_FAT | GO: 0007268~chemical synaptic transmission | 34 | 18.09 | 1.75E-16 | 4.00E-13 | ||
| GOTERM_BP_FAT | GO: 0055085~transmembrane transport | 42 | 22.34 | 6.43E-123.56E-11 | 1.16E-08 | ||
| GOTERM_BP_FAT | GO: 0006811~ion transport | 43 | 22.87 | 6.41E-08 | |||
| GOTERM_CC_FAT | GO: 0097458~neuron part | 62 | 32.98 | 3.04E-26 | 4.16E-23 | ||
| GOTERM_CC_FAT | GO: 0045202~synapse | 46 | 24.47 | 2.12E-23 | 2.89E-20 | ||
| GOTERM_CC_FAT | GO: 0044456~synapse part | 40 | 21.28 | 2.45E-21 | 3.35E-18 | ||
| GOTERM_CC_FAT | GO: 0043005~neuron projection | 42 | 22.34 | 6.90E-16 | 9.10E-13 | ||
| GOTERM_CC_FAT | GO: 0034702~ion channel complex | 21 | 11.17 | 7.61E-12 | 1.04E-08 | ||
| GOTERM_MF_FAT | GO: 0022836~gated channel activity | 19 | 10.11 | 2.27E-09 | 3.33E-06 | ||
| GOTERM_MF_FAT | GO: 0005216~ion channel activity | 20 | 10.64 | 1.96E-08 | 2.87E-05 | ||
| GOTERM_MF_FAT | GO: 0022838~substrate-specific channel activity | 20 | 10.64 | 3.51E-08 | 5.15E-05 | ||
| GOTERM_MF_FAT | GO: 0015267~channel activity | 20 | 10.64 | 1.08E-07 | 1.59E-04 | ||
| GOTERM_MF_FAT | GO: 0022803~passive transmembrane transporter activity | 20 | 10.64 | 1.12E-07 | 1.64E-04 | ||
Table 2: Go Analysis of DEGs in AD.
The complement and coagulation cascades, glycolysis/gluconeogenesis and metabolic pathways were all significantly impacted by down-regulated DEGs, while up-regulated DEGs were mostly abundant in the cell cycle, oocyte meiosis and progesterone-mediated oocyte maturation. KEGG pathway analysis revealed the opposite.
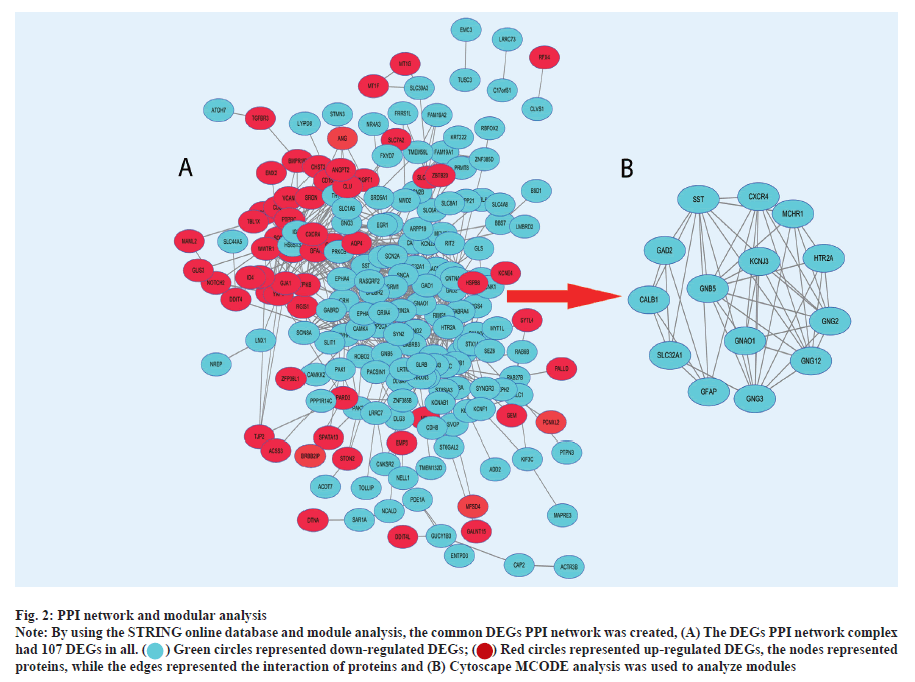
PPI network and modular analysis was shown here. In total, we imported 261 DEGs into the DEG PPI network complex, after optimization by ToppGene, including 191 down-regulated and 70 up-regulated genes (fig. 2A). The Cytoscape MCODE analysis then revealed that all 14 core nodes had up-regulated genes (fig. 2B). DAVID was used to re-examine KEGG pathway enrichment in order to analyze the potential route of these 14 chosen DEGs (p<0.05). Three important genes (Potassium Inwardly Rectifying Channel Subfamily J Member 5 (KCNJ5), Guanine Nucleotide Binding Protein subunit alpha O1 (GNAO1) and Guanine Nucleotide-Binding Protein subunit beta-5 (GNB5)) have shown a considerable enrichment in the retrograde endocannabinoid signaling pathway, according to the findings (Table 3).
Fig. 2: PPI network and modular analysis.
Note: By using the STRING online database and module analysis, the common DEGs PPI network was created, (A) The DEGs PPI network complex had 107 DEGs in all. 
 the nodes represented proteins, while the edges represented the interaction of proteins and (B) Cytoscape MCODE analysis was used to analyze modules.
the nodes represented proteins, while the edges represented the interaction of proteins and (B) Cytoscape MCODE analysis was used to analyze modules.
| Pathway ID | Name | Count | % | p-valve | Genes |
|---|---|---|---|---|---|
| hsa04723 | Retrograde endocannabinoid signaling | 15 | 3.4 | 5.63E-10 | GABRD GNAO1 GABRA4 GABRB3 GRIA4 GNG12 GRM1 KCNJ3 RIMS1 PRKCB SLC32A1 CNR1 GNB5 GNG2 GNG3 |
| hsa04727 | GABAergic synapse | 14 | 3.7 | 6.91E-10 | GABRD GNAO1 GABRA4 GABRB3 GABBR2 GNG12 PRKCB SLC32A1 GAD2 GLS GNB5 GNG2 GNG3 GAD1 |
Table 3: KEGG Pathway analysis of DEGs in AD.
The GEO database is a free public resource that contains a vast amount of gene expression data, most of them came from research, using gene expression profiling[7]. The use of gene microarray technology is widespread in many biological domains, such as diagnostic using molecules, drug research and studies on pathogenic mechanisms which can generate biological information. Microarray technology in conjunction with bioinformatics analysis tools offers a potent method for the investigation of hub genes, targets and functional pathways connected to AD.
To investigate additional beneficial prognostic biomarkers in AD, we used bioinformatics methods on these profile datasets (GSE5281, GSE48350 and GSE11882). In the current study, 263 AD specimens and 204 healthy specimens were both analyzed. The study collected 270 often altered DEGs logFC>0.5 and adjusted p value<0.05 using GEO2R and Venn diagram software, including 77 up-regulated DEGs logFC>0 and 193 down-regulated DEGs (log FC<0). Then, we optimized the DEGs through the GLAD4U and ToppGene databases. We obtained that changes were involved in BPs of DEGs i.e. regulation of protein activation cascade, defense response of related protein in GO term and that it also significantly enriched in cell cycles, particularly the mitotic one. Complement activation was reported to be associated with the changes in BPs. Significant MF enrichment was consistent with control of carbohydrate binding, monosaccharide binding, mannose binding, scavenger receptor activity and oxidoreductase activity. While GO pathway enrichment of DEGs was mostly associated with extracellular area, membrane attack complex and chromosomal alterations, changes in CCs (Table 4). KEGG pathway analysis revealed that the up-regulated DEGs were predominantly concentrated in progesterone-mediated oocyte maturation, the cell cycle and oocyte meiosis, while the complement and coagulation cascades, glycolysis/gluconeogenesis, and metabolic pathways were primarily enriched in the down-regulated DEGs. For the purpose of pathway analysis, down-regulated DEGs were significantly enriched in retrograde endocannabinoid signaling, GABAergic synapses and glutamatergic synapses, whereas up-regulated DEGs were significantly enriched in the Hippo signaling pathway and Transforming Growth Factor beta (TGF-β) signaling pathway (p<0.05). Next, the STRING online database and Cytoscape software were used to build the DEG PPI network complex, which consists of 14 core nodes (down-regulated). Furthermore, though the GLAD4U and ToppGene databases were optimized, we found that 3 out of 14 genes had significantly shown higher expression than the training samples. At the end, we re-analysed 15 genes by the using DAVID for KEGG pathway and finally found three genes (KCNJ5, GNAO1 and GNB5) (Table 5) enriched in the retrograde endocannabinoid signalling pathway, which had significance (p<0.05) and might be regarded as a new possible routes, to advance the effectiveness of treatment for AD patients.
| Category | GO terms | Count | p-value |
|---|---|---|---|
| BP | Trans-synaptic signaling | 34 | 1.57E-16 |
| Synaptic signaling | 34 | 1.57E-16 | |
| Anterograde trans-synaptic signaling | 34 | 1.57E-16 | |
| Chemical synaptic transmission | 34 | 1.57E-16 | |
| Transmembrane transport | 42 | 6.43E-12 | |
| CC | Neuron part | 62 | 3.04E-26 |
| Synapse | 46 | 2.12E-23 | |
| Synapse part | 40 | 2.45E-21 | |
| Neuron projection | 42 | 6.90E-16 | |
| Ion channel complex | 21 | 7.61E-112 | |
| MF | Gated channel activity | 19 | 2.27E-09 |
| Ion channel activity | 20 | 1.96E-08 | |
| Substrate-specific channel activity | 20 | 3.51E-08 | |
| Channel activity | 20 | 1.08E-07 | |
| Passive transmembrane transporter activity | 20 | 1.12E-07 |
Table 4:The Significant go terms (Top Five) enriched by the DEGs.
| Biomarker candidate | Name | Biological roles/Significance of the biomolecules |
|---|---|---|
| GNAO1 | Guanine Nucleotide Binding Protein subunit alpha O1, alpha-activating activity, polypeptide O | Mutations in GNAO1 are already noted to be associated with neurologic pathophysiology |
| KCNJ5 | K channel CNJ type 5 | KCNJ5 encode key subunits of GIRK |
| GNB5 | G protein subunit beta 5 | GNB5 encodes for the Guanine nucleotide-binding proteins |
Table 5: A List of common Biomarker candidates proposed for AD.
G Protein-Coupled Inwardly-Rectifying Potassium channel (GIRKs), whose key subunits are inward-rectifier potassium channels, Kir3.1 and Kir3.4, is encoded by KCNJ5[9]. The key to the valve of these neurotransmitters, such as Gamma-Aminobutyric Acid (GABA), dopamine, serotonin and adenosine, can also be constitutively active, by the Kir3/GIRK channel. Previous studies revealed that the Kir3/GIRK channel can interfere with pathological states involved in impairments in harmony, such as epilepsy and Down syndrome[10,11], both of which are highly related to AD[12]. Synaptic pathophysiology associated to AD has a hitherto unreported abnormal function of the Kir3/GIRK channel discovered in vitro[13]. Additionally, mounting data demonstrates that Kir3/GIRK channels are crucial for controlling neuronal excitability in preclinical stages of AD[14].
The heterotrimeric Guanine Nucleotide-Binding protein alpha subunit (Gαo), which is encoded by GNAO1 (polypeptide O, a guanine nucleotide-binding protein with alpha-activating activity), is essential for controlling neurotransmitter release, motility and brain development[15]. Gαo accounts for almost 1 % of the total brain membrane proteins. It is also coupled with different kinds of related G-Protein-Coupled Receptors (GPCRs)[16], containing Dopamine D2 (D2R) receptors, G-Protein Coupled Receptors for Gamma-Aminobutyric Acid (GABAB) receptor, α2 adrenergic and Adenosine A1 Receptor (A1R). In the central nervous system of mammals, Gαo is the most abundant postsynaptic membrane protein and it is observably localized having noticeably high expression in the brain[17]. According to recent studies, the phenotypic range of neurological illnesses linked to GNAO1 mutations, has been expanded by the identification of the syndrome of neurodevelopmental disorder with involuntary movements but no epileptic seizures (Neurodevelopmental Disorder with Involuntary Movements (NEDIM), Online Mendelian Inheritance in Man (OMIM): 617493)[18].
Guanine nucleotide-binding proteins (G-proteins) are heterotrimeric proteins made up of GNB5, which forms a heterodimer with the subunit and controls the subunit. GNB5 encodes these G-proteins. While GNB5-S is expressed at the tip of the dendrites of retinal ON-bipolar cells, GNB5-L is specifically expressed in photo receptor discs (rods and cones)[19]. By inactivating α-transducin bound Guanosine Triphosphate (GTP), Regulator of G-Protein Signalling 9 (RGS9)-GNB5-L and the transmembrane protein RGS9-1-Anchor Protein (R9AP) helps to switch off the visual response[20]. Additionally, Apolipoprotein E (APOE) 3/4 and APOE4/4 AD patients have down-regulated expression of the G-protein signaling molecule GNB5, according to a Serial Analysis of Gene Expression (SAGE) in the human hippocampus[21]. For potential drug therapies for neurodegenerative diseases like AD, G-protein-mediated signaling pathways, including GPCRs, have grown importance[22].
Although there are many studies on pathway and gene expression in AD, the number and methods of the studies are different. These hub genes combined with bioinformatics analysis, play a variety of functions in the onset and progression of AD. The current findings may offer a fresh thought for future investigation of AD. Nevertheless, the present study is limited by the lack of experiment in vivo and in vitro. In order to better understand AD pathophysiology and uncover new biomarkers or pharmacological targets for the development of better diagnostics and treatments, further molecular biology investigations should be carried out to confirm the expression and function of the DEGs at the protein level.
Ethical Approval:
All the experiments were approved by the Ethics Committee of the Shanghai Yangpu District Mental Health Center and followed the instructions for the Care and Use of Laboratory Animals published by the NIH (Publication No. 96-01).
Funding:
This study was supported by the Foundation of Shanghai University of Medicine and Health Sciences Foundation (20MC2020005).
Author’s contributions:
Yanting Zheng and Suli Huang contributed equally to this work.
Acknowledgements:
This study was supported by the grant from Yangpu District Mental Health Center, Shanghai, China.
Conflict of interests:
The authors declared no conflict of interest.
References
- Porteri C, Albanese E, Scerri C, Carrillo MC, Snyder HM, Martensson B, et al. The biomarker-based diagnosis of Alzheimer’s disease, 1-ethical and societal issues. Neurobiol Aging 2017;52:132-40.
[Crossref] [Google scholar] [PubMed]
- Waldrop MA, Karingada C, Storey MA, Powers B, Iammarino MA, Miller NF, et al. Gene therapy for spinal muscular atrophy: Safety and early outcomes. Pediatrics 2020;146(3):e20200729.
[Crossref] [Google scholar] [PubMed]
- Girard H, Potvin O, Nugent S, Dallaire-Théroux C, Cunnane S, Duchesne S, et al. Faster progression from MCI to probable AD for carriers of a single-nucleotide polymorphism associated with type 2 diabetes. Neurobiol Aging 2018;64:157-e11-17.
[Crossref] [Google scholar] [PubMed]
- Davis S, Meltzer PS. GEOquery: A bridge between the Gene Expression Omnibus (GEO) and BioConductor. Bioinformatics 2007;23(14):1846-7.
[Crossref] [Google scholar] [PubMed]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene ontology: Tool for the unification of biology. Nat Genet 2000;25(1):25-9.
[Crossref] [Google scholar] [PubMed]
- Chen J, Bardes EE, Aronow BJ, Jegga AG. ToppGene suite for gene list enrichment analysis and candidate gene prioritization. Nucleic Acids Res 2009;37(2):W305-11.
[Crossref] [Google scholar] [PubMed]
- Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 2002;30(1):207-10.
[Crossref] [Google scholar] [PubMed]
- Jourquin J, Duncan D, Shi Z, Zhang B. GLAD4U: Deriving and prioritizing gene lists from PubMed literature. BMC Genomics 2012;13(8):1-2.
[Crossref] [Google scholar] [PubMed]
- Yamada N, Asano Y, Fujita M, Yamazaki S, Inanobe A, Matsuura N, et al. Mutant KCNJ3 and KCNJ5 potassium channels as novel molecular targets in bradyarrhythmias and atrial fibrillation. Circulation 2019;139(18):2157-69.
[Crossref] [Google scholar] [PubMed]
- Kotajima-Murakami H, Ikeda K. Clinical study of GIRK channel inhibitors as candidate medicines for drug dependence. Nihon Yakurigaku Zasshi 2020;155(3):130-4.
[Crossref] [Google scholar] [PubMed]
- Ziegler GC, Röser C, Renner T, Hahn T, Ehlis AC, Weber H, et al. KCNJ6 variants modulate reward‐related brain processes and impact executive functions in attention‐deficit/hyperactivity disorder. Am J Med Genet B Neuropsychiatr Genet 2020;183(5):247-57.
[Crossref] [Google scholar] [PubMed]
- Nava-Mesa MO, Jiménez-Díaz L, Yajeya J, Navarro-Lopez JD. GABAergic neurotransmission and new strategies of neuromodulation to compensate synaptic dysfunction in early stages of Alzheimer’s disease. Front Cell Neurosci 2014;8:167.
[Crossref] [Google scholar] [PubMed]
- Nava-Mesa MO, Jiménez-Díaz L, Yajeya J, Navarro-Lopez JD. Amyloid-β induces synaptic dysfunction through G protein-gated inwardly rectifying potassium channels in the fimbria-CA3 hippocampal synapse. Front Cell Neurosci 2013;7:117.
[Crossref] [Google scholar] [PubMed]
- Djebari S, Iborra-Lázaro G, Temprano-Carazo S, Sánchez-Rodríguez I, Nava-Mesa MO, Múnera A, et al. G-Protein-gated inwardly rectifying potassium (Kir3/GIRK) channels govern synaptic plasticity that supports hippocampal-dependent cognitive functions in male mice. J Neurosci 2021;41(33):7086-102.
[Crossref] [Google scholar] [PubMed]
- Worley PF, Baraban JM, van Dop C, Neer EJ, Snyder SH. Go, a guanine nucleotide-binding protein: Immunohistochemical localization in rat brain resembles distribution of second messenger systems. Proc Natl Acad Sci USA 1986;83(12):4561-5.
[Crossref] [Google scholar] [PubMed]
- Evenseth LS, Gabrielsen M, Sylte I. The GABAB receptor-structure, ligand binding and drug development. Molecules 2020;25(13):3093.
[Crossref] [Google scholar] [PubMed]
- Aleman-Meza B, Loeza-Cabrera M, Peña-Ramos O, Stern M, Zhong W. High-content behavioral profiling reveals neuronal genetic network modulating Drosophila larval locomotor program. BMC Genet 2017;18(1):1-11.
[Crossref] [Google scholar] [PubMed]
- Feng H, Sjögren B, Karaj B, Shaw V, Gezer A, Neubig RR. Movement disorder in GNAO1 encephalopathy associated with gain-of-function mutations. Neurology 2017;89(8):762-70.
[Crossref] [Google scholar] [PubMed]
- Rao A, Dallman R, Henderson S, Chen CK. Gβ5 is required for normal light responses and morphology of retinal ON-bipolar cells. J Neurosci 2007;27(51):14199-204.
[Crossref] [Google scholar] [PubMed]
- Krispel CM, Chen CK, Simon MI, Burns ME. Prolonged photoresponses and defective adaptation in rods of Gβ5-/-mice. J Neurosci 2003;23(18):6965-71.
[Crossref] [Google scholar] [PubMed]
- Xu PT, Li YJ, Qin XJ, Kroner C, Green-Odlum A, Xu H, et al. A SAGE study of apolipoprotein E3/3, E3/4 and E4/4 allele-specific gene expression in hippocampus in Alzheimer disease. Mol Cell Neurosci 2007;36(3):313-31.
[Crossref] [Google scholar] [PubMed]
- Franco R, Martínez-Pinilla E, Navarro G, Zamarbide M. Potential of GPCRs to modulate MAPK and mTOR pathways in Alzheimer’s disease. Prog Neurobiol 2017;149:21-38.
[Crossref] [Google scholar] [PubMed]