- *Corresponding Author:
- Lu Liu
Department of Ultrasound, Zhuzhou Central Hospital, Zhuzhou, Hunan 412000, China
E-mail: anyjxllulu@163.com
| This article was originally published in a special issue, “Recent Progression in Pharmacological and Health Sciences” |
| Indian J Pharm Sci 2024:86(2) Spl Issue “87-93” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
In order to investigate the relationship between complement component 1q (A, B, C) and prognosis of colorectal cancer, we analyzed C1q (A, B, C) expression, clinical stage and methylation level in colorectal cancer from the Cancer Genome Atlas database and Gene Expression Omnibus database, including the prognosis of colorectal cancer. Complement 1q (A, B, C) were overexpressed (p=6.16e-07, p=4.01e-06 and p=3.28e-08, respectively) and showed low methylation levels (p=2.51e-11, p<1e-12 and p=1.64e-12, respectively) in colorectal cancer as per the cancer genome atlas database. The expression of C1q (A, B, C) were high in tumor tissues than normal tissues as per gene expression omnibus database (p=0.0044, p=8.9e-07 and p=5.2e-07, respectively). Additionally, C1q (A, B, C) and microsatellite instability have statistical significance (p=2.96e-08, p=1.08e-06 and p=4.18e-07, respectively). Finally, high expression of C1q (A, C) denotes poor survival in 5 y (p=0.026 and p=0.041, respectively) while high and low expression of C1q B have no significant difference survival in 5 y (p=0.082). However, the survival time is more than 3000 d and the prognosis of high expression of C1qB is poor. C1q (A, B, C) were overexpressed and showed low methylation levels in colorectal cancer. In addition, close correlation of C1q (A, B, C) with microsatellite instability has been explored and high expression of C1q (A, C) depicted poor survival in 5 y.
Keywords
Complement component 1q, mutations, methylation, colorectal cancer
Colorectal Cancer (CRC) is one of the common malignant tumors in the world[1]. About 10 % of the patients are diagnosed with CRC and cancerrelated deaths worldwide every year[2]. The diagnosis rate in women is higher than that of men. However, in women, incidence and mortality are approximately 25 % lower than in men[1]. With the development of medical technology, it is estimated that the incidence rate of CRC will be about 250 million worldwide by 2035. Sex and age are high risk factors of CRC[3]. However, the hereditary and environmental risk factors also play a part in the development of CRC. Approximately 10 %-20 % of patients have a family history[4]. According to studies, CRC has 12 %-35 % rate of heritability from twins and family[5].
Microsatellite Instability (MSI) is characterized by a high quantity of mutations in microsatellite locations[6]. The presence of MSI implies the disabling of Mismatch Repair (MMR) mechanisms[7]. MSI is recognized as one of the major carcinogenetic pathways and it has been confirmed that MSI mutation affects the prognosis of CRC[8]. At present, there are more and more target drugs for MSI mutations[9].
Colonoscopy has been well established as the gold standard for CRC screening with high sensitivity and specificity[1]. However, colonoscopy has some limitations like intestinal cleaning is required, manpower requirement, requiring experienced endoscopists and patient adherence[10]. Currently, with the development of molecular technology, proteins, Deoxyribonucleic Acid (DNA) (for the detection of mutations and methylation markers), Ribonucleic Acid (RNA) (mainly microRNAs) technology is becoming more and more mature. Discovery of new molecular targets is helpful to differentiate the precancerous lesions of colon cancer and improve prognosis.
Complement component 1q (C1q) contains three types of chains namely A, B and C which play a key role in immune complexes[11]. In addition to autoimmunity, C1q can also defense against infection. In prostate cancer, C1qA and C1qB shows high expression and prolongs the survival time[12]. C1q can induce apoptosis and has antiangiogenic effect in breast cancer research[13]. Therefore, we assume that the expression of C1q is related to the prognosis and may become a target molecule of CRC.
We analyzed the expression of C1q (A, B and C) and prognosis in CRC from The Cancer Genome Atlas (TCGA) database and Gene Expression Omnibus (GEO) database. Further, we have also clarified the correlation between C1q and MSI, to provide an evidence for finding new molecular targets for CRC.
Materials and Methods
Data collection and workflow illustration:
We obtained the expression levels, methylation levels and prognosis of C1q (A, B, C) for CRC from TCGA database (https://portal.gdc.cancer. gov). We have verified the expression of C1q (A, B, C) from GEO database (https://www.ncbi.nlm. nih.gov/geo/). MSI mutation was obtained from TCGA database (fig. 1).
University of Alabama at Birmingham Cancer data Analysis (UALCAN) online network:
We utilized UALCAN online network (http:// ualcan.path.uab.edu/) and studied the expression, methylation levels and prognosis of C1q (A, B, C) for CRC from TCGA database.
Gene expression profile and MSI mutation:
We obtained the expression profiling of C1q (A, B, C) by clusterProfiler package in R version 4.0.2 (http://www.r-project.org/) for series GSE37182 from GEO database. Further, we have obtained MSI mutation through cBioPortalData package in R version 4.0.2 from TCGA database. We used Spearman’s correlation analysis (ρ) to describe the correlation between quantitative variables without a normal distribution.
Statistical analysis:
The Student’s t-test (R function t-test) was used to determine whether there were significant differences between the two groups and p<0.05 was considered to be statistically significant. Statistical Package for Social Sciences (SPSS) 21.0 was used to draw the Receiver Operating Characteristic (ROC) curve and survival curve.
Results and Discussion
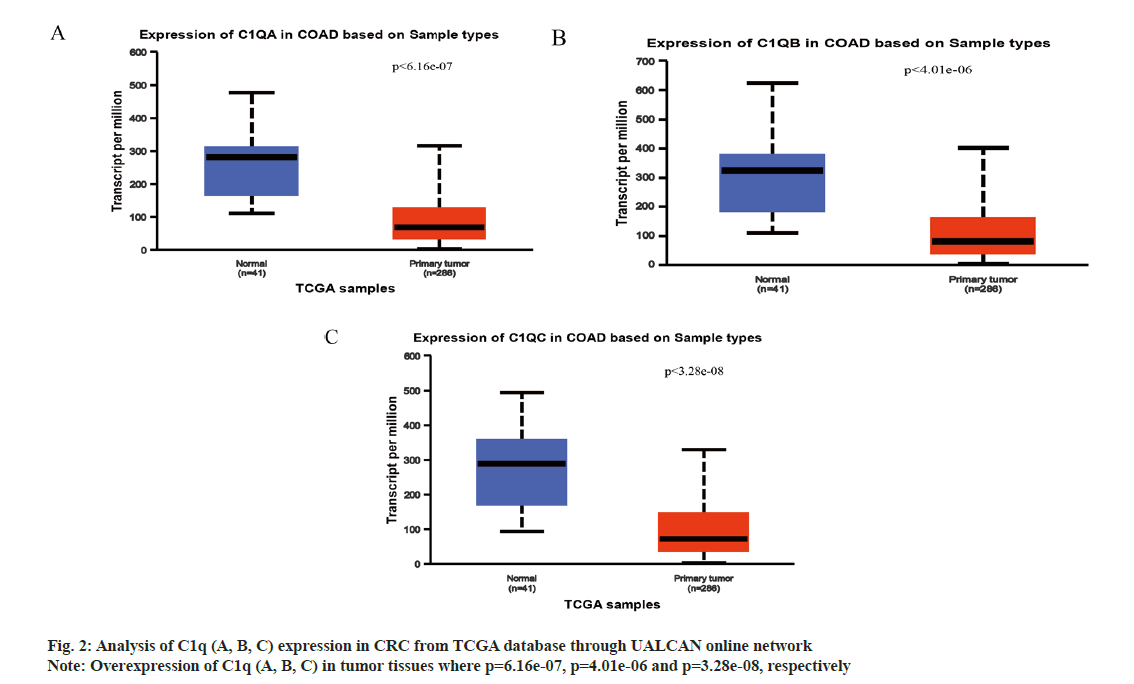
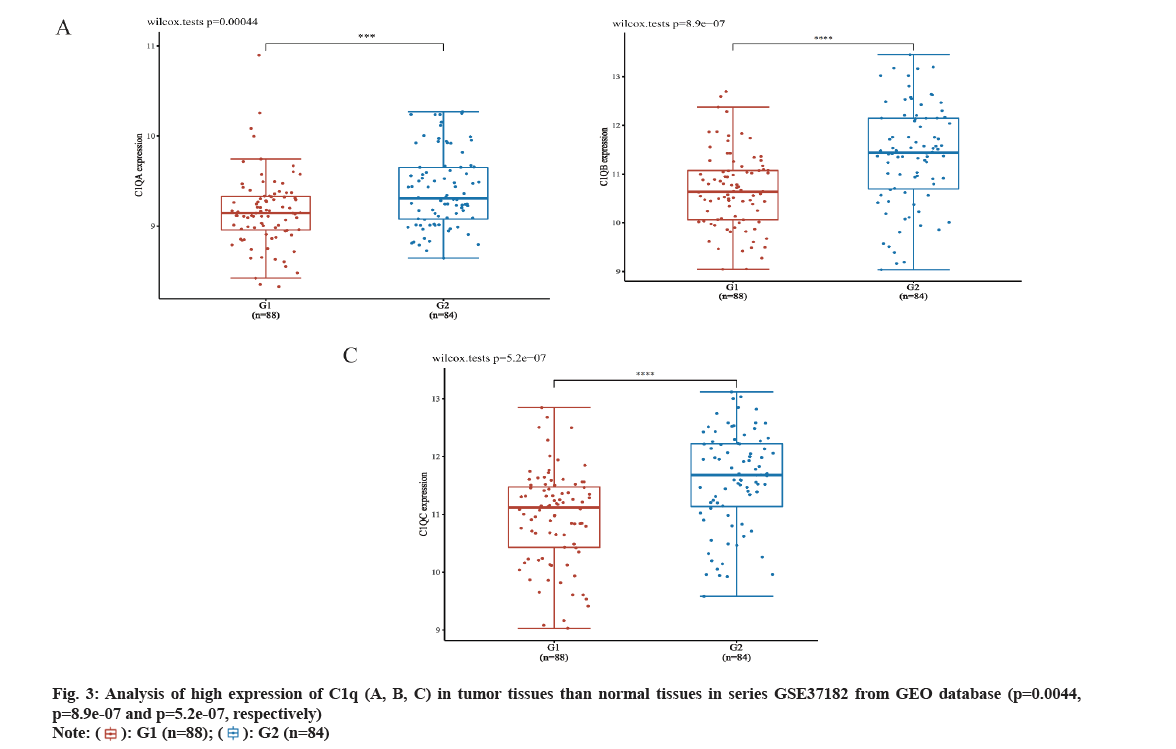
We analyzed C1q (A, B, C) expression in CRC from TCGA database through UALCAN online network. The C1q (A, B, C) were overexpressed in tumor tissues (p=6.16e-07, p=4.01e-06 and p=3.28e-08, respectively) (fig. 2A-fig. 2C). Subsequently, we verified the expression of C1q (A, B, C) in series GSE37182 from GEO database. We found that the expression of C1q (A, B, C) were high in tumor tissues than normal tissues (p=0.0044, p=8.9e-07 and p=5.2e-07, respectively) (fig. 3A-fig. 3C). At the same time, we analyzed the methylation of C1q (A, B, C) in CRC sample from TCGA database. We found that the expression of methylation levels of C1q (A, B, C) were low in tumor tissues than normal tissues in CRC (p<2.51e-11, p<1e-12 and p<1.64e-12, respectively) (fig. 4A-fig. 4C).
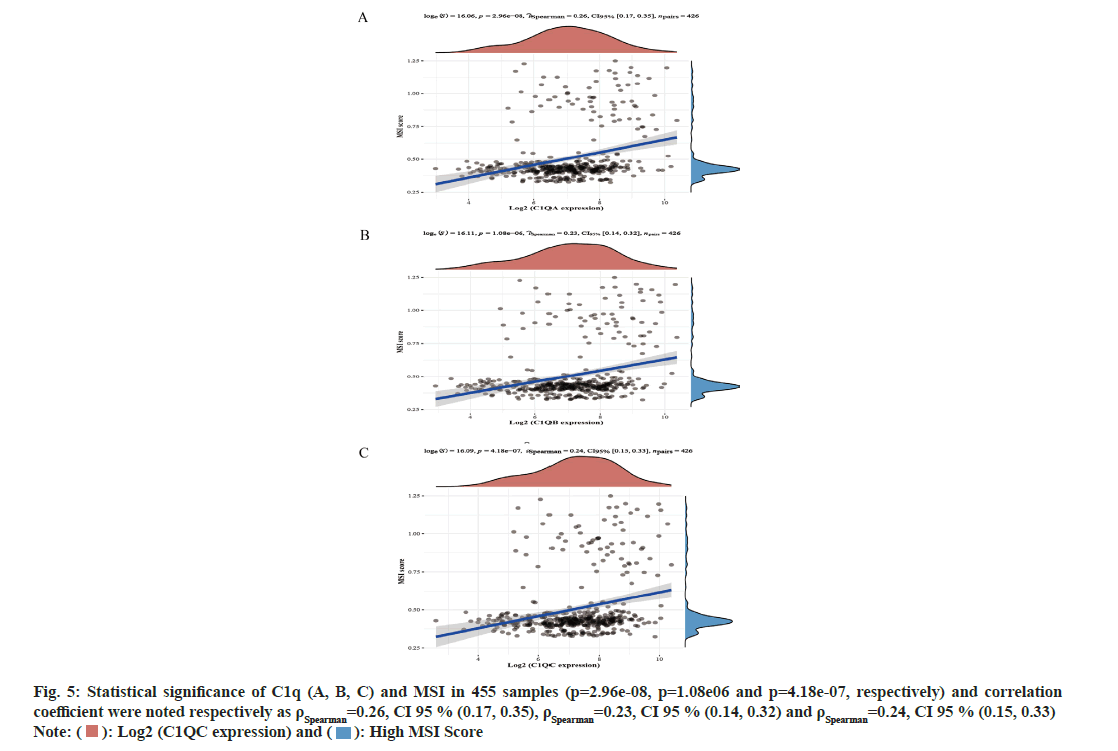
The expression of C1q (A, B, C) significantly decreased in normal tissues (p<0.05). In addition, the expression of C1q (A, B, C) were significantly different in Stage II, Stage III and Stage IV. For the correlation of C1qA, Stage II vs. Stage IV were compared, (p=2.2e-03) and Stage III vs. Stage IV were compared, where p=2.25e-02. Similarly for the correlation of C1qB, Stage II vs. Stage IV were compared (p=1.28e-03) and then Stage III vs. Stage IV was also compared (p=2.53e-02). While for the correlation of C1qC, Stage II vs. Stage IV was compared, (p=1.04e-03) and Stage III vs. Stage IV was also compared (p=1.19e-02). We analyzed the correlation between C1q (A, B, C) and MSI in CRC (fig. 5A-fig. 5C). In 455 samples, C1q (A, B, C) and MSI has showed statistical significance (p=2.96e-08, p=1.08e-06 and p=4.18e-07, respectively) and the correlation coefficient were noted respectively as ρSpearman=0.26, Confidence Interval (CI) 95 % (0.17, 0.35), ρSpearman=0.23, CI 95 % (0.14, 0.32), ρSpearman=0.24, CI 95 % (0.15, 0.33).
Fig. 5: Statistical significance of C1q (A, B, C) and MSI in 455 samples (p=2.96e-08, p=1.08e06 and p=4.18e-07, respectively) and correlation coefficient were noted respectively as ρSpearman=0.26, CI 95 % (0.17, 0.35), ρSpearman=0.23, CI 95 % (0.14, 0.32) and ρSpearman=0.24, CI 95 % (0.15, 0.33)
Note: ( ): Log2 (C1QC expression) and (
): Log2 (C1QC expression) and ( ): High MSI Score
): High MSI Score
We analyzed C1q (A, B, C) expression and impact of C1q (A, B, C) on the prognosis of CRC from TCGA database. Among 279 samples, 71 samples showed low expression while 208 samples showed high expression of C1q (A, B, C). High expression of C1q (A and C) depicted poor survival in 5 y of time (p=0.026 and p=0.041, respectively) while high and low expression of C1q B have no significant difference of survival in 5 y of time (p=0.082) (fig. 6A-fig. 6C). However, the survival time is more than 3000 d and the prognosis of high expression of C1qB is poor.
In our study, while analyzing C1q (A, B, C) expression, methylation and prognosis of CRC cancer through TCGA database and GEO database, we found the overexpression of C1q (A, B, C) in tumor tissues. At this time, the methylation levels of C1q (A, B, C) were low in tumor tissues. The prognosis of C1q with overexpression and low methylation significantly had poor prognosis than that of C1q (A, B, C) with low expression and high methylation. Subsequently, we analyzed the correlation between C1q (A, B, C) and MSI, where we found that C1q (A, B, C) and MSI are closely related to each other. Therefore, we consider C1q (A, B, C) as a novel target molecule in CRC.
C1q can synthesize matrix proteins that can promote tumor growth and metastasis in tumor microenvironment[14]. It is well known that C1q is associated with infection and autoimmunity[13]. At present, some studies indicate that C1q as a cancerpromoting factor. It can show impact on tumor biology including vascularization and metastatic spread[15].
Further, in various cancers including prostate cancer, ovarian cancer and melanoma, the expression of C1q has been proven to relate with the correlation and cancer prognosis[16]. C1q (A, B, C) overexpression plays a positive role that can prolong the Overall Survival (OS) in osteosarcoma[17]. At this time, Bandini et al.[18] confirmed that the overexpression of C1q (A, B, C) shows good prognosis in breast cancer which was contrary to our results. In our study, the overexpression of C1q (A, B, C) plays poor role in prognosis of CRC. However, the C1q levels are increased when compared with normal tissues and these levels are negatively associated with survival of glioblastoma multiforme[19]. In prostate cancer also, C1q plays poor role for prognosis[12].
It is well known that MSI mutation affects the prognosis of CRC. MSI detection has been widely used in clinical practice in patients with CRC, while the exploration in other tumors is still continuing[20]. MSI may also play a role in endometrial, ovarian, skin, brain and upper gastrointestinal tumors[21]. In clinical study, the results showed that the OS and Disease-Free Survival (DFS) of MSI-positive tumor patients were better and were independent of prognostic factors comparatively at different stages[22,23]. In addition, when stage I/II CRC patients were compared with MSI-High (MSI-H) patients showed worse pathological types and survival outcomes[22,23]. In patients with CRC, MSI is also a predictor of ineffective 5-fluorouracil (5-FU) based adjuvant chemotherapy[24]. In addition, other studies suggest that the level of MMR indicates the effectiveness of receiving Programmed Cell Death Protein 1 (PD-1)/PD Ligand 1 (PD-L1) immune checkpoint inhibitors[25].
In our study, we found that C1q (A, B, C) and MSI are closely related and C1q (A, B, C) can impact the prognosis of CRC. Therefore, we speculate that C1q may become a new target molecule for predicting CRC. In future, CRC tissues and cells might be useful to detect the expression of C1q and methylated tissues for validation. It is suggested that C1q can be applied in clinical practice to predict the prognosis of CRC.
Conflict of interests:
The authors declared no conflict of interest.
References
- Dekker E, Tanis PJ, Vleugels JL, Kasi PM, Wallace M. Colorectal cancer. Lancet 2019;394(10207):1467-80.
[Crossref] [Google Scholar] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68(6):394-424.
[Crossref] [Google Scholar] [PubMed]
- Arnold M, Sierra MS, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017;66(4):683-91.
[Crossref] [Google Scholar] [PubMed]
- Henrikson NB, Webber EM, Goddard KA, Scrol A, Piper M, Williams MS, et al. Family history and the natural history of colorectal cancer: Systematic review. Genet Med 2015;17(9):702-12.
[Crossref] [Google Scholar] [PubMed]
- Czene K, Lichtenstein P, Hemminki K. Environmental and heritable causes of cancer among 9.6 million individuals in the Swedish family-cancer database. Int J Cancer 2002;99(2):260-6.
[Crossref] [Google Scholar] [PubMed]
- Vilar E, Gruber SB. Microsatellite instability in colorectal cancer-the stable evidence. Nat Rev Clin Oncol 2010;7(3):153-62.
[Crossref] [Google Scholar] [PubMed]
- Overman MJ, Bergamo F, McDermott RS, Aglietta M, Chen F, Gelsomino F, et al. Nivolumab in patients with DNA mismatch repair-deficient/microsatellite instability-high (dMMR/MSI-H) metastatic colorectal cancer (mCRC): Long-term survival according to prior line of treatment from CheckMate-142. Lancet 2017;18(9):1182-91.
[Crossref] [Google Scholar] [PubMed]
- Gian LD, Lorena B, Cinzia A, Gioacchino L, Federica G, Francesca N. Microsatellite instability in colorectal cancer. Acta Biomed 2018;89(9):97-101.
[Crossref] [Google Scholar] [PubMed]
- Yamamoto H, Imai K. Microsatellite instability: An update. Arch Toxicol 2015;89(6):899-921.
[Crossref] [Google Scholar] [PubMed]
- Shaukat A, Rector TS, Church TR, Lederle FA, Kim AS, Rank JM, et al. Longer withdrawal time is associated with a reduced incidence of interval cancer after screening colonoscopy. Gastroenterology 2015;149(4):952-7.
[Crossref] [Google Scholar] [PubMed]
- van de Bovenkamp FS, Dijkstra DJ, van Kooten C, Gelderman KA, Trouw LA. Circulating C1q levels in health and disease, more than just a biomarker. Mol Immunol 2021;140:206-16.
[Crossref] [Google Scholar] [PubMed]
- Hong Q, Sze CI, Lin SR, Lee MH, He RY, Schultz L, et al. Complement C1q activates tumor suppressor WWOX to induce apoptosis in prostate cancer cells. PloS One 2009;4(6):e5755.
[Crossref] [Google Scholar] [PubMed]
- Azzato EM, Lee AJ, Teschendorff A, Ponder BA, Pharoah P, Caldas C, et al. Common germ-line polymorphism of C1QA and breast cancer survival. Br J Cancer 2010;102(8):1294-9.
[Crossref] [Google Scholar] [PubMed]
- Bulla R, Tripodo C, Rami D, Ling GS, Agostinis C, Guarnotta C, et al. C1q acts in the tumour microenvironment as a cancer-promoting factor independently of complement activation. Nat Commun 2016;7(1):1-11.
[Crossref] [Google Scholar] [PubMed]
- Espericueta V, Manughian-Peter AO, Bally I, Thielens NM, Fraser DA. Recombinant C1q variants modulate macrophage responses but do not activate the classical complement pathway. Mol Immunol 2020;117:65-72.
[Crossref] [Google Scholar] [PubMed]
- Revel M, Sautès-Fridman C, Fridman WH, Roumenina LT. C1q+ macrophages: Passengers or drivers of cancer progression. Trends Cancer 2022;8(7):517-26.
[Crossref] [Google Scholar] [PubMed]
- Chen LH, Liu JF, Lu Y, He XY, Zhang C, Zhou HH. Complement C1q (C1qA, C1qB, and C1qC) may be a potential prognostic factor and an index of tumor microenvironment remodeling in osteosarcoma. Front Oncol 2021;11:1-15.
[Crossref] [Google Scholar] [PubMed]
- Bandini S, Macagno M, Hysi A, Lanzardo S, Conti L, Bello A, et al. The non-inflammatory role of C1q during Her2/neu-driven mammary carcinogenesis. Oncoimmunology 2016;5(12):e1253653.
[Crossref] [Google Scholar] [PubMed]
- Bouwens TA, Trouw LA, Veerhuis R, Dirven CM, Lamfers ML, Al-Khawaja H. Complement activation in glioblastoma multiforme pathophysiology: Evidence from serum levels and presence of complement activation products in tumor tissue. J Neuroimmunol 2015;278:271-6.
[Crossref] [Google Scholar] [PubMed]
- Boland CR, Goel A. Microsatellite instability in colorectal cancer. Gastroenterology 2010;138(6):2073-87.
[Crossref] [Google Scholar] [PubMed]
- Nakamura Y, Kawazoe A, Lordick F, Janjigian YY, Shitara K. Biomarker-targeted therapies for advanced-stage gastric and gastro-oesophageal junction cancers: An emerging paradigm. Nat Rev Clin Oncol 2021;18(8):473-87.
[Crossref] [Google Scholar] [PubMed]
- Ros J, Saoudi N, Salvà F, Baraibar I, Alonso G, Tabernero J, et al. Ongoing and evolving clinical trials enhancing future colorectal cancer treatment strategies. Expert Opin Investig Drugs 2022;31(3):235-47.
[Crossref] [Google Scholar] [PubMed]
- Roset M, Amonkar M, Patel R, Lara N, Kothari S. Real-world treatment patterns and clinical outcomes for standard of care regimens in patients with deficient MMR or MSI-high metastatic colorectal and non-colorectal cancer: A retrospective chart review study in France. Adv Ther 2022;39(3):1215-29.
[Crossref] [Google Scholar] [PubMed]
- Copija A, Waniczek D, Witkos A, Walkiewicz K, Nowakowska-Zajdel E. Clinical significance and prognostic relevance of microsatellite instability in sporadic colorectal cancer patients. Int J Mol Sci 2017;18(1):1-12.
[Crossref] [Google Scholar] [PubMed]
- Zhang Y, Zhang F, Zhao L, Fu X, Shang Y, Gao Q. Long-term survival of a patient with microsatellite-stable refractory colorectal cancer with regorafenib and PD-1 inhibitor sintilimab: A case report and review of literature. BMC Gastroenterol 2021;21(1):1-6.
[Crossref] [Google Scholar] [PubMed]
 ): G1 (n=88); (
): G1 (n=88); (  ): G2 (n=84)
): G2 (n=84) 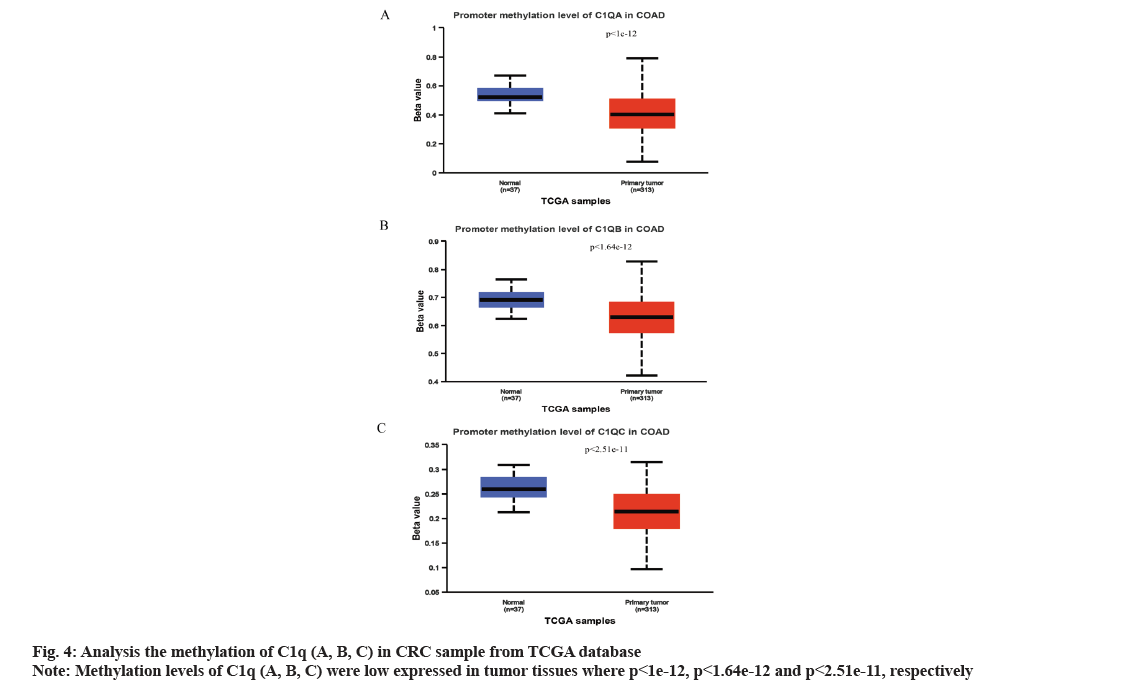
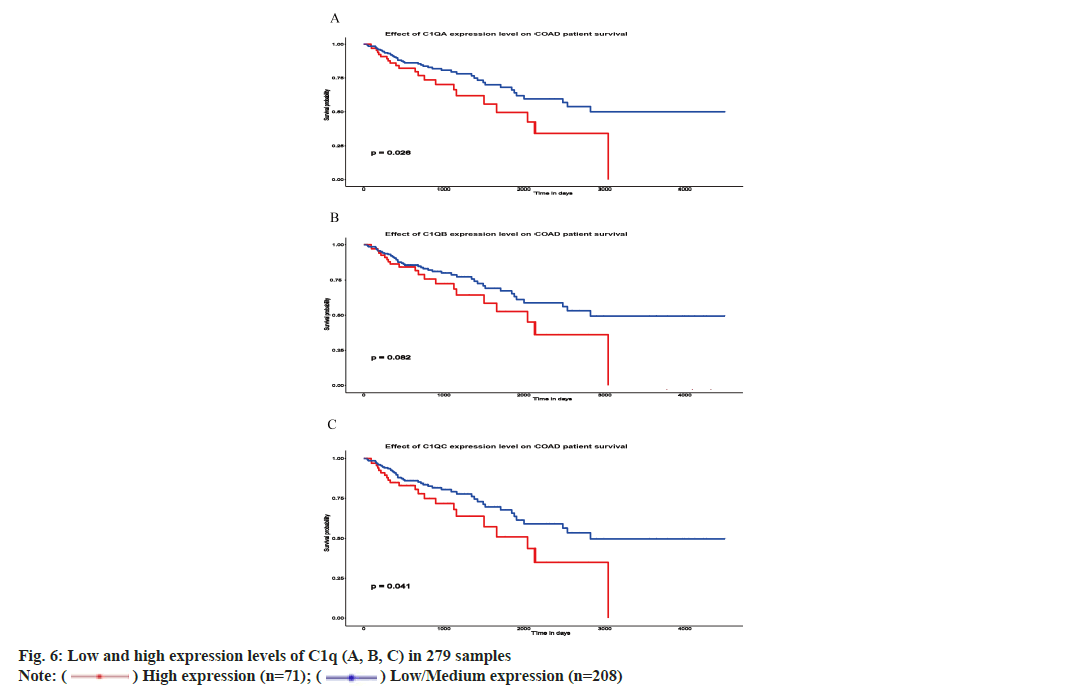
 ) High expression (n=71); (
) High expression (n=71); ( ) Low/Medium expression (n=208)
) Low/Medium expression (n=208) 



