- *Corresponding Author:
- S. J. Shangguan
Anesthesiology Department, 1Joint Surgery and Traumatic Orthopaedics, Affiliated Hospital of Shandong Medical College, Linyi, 276000, China
E-mail: shangguanshijin08@163.com
| This article was originally published in a special issue,“Therapeutic Perspectives in Biomedical Research and Pharmaceutical Sciences and their Nursing Methods” |
| Indian J Pharm Sci 2021:83(4)Spl issue “198-206” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
To investigate the effect of etomidate on the proliferation, migration and invasion of glioma cells and its possible mechanism is the main objective. Glioma cells SW1088 were cultured in vitro and interfered with different doses of etomidate. Cell proliferation was detected by cell counting kit-8 method and cell migration and invasion were detected by Transwell. Long chain non-coding RNA homeobox A11 antisense RNA over expression vector or microRNA-515-5p mimetic or co-transfecting long chain non-coding RNA homeobox A11 antisense RNA over expression vector and microRNA-515-5p mimetic was transfected to SW1088 cells. After etomidate treatment of glioma SW1088 cells, the expression of microRNA-515-5p increased, but A value of the cells, the number of migration and invasion, the protein expression levels of proliferating cell nuclear antigen, matrix metalloproteinase-2, matrix metalloproteinase-9 and long chain non-coding RNA homeobox A11 antisense RNA decreased (p<0.05), while they increased after 400 μmol/l etomidate interfered with SW1088 cells of over expressing long chain non-coding RNA homeobox A11 antisense RNA (p<0.05). Over expression of microRNA-515-5p reduced the A value of SW1088 cells, the number of migration and invasion and the expression levels of proliferating cell nuclear antigen, matrix metalloproteinase-2 and matrix metalloproteinase-9 proteins. Also, it could reverse the over expression of long chain non-coding RNA homeobox A11 antisense RNA on the proliferation, migration and invasion of SW1088 cells treated with etomidate. Etomidate may inhibit the proliferation, migration and invasion of glioma cells by regulating the long chain non-coding RNA homeobox A11 antisense RNA/microRNA-515- 5p axis.
Keywords
Etomidate, glioma, long noncoding RNA, homeobox A11 antisense RNA, microRNA-515-5p, cell proliferation
Glioma is a common malignant tumor and its treatment is mainly surgery. Surgery cannot be separated from the use of anesthetics. Recent studies have shown that anesthetics can affect the malignant phenotype of tumor cells and then affect the prognosis of patients[1]. For example, Xu et al.[2] showed that sevoflurane can inhibit proliferation, colony formation, migration and invasion of glioma cell in vitro by regulating circ_0012129/micro RNA (miR)-761/TGIF2 axis to promote apoptosis and hinder tumor growth in vivo. Etomidate is a commonly used intravenous anesthetic in clinic and its anesthetic effect is good. Studies have shown that etomidate can reduce the proliferation, invasion and migration of gastric cancer cells by up-regulating miR-211-5p and then inhibiting roundabout homolog 1 precursor (ROBO1) expression[3] and can also inhibit the proliferation, invasion and promote apoptosis of lung cancer A549 cells by inhibiting phosphatidylinositol 3‑kinase (PI3K)/protein kinase B (AKT) (PI3K/AKT) signaling pathway[4]. At present, however, the influence of etomidate on malignant phenotype of glioma cell and its molecular mechanism are unknown.
Long chain non-coding RNA (lncRNA) and MicroRNA (miRNA) are two kinds of small non-coding RNA involved in regulating malignant phenotype of tumor cells and lncRNA can target miRNA to play biological functions[5]. The homeobox A11 antisense RNA (HOXA11-AS) belongs to LncRNA. It is highly expressed in glioma tissues, promoting the migration, invasion and proliferation of glioma and hindering cell apoptosis to promote the development of glioma[6]. Online prediction software of Starbase target gene shows that LncRNA HOXA11-AS may target miR-515-5p. Studies have shown that miR-515-5p has low expression in hepatocellular carcinoma[7], bladder cancer[8] and lung squamous cell carcinoma[9]. Up-regulation of miR- 515-5P expression can inhibit the malignant phenotype of tumor cells and the development of tumor. At present, the influence of miR-515-5p on the occurrence and development of glioma is unknown. Therefore, in this study, human glioma cell lines (SW1088) cells were taken as the research object and the LncRNA HOXA11-AS/miR-515-5p axis was taken as the breakthrough point. The influence of etomidate on the proliferation, migration and invasion of SW1088 cells and its molecular mechanism were mainly explored.
Materials and Methods
Cells and reagents:
SW1088 cell line, Shanghai Cell Bank, Chinese Academy of Sciences; Etomidate, purity 99.99 %, Sigma Company, USA; Fetal bovine serum (FBS), Hangzhou Sijiqing; Roswell Park Memorial Institute Medium (RPMI) 1640 culture medium, cell counting kit-8 (CCK-8) and bicinchoninic acid (BCA) protein detection kit, Beijing Solarbio; LipofectamineTM 2000 kit, Invitrogen Company, USA; overexpression vector (pcDNA-LncRNAHOXA11-AS), empty vector (pcDNA), Wild type (WT)-LncRNA HOXA11-AS, mutant (MUT)-LncRNA HOXA11-AS, miR-515-5p mimics, microRNA negative control (miR-NC) and polymerase chain reaction (PCR) premiers, Genepharma Company, Shanghai; rabbit anti-human proliferating cell nuclear antigen (PCNA), matrix metalloproteinase-2 (MMP-2), matrix metalloproteinase-9 (MMP-9) and β-actin antibody, Santa Cruz Company, USA; RNA extraction kit, reverse transcription kit and PCR kit, Dalian Baoshengwu; dual-luciferase activity detection kit, Shanghai Beyotime Biotechnology.
Methods:
Cell culture and transfection: The resuscitated SW1088 cells were cultured in RPMI 1640 medium containing 10 % FBS. SW1088 cells in logarithmic growth phase were inoculated in the 6-well plate (2.5×105 cells/well). PcDNA-NC, pcDNA-LncRNA HOXA11-AS, miR-515-5p mimics, miR-NC, cotransfecting pcDNA-LncRNA HOXA11-AS and miR- 515-5p mimics, pcDNA-LncRNA HOXA11-AS and miR-NC were transfected by LipofectamineTM 2000 liposome method respectively. The culture medium was changed and the culture was maintained for another 24 h. After that cells were collected. Then, the transfection effect was tested and the cells were collected for later use.
Cells grouping: Untransfected SW1088 cells were divided into negative control (NC) group and etomidate group with different doses (100, 200, 400 and 800 μmol/l). Among them, cells of NC group were cultured in conventional medium and cells of etomidate group were cultured in medium containing 100, 200, 400 and 800 μmol/l etomidate respectively. Cells of transfected pcDNA-NC, pcDNA-LncRNA HOXA11-AS, cotransfecting pcDNA-LncRNA HOXA11-AS and miR- 515-5p mimics, pcDNA-LncRNA HOXA11-AS and miR-NC were all cultured in a medium containing 400 μmol/l etomidate. They were divided into pcDNANC+ 400 μmol/l etomidate group, pcDNA-LncRNA HOXA11-AS+400 μmol/l etomidate group, mir- 515-5p+pcDNA-LncRNA HOXA11-AS+400 μmol/l etomidate group and miR-NC+pcDNA LncRNA HOXA11-AS+400 μmol/l etomidate group. Cells of transfected miR-515-5p mimics and miR-NC were all cultured in normal medium and were divided into miR- 515-5p group and miR-NC group.
Detection of cell proliferation by CCK-8: The untransfected and transfected cells were inoculated in the 96-well plate (2.5×104 cells/well) and treated for 24 h in groups as shown in cells grouping. After adding 10 μl CCK-8, they were incubated for 2 h and absorbance (a) at 450 nm of microplate reader was measured.
Detection of cell migration and invasion by transwell: The untransfected and transfected cells were all inoculated in the 6-well plate (2.5×105 cells/well) and treated in groups as shown in cells grouping for 24 h. The cells were collected and the density was adjusted to 5×104 cells/well. Invasion experiment: transwell chamber was placed in the 24-well plate. Matrigel was spread on the upper chamber and 100 μl cell suspensions were added after air drying, 500 μl culture medium was added to the lower chamber. After culturing for 24 h, the culture medium was discarded. After fixing with paraformaldehyde and staining with crystal violet, the cells were observed under microscope. 5 visual fields were taken at random and counting was carried out. The migration experiment was the same as the invasion experiment except that matrigel was not applied.
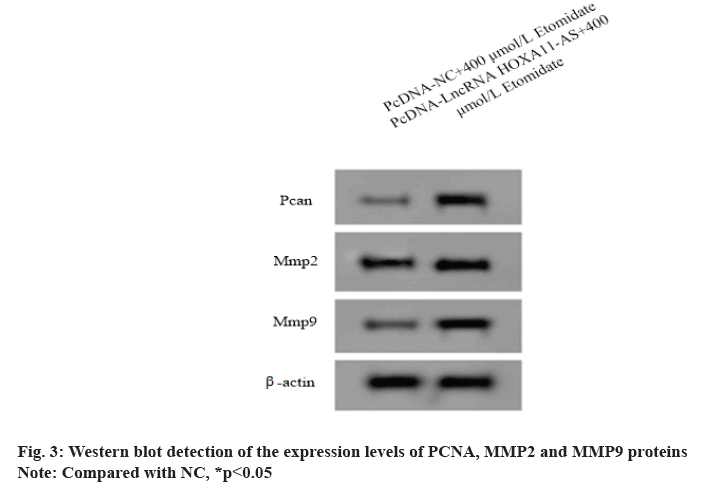
Western blot detection of the expression levels of PCNA, MMP2 and MMP9 proteins:
The untransfected and transfected cells were inoculated in the 6-well plate (2.5×105 cells/well) and treated in groups as shown in cells grouping for 24 h. Then, radioimmunoprecipitation assay (RIPA) reagent was used to extract the total protein in the cells and BCA method was used to measure the protein content. After sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), membrane transfer and 5 % skimmed milk powder were incubated at 4° overnight in PCNA, MMP2, MMP9 and β-actin (1:1000) primary antibody respectively and then incubated at 37° for 1 h in goat anti-rabbit secondary antibody (1:2000). The expression levels of PCNA, MMP2 and MMP9 relative to β-actin were analyzed by Image J software.
Quantitative reverse transcription polymerase chain reaction (RT-qPCR) experiment:
The untransfected and transfected cells were all inoculated in the 6-well plate (2.5×105 cells/well) and treated in groups as shown in cells grouping for 24 h. Then the total RNA in the cells was extracted by RNA extraction kit and then amplified by PCR after reverse transcription. The amplification procedure was 95° for 3 min, 95° for 10 s, 58° for 30 s and 72° for 30 s, with a total of 35 cycles. Sequence of primers: for LncRNA HOXA11-AS, upstream 5'-ACGCTAGGCACCACTTTGTT-3' and downstream 5'-CCGGCTACTAGTCAGTGTG-3'; for glyceraldehyde 3-phosphate dehydrogenase (GAPDH), upstream 5'-CCCACTCCTCCACCTTTGAC-3 and downstream 5'-GGATCTCGCTCCTGGAAGATG-3'; for miR-515-5p, upstream 5'-TTCTCCAAAAGAAAGCACTTTCTG-3' and downstream 5'-TGGTGTCG TGGAGTCG-3'; for U6, upstream 5'-TCCGGGTGATGCTTTTCCTAG-3' and downstream 5'-CGCTTCACGAATTTGCGTGTCAT-3'. The expression levels of LncRNA HOXA11-AS relative to GAPDH and miR-515-5p and relative to U6 were calculated by 2-△△Ct.
Dual-luciferase reporter gene experiment: SW1088 cells were inoculated in the 6-well plate (2.5×105 cells/ well). WT-LncRNA HOXA11-AS and miR-515-5p mimics or miR-NC, MUT-LncRNA HOXA11-AS and miR-515-5p mimics or miR-NC were co-transfected by LipofectamineTM 2000 liposome for 6 h method respectively. The culture medium was changed. After that, the cells were cultured for another 24 h. Then, they were collected for 6 h and centrifugation (3500 r/min, 10 min). After that, supernatant was taken to detect luciferase activity.
Statistical analysis:
SPSS 22.0 software was used to analyze the experimental data. The measurement data is expressed by mean±standard deviation (x̅±s). After normality test and variance homogeneity test, the comparison between the two groups was carried out by independent sample t test. One-way analysis of variance (ANOVA) was used for comparison among groups and lysergic acid diethylamide (LSD) t test was used for pairwise comparison among groups. p<0.05 indicated that the difference was statistically significant.
Results and Discussion
Effects of etomidate at different concentrations on the proliferation of SW1088 cells were observed. After the Glioma SW1088 cells were treated with etomidate at 100, 200, 400 and 800 μmol/l for 24 h, the cell A value and PCNA protein expression decreased (p<0.05), indicating that etomidate could inhibit the proliferation of SW1088 cells (fig. 1 and Table 1).
| Group | PCNA | A value |
|---|---|---|
| NC | 0.82±0.07 | 1.156±0.11 |
| 100 μmol/l etomidate | 0.62±0.05* | 0.943±0.08* |
| 200 μmol/l etomidate | 0.49±0.04* | 0.712±0.06* |
| 400 μmol/l etomidate | 0.35±0.03* | 0.532±0.04* |
| 800 μmol/l etomidate | 0.33±0.02* | 0.553±0.04* |
| F | 181.180 | 126.996 |
| p | 0.000 | 0.000 |
Note: Compared with the NC group, *p<0.05
Table 1: Effects of Different Concentrations of Etomidate on the Proliferation of Sw1088 Cells (X̅±S, N=9)
Effects of etomidate at different concentrations on migration and invasion of SW1088 cells were observed. After Glioma SW1088 cells were intervened with etomidate at 100, 200, 400 and 800 μmol/l for 24 h, the number of migration and invasion decreased (p<0.05), the expression levels of MMP2 and MMP9 proteins decreased (p<0.05), indicating that etomidate could inhibit migration and invasion of SW1088 cells (fig. 2 and Table 2).
| Group | MMP2 | MMP9 | Number of cell migration | Number of cell invasion |
|---|---|---|---|---|
| NC | 0.93±0.08 | 0.79±0.07 | 197±10.02 | 149±11.02 |
| 100 μmol/l etomidate | 0.81±0.07* | 0.65±0.05* | 167±11.19* | 113±9.03* |
| 200 μmol/l etomidate | 0.61±0.05* | 0.49±0.03* | 132±11.05* | 82±4.10* |
| 400 μmol/l etomidate | 0.45±0.04* | 0.40±0.03* | 93±8.11* | 69±4.35* |
| 800 μmol/l etomidate | 0.43±0.02* | 0.38±0.02* | 101±8.16* | 64±4.11* |
| F | 138.076 | 143.297 | 181.658 | 222.055 |
| p | 0.000 | 0.000 | 0.000 | 0.000 |
Note: Compared with the NC group, *p<0.05
Table 2: Effect of Etomidate on Migration and Invasion of Sw1088 Cells (X̅±S, N=9)
Effects of etomidate at different concentrations on the expression levels of LncRNA HOXA11-AS and miR- 515-5p in SW1088 cells were observed. After Glioma SW1088 cells were treated with etomidate at 100, 200, 400 and 800 μmol/l for 24 h, LncRNA HOXA11- AS decreased (p<0.05), while miR-515-5p increased (p<0.05), indicating that etomidate could inhibit migration and invasion of SW1088 cells (Table 3).
| Group | LncRNA HOXA11-AS | miR-515-5p |
|---|---|---|
| NC | 1.00±0.06 | 1.00±0.10* |
| 100 μmol/l etomidate | 0.83±0.07* | 1.52±0.12* |
| 200 μmol/l etomidate | 0.71±0.06* | 1.83±0.15* |
| 400 μmol/l etomidate | 0.57±0.04* | 2.24±0.18* |
| 800 μmol/l etomidate | 0.42±0.03* | 2.33±0.13* |
| F | 155.743 | 139.458 |
| p | 0.000 | 0.000 |
Note: Compared with the NC group, *p<0.05
Table 3: Effect of Etomidate on the Expression Levels of Lncrna Hoxa11-as and Mir-515-5p (X̅±S, N=9)
Over expression of LncRNA HOXA11-AS could weaken the effect of etomidate on the proliferation, migration and invasion of SW1088 cells. The expression levels of LncRNA HOXA11-AS in SW1088 cells transfected with pcDNA-LncRNA HOXA11-AS and pcDNA-NC were 1.89±0.13 and 1.00±0.08 respectively with significant difference (p<0.05), indicating that SW1088 cells overexpressing LncRNA HOXA11-AS were successfully constructed.
After interference of SW1088 cells overexpressing LncRNA HOXA11-AS with 400 μmol/l etomidate, the cell A value and the number of migration and invasion increased (p<0.05) and the expression levels of PCNA, MMP2 and MMP9 proteins in SW1088 cells increased (p<0.05), indicating that overexpression of etomidate could reduce the proliferation of SW1088 cells (fig. 3 and Table 4).
| Group | LncRNA HOXA11-AS | PCNA | MMP2 | MMP9 | A value | Number of cell migration | Number of cell invasion |
|---|---|---|---|---|---|---|---|
| pcDNA-NC+400 μmol/l etomidate | 1.00±0.08 | 0.34±0.03 | 0.47±0.03 | 0.42±0.04 | 0.527±0.04 | 97±6.13 | 64±3.28 |
| pcDNA-LncRNA HOXA11-AS+400 μmol/l etomidate | 1.89±0.13* | 0.83±0.06* | 0.90±0.08* | 0.81±0.07* | 0.987±0.08* | 167±12.17* | 124±8.12* |
| t | 17.492 | 21.913 | 15.098 | 14.512 | 15.429 | 15.411 | 20.554 |
| p | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
Note: Compared with pcDNA-NC+400 μmol/l etomidate group, *p<0.05
Table 4: Lncrna Hoxa11-as Could Weaken the Effect of Etomidate on the Proliferation, Migration and Invasion of Sw1088 Cells (X̅±S, N=9)
Targeted regulation of miR-515-5p with LncRNA HOXA11-AS is performed. The online target gene prediction software Starbase showed that there were continuous binding sites between LncRNA HOXA11- AS and miR-515-5p nucleotides, as shown in fig. 4. MiR-515-5p could reduce the luciferase activity of WT-LncRNA HOXA11-AS (p<0.05), but had no significant effect on the luciferase activity of MUTLncRNA HOXA11-AS (p>0.05), indicating that LncRNA HOXA11-AS could bind to miR-515-5p in a targeted manner, as shown in Table 5. The expression levels of miR-515-5p in SW1088 cells transfected with pcDNA-LncRNA HOXA11-AS and pcDNA-NC were 0.44±0.03 and 1.00±0.07 respectively with significant difference (p<0.05), which further indicated that LncRNA HOXA11-AS negatively regulated miR-515- 5p expression.
| Group | Relative luciferase activity | |
|---|---|---|
| WT-LncRNA HOXA11-AS | MUT-LncRNA HOXA11-AS | |
| miR-NC | 1.00±0.10 | 1.00±0.07 |
| miR-515-5p | 0.43±0.03* | 0.97±0.06 |
| t | 16.379 | 0.976 |
| p | 0.000 | 0.344 |
Note: Compared with the miR-NC group, *p<0.05
Table 5: Detection Results of Dual-Luciferase Activity (X̅±S, N=9)
Over expression of miR-515-5p reversed the effect of LncRNA HOXA11-AS on the proliferation, migration and invasion of SW1088 cells treated with etomidate. The expression levels of miR-515-5p in SW1088 cells transfected with miR-515-5p mimics and miRNC were 2.27±0.14 and 1.00±0.11 respectively with significant difference (p<0.05), indicating that SW1088 cells overexpressing miR-515-5p were successfully constructed. Compared with the miR-NC group, the A value and the number of migration and invasion of SW1088 cells in the miR-515-5p group decreased (p<0.05) and the expression levels of PCNA, MMP2 and MMP9 proteins decreased (p<0.05), indicating that overexpression of miR-515-5p could inhibit the proliferation, migration and invasion of SW1088 cells. Compared with the miR-NC+LncRNA HOXA11- AS+400 μmol/l etomidate group, the A value and the migration and invasion number of SW1088 cells in miR- 515-5p+miR-NC+LncRNA HOXA11-AS+400 μmol/l etomidate group decreased (p<0.05). The expression levels of PCNA, MMP2 and MMP9 proteins decreased (p<0.05), indicating that over expression of miR-515- 5p could reverse the influence of over expressing LncRNA HOXA11-AS on the proliferation, migration and invasion of SW1088 cells treated with etomidate (fig. 5 and Table 6).
| Group | miR-515-5p | PCNA | MMP2 | MMP9 | A value | Number of cell migration | Number of cell invasion |
|---|---|---|---|---|---|---|---|
| miR-NC | 1.00±0.11 | 0.83±0.07 | 0.95±0.06 | 0.76±0.06 | 1.147±0.08 | 204±15.47 | 141±8.03 |
| miR-515-5p | 2.27±0.14* | 0.37±0.02* | 0.44±0.03* | 0.32±0.02* | 0.613±0.04* | 115±7.38* | 62±4.02* |
| miR-NC+pcDNA-LncRNAHOXA11-AS+400 μmol/l etomidate | 1.25±0.10 | 0.85±0.07 | 0.91±0.09 | 0.83±0.06 | 0.994±0.07 | 172±11.51 | 121±8.02 |
| miR-515-5p+pcDNA-LncRNAHOXA11-AS+400 μmol/l etomidate | 2.04±0.15# | 0.41±0.04# | 0.47±0.03# | 0.33±0.02# | 0.586±0.04# | 104±7.11# | 57±3.26# |
| F | 209.084 | 206.949 | 201.667 | 335.100 | 193.581 | 169.493 | 410.677 |
| p | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 | 0.000 |
Note: Compared with the miR-NC group, *p<0.05; compared with the miR-NC+pcDNA-LncRNA HOXA11-AS+400 μmol/l etomidate, #p<0.05
Table 6: Mir-515-5p Reversed the Effect of Lncrna Hoxa11-as on The Proliferation, Migration and Invasion of Sw1088 Cells Treated with Etomidate (X̅±S, N=9)
Surgery is the main treatment for glioma. Studies have shown that the use of anesthetics can affect the growth and invasion of tumor cells, which is also one of the factors affecting the prognosis of patients[10]. Etomidate is a commonly used intravenous anesthetic, which can also inhibit malignant phenotype of tumor cells. It has been reported that etomidate can target and regulate miR-3666/Ubiquilin 1 (UBQLN1) to weaken the proliferation ability of breast cancer cells and aggravate apoptosis[11]; etomidate can stimulate the opening of chloride ion channel on the cell surface by binding with γ-aminobutyric acid receptor on the cell surface, thus blocking PI3K/Akt signaling pathway and inhibiting the migration of pituitary tumor cells[12]. At present, however, the effect of etomidate on malignant phenotype of glioma cell and its mechanism have not been reported.
In this study, etomidate (100, 200, 400, 800 μmol/l) decreased the A value and the migration and invasion number of glioma SW1088 cells, indicating that it inhibited the proliferation, migration and invasion of glioma cells. Cell proliferation, migration and invasion are regulated by many genes, among which PCNA is a proliferating cell nuclear antigen. The higher the expression level is, the stronger the cell proliferation ability[13]. MMP2 and MMP9 belong to the family members of MMPs. They participate in the degradation of extracellular matrix and promote the migration and invasion of tumor cells[14]. In this study, etomidate could effectively inhibit the expression levels of PCNA, MMP2 and MMP9 in glioma cells, suggesting that etomidate may directly or indirectly regulate the expression levels of proteins related to proliferation, migration and invasion to affect the malignant phenotype of glioma cells.
LncRNA is a small non-coding RNA widely existing in eukaryotes, which is closely related to the occurrence and development of tumors. As an lncRNA, HOXA11- AS is involved in various tumor development processes. Studies have shown that LncRNA HOXA11-AS is highly expressed in nasopharyngeal carcinoma tissues and cisplatin-resistant nasopharyngeal carcinoma cells and it realizes target binding with miR-454-3p and promotes the expression of c-Met to enhance the vitality of nasopharyngeal carcinoma cells and cisplatin resistance. Thus, it can be used as a molecular target to reverse cisplatin resistance in nasopharyngeal carcinoma[15]; the expression of LncRNA HOXA11- AS in colon cancer tissues is significantly higher than that in non-cancer tissues. The high expression of LncRNA HOXA11-AS is closely related to clinical and pathological features such as late clinical stage, tumor size and tumor recurrence. LncRNA HOXA11-AS promotes the metastasis of colon cancer as a molecular sponge of miR-149-3p[16]; the expression of LncRNA HOXA11-AS is up-regulated in gastric cancer tissues and cell lines. It activates β-catenin signaling pathway by targeting miR-148a/WNT1 axis, promotes migration and invasion of gastric cancer cells and can be used as a molecular target for gastric cancer treatment[17]. It has been reported that LncRNA HOXA11-AS is over expressed in neuroglioma and is related to the late stage and poor prognosis of brain glioma. It promotes the growth and metastasis of neuroglioma by targeting miR-130a-5p/HMGB2 axis[18]. In this study, etomidate could inhibit the expression of LncRNA HOXA11- AS in glioma cells and over-expression of LncRNA HOXA11-AS could weaken the inhibitory effect of etomidate on proliferation, migration and invasion of glioma cells, suggesting that etomidate may regulate the malignant phenotype of glioma cells by down regulating LncRNA HOXA11-AS. To further explore that etomidate may regulate malignant phenotype of glioma cell by down-regulating LncRNA HOXA11-AS, this study confirmed LncRNA HOXA11-AS targeted to negatively regulate miR-515-5p. The expression of miR- 515-5p decreased in many kinds of tumors, playing the role of tumor suppressor gene. The expression of miR- 515-5p in breast cancer tissues and cells decreased. The proliferation, migration and invasion ability of breast cancer cells in vitro could be reduced by targeted upregulation of miR-515-5p expression and the growth of breast cancer tumors in vivo could be inhibited[19]; circ_0067997 was up-regulated in gastric cancer. It could play the role of miR-515-5p molecular sponge to promote the expression of X-linked inhibitor of apoptosis protein (XIAP) and then promote the vitality, colony formation and invasion of gastric cancer cells. Circ_0067997/miR-515-5p/XIAP axis provides a new idea for elucidating the molecular mechanism of gastric cancer[20]. This study showed that overexpression of miR-515-5p could significantly reduce the proliferation, migration and invasion of glioma cell, suggesting that miR-515-5p also played an anti-oncogene role in the occurrence and development of glioma and it can be used as a molecular target for glioma treatment. The research also showed that etomidate could promote the expression of miR-515-5p in glioma cells and over-expression of miR-515-5p reversed the effect of over-expression of LncRNA HOXA11-AS on the proliferation, migration and invasion of glioma cells treated with etomidate, thus suggesting that etomidate could inhibit the proliferation of glioma cells by targeting down-regulation of LncRNA HOXA11-AS and then up-regulation of miR-515-5p.
To sum up, etomidate has a certain inhibitory effect on proliferation, migration and invasion of glioma cells and its mechanism may be related to the regulation of LncRNA HOXA11-AS/miR-515-5p axis.
Conflicts of interest:
The authors declared no conflict of interest.
References
- Huang W, Li ZH, Wang DX. The influence of common anesthesia methods drug selection and intraoperative management on the prognosis of elderly patients in tumor surgery. J Clin Anesthesiol 2018;34(7):716-20.
- Xu W, Xue R, Xia R, Liu WW, Zheng JW, Tang L, et al. Sevoflurane impedes the progression of glioma through modulating the circular RNA has_circ_0012129/miR-761/TGIF2 axis. Eur Rev Med Pharmacol Sci 2020;24(10):5534-48.
- Wang DM, Xu BY, Yang T, He J, Zhao HP, Lu S, et al. Etomidate inhibits proliferation, migration and invasion of gastric cancer cells by regulating the expression of microRNA-211-5p/ROBO1. Chin Pharm J 2020;55(20):1686-95.
- Du JZ, Dai QX, Rong FJ, Xu X. Effects of etomidate on PI3K/AKT pathway and apoptosis of non-small cell lung cancer A549 cells. Hebei Med 2020;26(7):1066-71.
- Yin D, Lu X. Silencing of long non-coding RNA HCP5 inhibits proliferation, invasion, migration and promotes apoptosis via regulation of miR-299-3p/SMAD5 axis in gastric cancer cells. Bioengineered 2021;12(1):225-39.
- Jiang J, Wang X, Gao G, Liu X, Chang H, Xiong R, et al. Silencing of lncRNA HOXA11-AS inhibits cell migration, invasion, proliferation and promotes apoptosis in human glioma cells via upregulating microRNA-125a: in vitro and in vivo studies. Am J Transl Res 2019;11(10):6382-92.
- Guo F, Li S, Guo C, Xu X, Zhou X, Ma D, Cao Z, Bing Z, Cui Y. Circular RNA circMAGI3 accelerates the glycolysis of non-small cell lung cancer through miR-515-5p/HDGF. Am J Transl Res 2020;12(7):3953-63.
- Dai G, Huang C, Yang J, Jin L, Fu K, Yuan F, et al. LncRNA SNHG3 promotes bladder cancer proliferation and metastasis through miR‐515‐5p/GINS2 axis. J Cell Mol Med 2020;24(16):9231-43.
- Ye P, Lv X, Aizemaiti R, Cheng J, Xia P, Di M. H3K27ac‐activated LINC00519 promotes lung squamous cell carcinoma progression by targeting miR‐450b‐5p/miR‐515‐5p/YAP1 axis. Cell Prolif 2020;53(5):e12797.
- Wu B, Zheng Z. The effect of anesthesia on immune function of tumor patients and tumor recurrence and metastasis after operation. Chin J Colorectal Dis 2018;7(3):270-5.
- Li F, Kong JF. Mechanism of etomidate inhibiting proliferation and induction of apoptosis in breast cancer cells by regulating miR-3666/UBQLN1 gene expression. Genomics Appl Biol 2020;39(2):326-33.
- Zhu M, Liu YH, Yu CJ. Effect of etomidate on migration ability of pituitary tumor cells. Chin J Clin Pharmacol 2018;34(11):1357-60.
- Fan HN, Liao XH, Zhang J, Zheng HM. Macrophages promote cell proliferation in colorectal cancer via IL-1β-mediated downregulation of miR-28-3p. J Biol Regul Homeost Agents 2020;34(5):1657-68.
- Cai P, Wu M, Zhang B, Wu S, Wei H, Wei L. Long non‑coding RNA SNHG12 regulates cell proliferation, invasion and migration in endometrial cancer by targeting miR‑4429. Mol Med Rep 2020;22(4):2842-50.
- Lin FJ, Lin XD, Xu LY, Zhu SQ. Long Noncoding RNA HOXA11-AS modulates the resistance of nasopharyngeal carcinoma cells to cisplatin via miR-454-3p/c-Met. Mol Cells 2020;43(10):856-69.
- Chen D, Zhang M, Ruan J, Li X, Wang S, Cheng X, et al. The long non-coding RNA HOXA11-AS promotes epithelial mesenchymal transition by sponging miR-149-3p in Colorectal Cancer. J Cancer 2020;11(20):6050-8.
- Guo T, Yuan X, Liu DF, Peng SH, Xu AM. LncRNA HOXA11-AS promotes migration and invasion through modulating miR-148a/WNT1/β-catenin pathway in gastric cancer. Neoplasma 2020;67(3):492-500.
- Xu CH, Xiao LM, Liu Y, Chen LK, Zheng SY, Zeng EM, et al. The lncRNA HOXA11-AS promotes glioma cell growth and metastasis by targeting miR-130a-5p/HMGB2. Eur Rev Med Pharmacol Sci 2019;23(1):241-52.
- Huang FJ, Dang JQ, Zhang S, Cheng ZY. Circular RNA hsa_circ_0008039 promotes proliferation, migration and invasion of breast cancer cells through upregulating CBX4 via sponging miR-515-5p. Eur Rev Med Pharmacol Sci 2020;24(4):1887-98.
- Zhang H, Wang X, Huang H, Wang Y, Zhang F, Wang S. Hsa_circ_0067997 promotes the progression of gastric cancer by inhibition of miR-515-5p and activation of X chromosome-linked inhibitor of apoptosis (XIAP). Artif Cells Nanomed Biotechnol 2019;47(1):308-18.