- *Corresponding Author:
- Qi Zhou
Department of Oncology, Chongqing University Fuling Central Hospital, Shapingba, Chongqing 400038, China
E-mail: qizhou112@163.com
| This article was originally published in a special issue, “Advanced Targeted Therapies in Biomedical and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2023:85(1) Spl Issue “108-118” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Colorectal cancer is one of the major public health issues worldwide due to its high mortality and morbidity. Angiogenesis is the fundamental step of tumors transition from dormant to malignant state and it is recognized as one of the hallmarks of cancer. High mobility group AT-hook 2 encodes a small non-histone chromatinassociated protein that can modulate transcription by altering the chromatin architecture. Currently the mechanism by which high mobility group AT-hook 2 involved in angiogenesis in colorectal cancer has not been clarified. The expression of High mobility group AT-hook 2 was analyzed by immunohistochemistry, Western blotting and bioinformatics methods. cBioPortal and mExpress online tools were applied to explore the copy-number variation and methylation of high mobility group AT-hook 2 in colorectal cancer patients. Single cell data from gene expression omnibus was used to examine the specific cell type that contributed to the high mobility group AT-hook 2 expression in colorectal cancer. Lentivirus was used to knock down high mobility group AT-hook 2 in colorectal cancer cells and human umbilical vein endothelial cells was used to study angiogenesis. In the current study, we first detected the expression pattern of high mobility group AT-hook 2 in colorectal cancer patients and evaluated its clinical values and for the first time showed that copy number amplification contributed to the up regulation of high mobility group AT-hook 2 in colorectal cancer patients. By analyzing colorectal cancer single cell data we found that high mobility group AT-hook 2 was specifically up regulated in the colorectal epithelial cells. Furthermore, knocking down of high mobility group AT-hook 2 in colorectal cancer epithelial cells suppresses angiogenesis via dual regulation of vascular endothelial growth factor-A and semaphorin 3A in colorectal cancer through inactivating vascular endothelial growth factor receptor 2 pathway in human umbilical vein endothelial cells. High mobility group AT-hook 2 might be a promising prognostic marker and target for treating advanced colorectal cancer patients.
Keywords
High mobility group AT-hook 2, colorectal cancer, angiogenesis, semaphorin 3A, vascular endothelial growth factor A
Colorectal Cancer (CRC) is one of the major public health issues worldwide due to its high mortality and morbidity[1]. Although the survival outcome of CRC patients has dramatically increased due to the improvement of diagnostic and therapeutic strategies, up to 20 % of CRC patients indicate metastatic phenotype upon initial diagnosis and at least 30 % of them will die of distal metastasis[2], therefore CRC patients at advanced stages suffer from undesired 5 y survival rate (<10 %)[3,4].
Angiogenesis is the fundamental step of tumors transition from dormant to malignant state by providing O2 and nutrients to tumor cells and is widely believed to be indispensable for distal metastasis which is responsible for the high mortality of CRC[4,5]. Rooted in the belief that blocking vessel supply starves tumors to death[5,6] thus timely inhibition of angiogenesis can prevent further growth and metastasis therefore anti-angiogenic treatments have been widely used for CRC treatment[7-10]. Clinically different strategies for angiogenesis intervention are based on modulating key steps of the angiogenic process, including endothelial cell proliferation cell-matrix adhesion and metastasis[11]. It is well known that Vascular Endothelial Growth Factor A (VEGF-A) promoted tumor angiogenesis via banding to Vascular Endothelial Growth Factor Receptor 2 (VEGFR2) on the vascular endothelial cells[12]. Indeed the monoclonal anti-VEGF antibody bevacizumab[13,14] and the second-generation multi-targeted Receptor Tyrosine Kinase Inhibitors (RTKIs) sunitinib[15] and sorafenib[16] have prolonged the life of numerous cancer patients. However, drug resistance would be inevitably established out of unknown mechanisms. On the other hand the adverse effects such as increased risk of stroke and arterial events, gastrointestinal bleeding, gastrointestinal perforation, delayed wound healing were increased thus the usage of anti-angiogenesis therapy was confined with its side effects[17,18]. Thus, uncover novel anti-tumor angiogenesis mechanisms in CRC would provide potential target for clinical interventions.
High Mobility Group AT-Hook 2 (HMGA2) is a chromatin-associated proteins that acts as an architectural transcription factor which modulate the transcription of target genes through directly binding to Deoxyribonucleic Acid (DNA) sequences and altering the structure of DNA[19-21]. Previous studies showed that HMGA2 played important roles in the proliferation, metastasis, DNA damage responses and cancer stem cell maintaining[22]. However the function of HMGA2 in angiogenesis in CRC remained unclear. Tumor Microenvironment (TME) was important for angiogenesis was maintained by cancer cells and stromal cells via the secretion of various cytokines, growth factors and other effector molecules[23]. In the current study, through bioinformatics and Immunohistochemistry (IHC) analysis, we found that HMGA2 was significantly overexpressed especially in patients at advanced stage and associated with undesired disease progression. Specifically HMGA2 expression was positively correlated with the density of Cluster of Differentiation 31 (CD31) (marker of angiogenesis) in CRC patient tissues. Our data for the first time indicated that copy number amplification played a role in regulating HMGA2 expression in Colon Adenocarcinoma (COAD) manifested as gain of copy number denoted high expression of HMGA2. Single cell data analysis indicated that HMGA2 was specifically up regulated in the colorectal epithelial cells. Mechanical study showed that knocking down of HMGA2 suppresses angiogenesis via dual regulation of VEGF-A and Semaphorin 3A (SEMA3A) in CRC through inactivating VEGFR2 pathway in HUVECs. Taken together our study for the first time showed that HMGA2 was up regulated 109in colorectal epithelial cells due to copy number amplification and knock down HMGA2 in CRC epithelial cells could inhibit the proliferation and metastasis of HUVECs via compromising VEGFR2 signaling.
Materials and Methods
Samples:
Surgically resected specimens were collected from 154 CRC cases and paired adjacent normal tissues at three hospitals affiliated with the Third Military Medical University and Fu Ling Central Hospital from 2004 to 2007. The specimens were fixed with 4 % neutral buffered paraformaldehyde and embedded in paraffin; after staining with haematoxylin and eosin, the specimens were analyzed by a senior pathologist. A semi-automated tissue microarray (Beecher Instruments, Sun Prairie, Wisconsin , United States of America (USA)) was used to construct tissue microarrays (1 mm needle diameter) by punching out one or two tissue cores from each specimen to obtain two paraffin blocks containing 154 CRC specimens and paired adjacent normal tissues. The study protocol was approved by the Ethical Committee of Southwest Hospital Third Military Medical University.
Cell lines:
Normal colon epithelial cell line Fetal Human Cells (FHC) and Human Umbilical Vein Endothelial Cells (HUV-ECs) were purchased from the American type culture collection (Manassas, Virginia, USA). HCT-116, LS174T, HT-29, SW480, SW620, RKO, LoVo, HCT-8 and Cancer coli-2 (Caco2) CRC cell lines were purchased from cell bank of Shanghai Institute of cell biology, Chinese Academy of Sciences. LS174T, RKO, and Caco2 cells were cultured in eagle’s minimum essential medium; FHC cells were cultured in Dulbecco’s Modified Eagle’s Medium:F12 (Gibco); HCT-116 and HT-29 and HCT-8 cells were cultured in McCoy’s 5a medium; SW480 and SW620 cells were cultured in Leibovitz’s L-15 medium; and HUV-ECs and LoVo cells were cultured in F-12K medium. All media were purchased from Gibco (Grand Island, NY, USA) and were supplemented with 10 % Fetal Bovine Serum (FBS) (Gibco). Cells were cultured at 37° and 5 % Carbon dioxide (CO2).
IHC:
Formalin-fixed, paraffin-embedded tissue specimens and xenografts were cut into 5 μm-thick sections that were analyzed by IHC using the Real Envision detection system (Dako, Carpinteria, California, USA) according to the manufacturer’s protocol. The following primary antibodies were used; rabbit monoclonal anti-HMGA2 (1:2000; OriGene, Rockville, Maryland, USA), rabbit polyclonal anti CD31 (1:500; Abcam, Cambridge, Massachusetts (MA), USA), rabbit polyclonal anti-VEGFA (1:1000; Abcam) and rabbit polyclonal anti-SEMA3A (1:500; Abcam). The percentage of immune positive cells (staining percentage) was determined under the microscope and scored as follows; 0 means no positive cells; 1 was 1 %-25 % positive cells; 2 was 26 %-50 % positive cells; 3 was 51 %-75 % positive cells; and 4 means ≥76 % positive cells. Intensity was scored as 0=none, 1=weak, 2=moderate and 3=strong.
Comprehensive score=Staining percentage×intensity HMGA2 expression was classified into low/high expression group according to the median of total samples.
Western Blotting (WB):
WB was performed as previously described. The primary antibodies used in this study were; rabbit monoclonal anti-HMGA2 (1:2000; OriGene); rabbit polyclonal anti-VEGFA (1:1000; Abcam); rabbit polyclonal anti-SEMA3A (1:1000); rabbit monoclonal anti-VEGFR-2 (1:1000), rabbit monoclonal anti-phosphorylated (p-) VEGFR- 2(Y1175) (1:1000), rabbit monoclonal anti- Extracellular Signal-Regulated Kinase (ERK)1/2 (1:1000), rabbit monoclonal anti-p-ERK1/2 (1:1000), rabbit monoclonal anti- Protein kinase B (AKT) (1:1000), rabbit monoclonal anti-p-AKT (1:1000) and rabbit polyclonal anti-β-actin (1:3000) all from Cell Signaling Technology; Danvers, MA, USA).
Plasmid construction and stable transfection of cell lines:
HMGA2 short hairpin Ribonucleic acid (shRNA) lentiviral particles were constructed by Genechem Co. (Shanghai, China). The shRNA sequence targeting HMGA2 and a non-targeting scrambled sequence were as follows: shHMGA2#1, GAAATGGCCACAACAAGTTGT; shHMGA2#2, AGTCCCTCTAAAGCAGCTCAA and shHMGA2#3, TCCTCACAAGAGTCTGCCGAA. HCT-116 and HT-29 cells were cultured in 24-well plates at 5×104/well and infected with lentivirus containing the shRNA constructs for 24 h without FBS. Fresh culture medium containing 1 μg/ml puromycin was added to select stably transfected cell lines. The knockdown efficiency of HMGA2 was confirmed by WB.
Conditioned medium preparation and treatment for HUVECs:
Equal numbers of HCT-116 and HT-29 cells infected with lentivirus containing scrambled or HMGA2 shRNA were seeded in 100 mm dishes and allowed to attach. The medium was centrifuged at 1000 rpm for 3 min to remove any cell contaminants and termed as Conditional Medium (CM). As for the treatment of HUVECs, CM collected from scrambled or HMGA2 shRNA HCT116 cells were subsequently subjected to the culture of HUVECs.
Cell proliferation migration and invasion assays:
HUVECs proliferation was analyzed using Cell Counting Kit-8 (Dojindo Laboratories, Kumamoto, Japan) according to the manufacturer’s protocol. Briefly, 5000 cells per well were seeded in 96-well plate and cultured with normal medium and change with control or HMGA2 KD CM after 24 h. The absorbance was determined before changing with CM to confirm the identical cell number. The absorbance was determined at 24 h and 48 h CM changing. For cell migration assay CM was added respectively and wound closure (migration) was evaluated at 0 h and 24 h. As for invasion assay, the 8.0 μm pore transwell membrane was coated with 1 mg/ml Matrigel (BD Biosciences, Franklin Lakes, New Jersey (NJ), USA). The lower chamber was filled with F-12K medium containing 10 % FBS and the upper chamber was filled with a cell suspension containing 1×104 cells in CM. Cells were allowed to migrate at 37° and 5 % CO2 for 48 h. Invasion cells were fixed and stained with crystal violet and counted under a light microscopy in five randomly selected fields. Results were expressed as the average number of cells per field. The experiments were performed three times independently.
In vivo tumor formation assay:
Male nude mice (4 w to 6 w old) were purchased from the center of experimental animals of Third Military Medical University (Chongqing, China). The mice were subcutaneously inoculated with HCT- 116 cells (1×106) infected with lentivirus expressing HMGA2 or scrambled shRNA. Tumor-bearing mice were euthanized 4 w after transplantation. The width and length of tumors were measured with calipers and tumor volume was calculated according to the formula: volume (3 mm)=width 2 mm×length 0.4 mm. All experimental animal procedures were approved by the Institutional Animal Care and Use Committee of Third Military Medical University, China.
Immunofluorescence microscopy:
Frozen sections of fresh xenografts tissue were fixed with 4 % paraformaldehyde and treated with triton-100 and then incubated with rabbit polyclonal anti-CD31 (1:500) and rabbit monoclonal anti- HMGA2 (1:2000) antibodies followed by the secondary antibodies. Nuclei were counterstained with 4',6-Diamidino-2-Phenylindole (DAPI) (Sigma- Aldrich, St. Louis, MO, USA) and slides were visualized with a laser confocal scanning microscope (SP-5; Leica Microsystems, Wetzlar, Germany).
Enzyme-Linked Immunosorbent Assay (ELISA):
Secretion of VEGF-A, Angiostatin (ANG)-2, VEGF-C, Hepatocyte Growth Factor (HGF), endothelin-1, Platelet-Derived Growth Factor-BB (PDGF-BB) and SEMA3A was determined using ELISA kits (Senxiong Biotech, Beijing, China) according to the manufacturer’s instructions and normalized to the total number of cells.
Bioinformatics analysis and public data base:
The Cancer Genome Atlas (TCGA) COAD raw counts data was download use GDC client from https://portal.gdc.cancer.gov/. Raw data was processed in R (version 4.0.2) using Dseq2 and EdgeR package and figures were plotted using ggplot2.
Single cell analysis: The single cell data set of 9 CRC patients was retrieved from Gene Expression Omnibus (GEO) GSE166555 and standard pipeline was applied for data processing using R. All clusters of cells in CRC were annotated by single R and cell marker according to the composition of the marker genes.
Statistical analysis:
Statistical analyses were performed using Statistical Package for the Social Sciences (SPSS) v.17.0 software (SPSS Inc., Chicago, Illinois (IL), USA). Kaplan–Meier survival plots and the log-rank test were used to compare patient survival rates. Differences between experimental groups and controls were assessed with the Student’s test. p<0.05 was considered statistically significant.
Results and Discussion
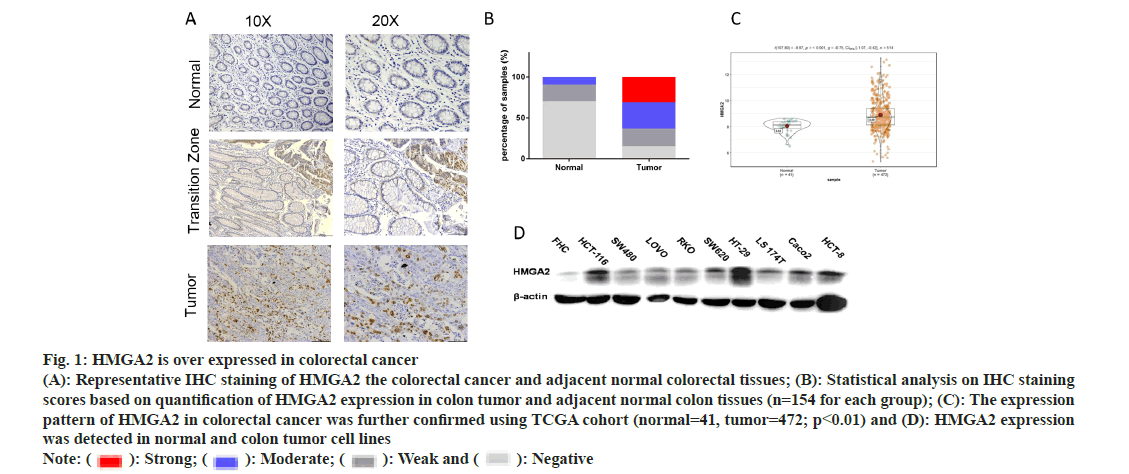
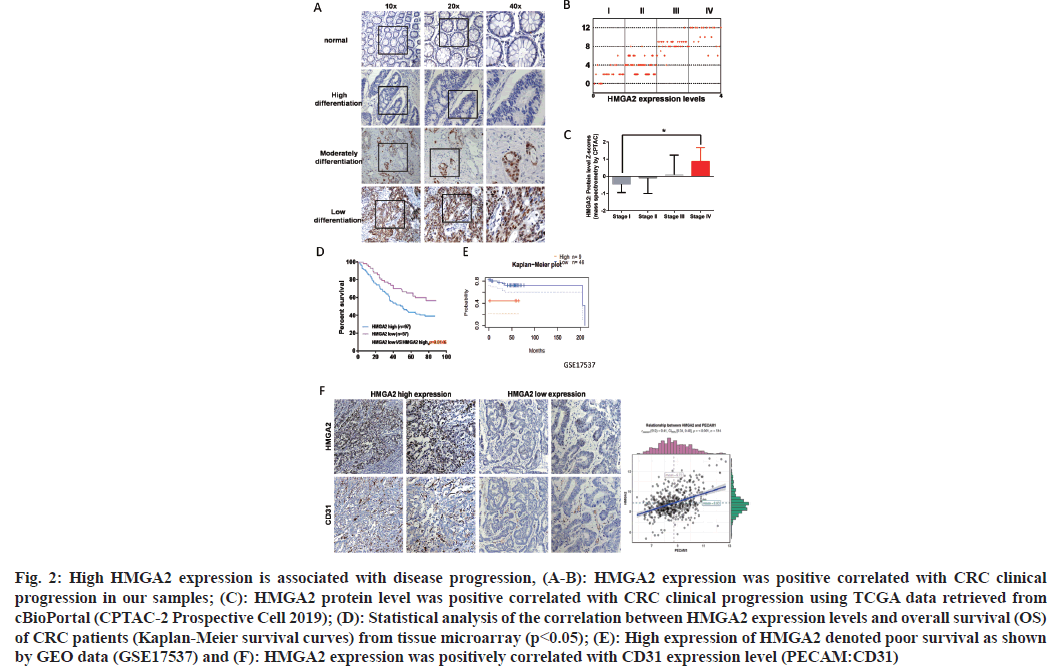
To investigate the potential role of HMGA2 in CRC, we first detected the expression pattern of HMGA2 in CRC patient’s sample and cell lines. HMGA2 expression was markedly higher in CRC patient’s sample and cell lines as detected by IHC and WB (fig. 1A-fig. 1D) and this observation were further confirmed by analyzing the TCGA CRC cohort (fig. 1C). Thus we speculated whether this abnormal expression could reflect clinical progression. We collected 154 CRC patients sample with clinical information and found that patients featured by low differentiation and advanced clinical stage showed high expression of HMGA2 (fig. 2A and fig. 2B) which was further confirmed using public datasets (fig. 2C). Furthermore high HMGA2 expression predicted worse prognosis (p<05; fig. 2D and fig. 2E). Tumor angiogenesis is a vital supporter of tumor invasion and metastasis, which is responsible for the high mortality and poor prognosis[24]. The expression of CD31 could partially reflect the formation of micro vascular and denoted increased angiogenesis[25] and we found that HMGA2 protein level was positively correlated with CD31 expression in serial sections of CRC tissues from 154 cases and TCGA CRC datasets (fig. 2F).
Fig. 1: HMGA2 is over expressed in colorectal cancer (A): Representative IHC staining of HMGA2 the colorectal cancer and adjacent normal colorectal tissues; (B): Statistical analysis on IHC staining scores based on quantification of HMGA2 expression in colon tumor and adjacent normal colon tissues (n=154 for each group); (C): The expression pattern of HMGA2 in colorectal cancer was further confirmed using TCGA cohort (normal=41, tumor=472; p<0.01) and (D): HMGA2 expression was detected in normal and colon tumor cell lines Note: ( ): Strong; (
): Strong; ( ): Moderate; (
): Moderate; ( ): Weak and (
): Weak and ( ): Negative
): Negative
Fig. 2: High HMGA2 expression is associated with disease progression, (A-B): HMGA2 expression was positive correlated with CRC clinical progression in our samples; (C): HMGA2 protein level was positive correlated with CRC clinical progression using TCGA data retrieved from cBioPortal (CPTAC-2 Prospective Cell 2019); (D): Statistical analysis of the correlation between HMGA2 expression levels and overall survival (OS) of CRC patients (Kaplan-Meier survival curves) from tissue microarray (p<0.05); (E): High expression of HMGA2 denoted poor survival as shown by GEO data (GSE17537) and (F): HMGA2 expression was positively correlated with CD31 expression level (PECAM:CD31)
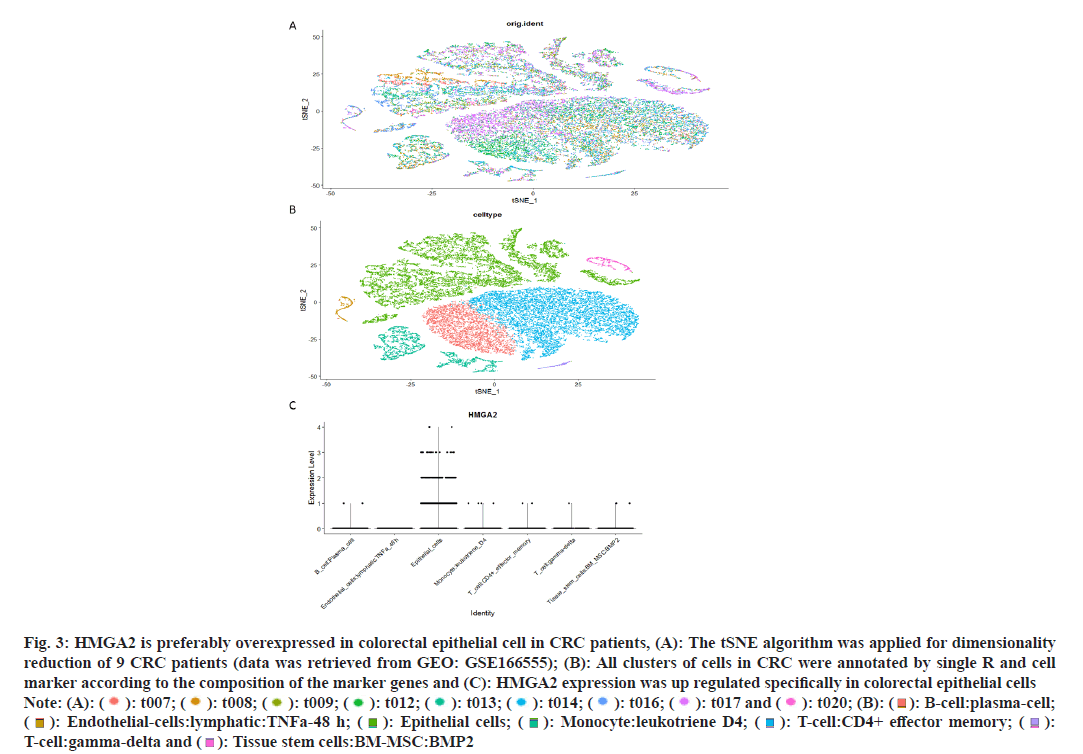
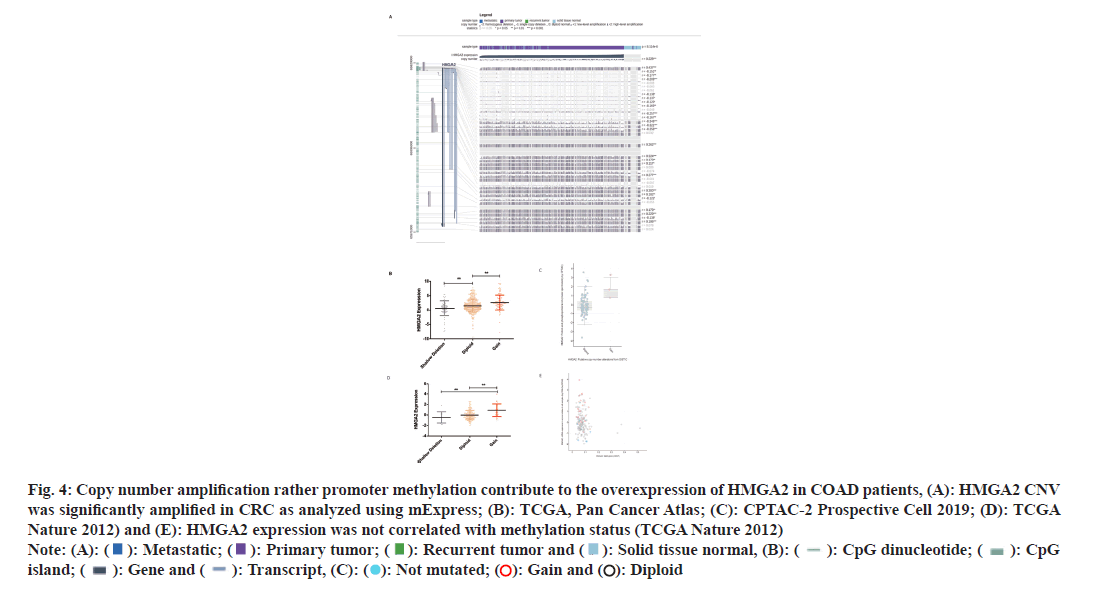
Due to the heterogeneity origins of CRC[26,27] we analyzed a CRC single cell data set (including 9 CRC patient samples) to unveil which type of cell contribute to the high expression of HMGA2. After t-distributed Stochastic Neighbor Embedding (tSNE) reduction and cell type annotation (fig. 3A and fig. 3B) we found that colorectal epithelial cells showed high expression of HMGA2 among 7 types of cells (fig. 3C). To further investigate the possible mechanisms contributed to the abnormal expression of HMGA2 in CRC, we analyzed online data sets and found Copy-Number Variation (CNV) was significantly disrupted in CRC (fig. 4A) and gain of copy number denoted high expression of HMGA2 by analyzing TCGA data sets (fig. 4B-fig. 4D), even though promoter methylation was observed it was not correlated with gene expression (fig. 4E). Thus the above results manifested that overexpression of HMGA2 specifically in colorectal epithelial cells was associated with undesired disease progression which could be attributed to angiogenesis and copy number amplification was responsible for the abnormal expression of HMGA2 in CRC.
Fig. 3: HMGA2 is preferably overexpressed in colorectal epithelial cell in CRC patients, (A): The tSNE algorithm was applied for dimensionality reduction of 9 CRC patients (data was retrieved from GEO: GSE166555); (B): All clusters of cells in CRC were annotated by single R and cell marker according to the composition of the marker genes and (C): HMGA2 expression was up regulated specifically in colorectal epithelial cells Note: (A): ( ): t007; (
): t007; ( ): t008; (
): t008; (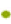 ): t009; (
): t009; ( ): t012; (
): t012; (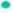 ): t013; (
): t013; ( ): t014; (
): t014; ( ): t016; (
): t016; ( ): t017 and (
): t017 and ( ): t020; (B): (
): t020; (B): ( ): B-cell:plasma-cell; (
): B-cell:plasma-cell; ( ): Endothelial-cells:lymphatic:TNFa-48 h; (
): Endothelial-cells:lymphatic:TNFa-48 h; ( ): Epithelial cells; (
): Epithelial cells; ( ): Monocyte:leukotriene D4; (
): Monocyte:leukotriene D4; ( ): T-cell:CD4+ effector memory; (
): T-cell:CD4+ effector memory; ( ): T-cell:gamma-delta and (
): T-cell:gamma-delta and ( ): Tissue stem cells:BM-MSC:BMP2
): Tissue stem cells:BM-MSC:BMP2
Fig. 4: Copy number amplification rather promoter methylation contribute to the overexpression of HMGA2 in COAD patients, (A): HMGA2 CNV was significantly amplified in CRC as analyzed using mExpress; (B): TCGA, Pan Cancer Atlas; (C): CPTAC-2 Prospective Cell 2019; (D): TCGA Nature 2012) and (E): HMGA2 expression was not correlated with methylation status (TCGA Nature 2012) Note: (A): ( ): Metastatic; (
): Metastatic; ( ): Primary tumor; (
): Primary tumor; ( ): Recurrent tumor and (
): Recurrent tumor and ( ): Solid tissue normal, (B): (
): Solid tissue normal, (B): ( ): CpG dinucleotide; (
): CpG dinucleotide; ( ): CpG island; (
): CpG island; ( ): Gene and (
): Gene and ( ): Transcript, (C): (
): Transcript, (C): ( ): Not mutated; (
): Not mutated; ( ): Gain and (
): Gain and ( ): Diploid
): Diploid
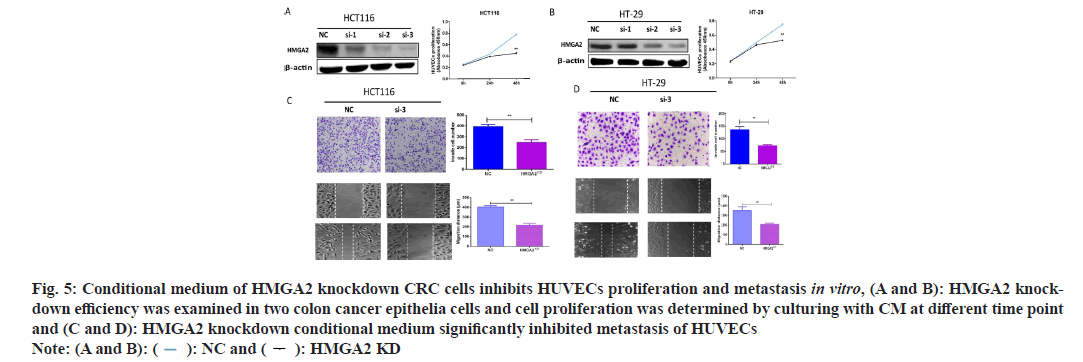
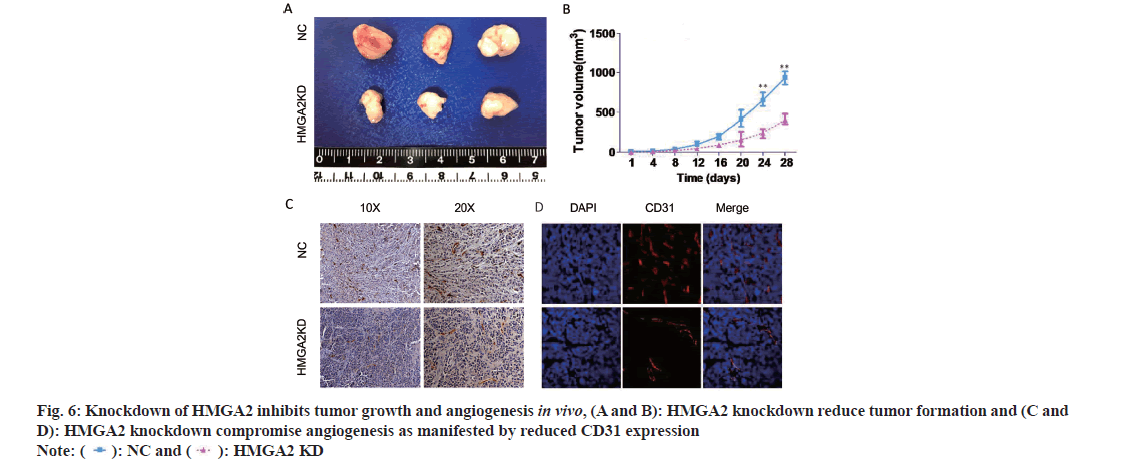
Our results demonstrated that high HMGA2 expression was positively associated with CD31 expression (marker of angiogenesis) in serial sections of CRC tissues from 154 cases and TCGA CRC datasets (fig. 2F) and it was specifically up regulated in colorectal epithelial cells (fig. 3C). Therefore we speculated that cancerous colorectal epithelial cells with HMGA2 overexpression might regulate vascular endothelial cells behavior by intercellular communication. Thus we collected the CM from scrambled or HMGA2 shRNA lentivirus infected colorectal epithelial cells and treated HUVECs as previously described[3]. The results showed that HMGA2 shRNA CM from two CRC cell lines compromised cell proliferation (fig. 5A and fig. 5B) and inhibited the invasion and migration of HUVECs (fig. 5C and fig. 5D). To further confirm the inhibition effects of HMGA2 knock down on tumor formation in vivo, HCT-116, NC and HMGA2, KD cells were subcutaneously transplanted to nude mice. After 28 d, mice were euthanized and tumors were harvested, weighed, and analyzed. The results showed that HMGA2 KD reduced the tumor formation (fig. 6A and fig. 6B). Tumor angiogenesis was determined by CD31 expression in tumor specimens through IHC and IF. The result demonstrated that HMGA2 KD inhibited tumor angiogenesis in mice (fig. 6C and fig. 6D). Collectively, these results indicated that knock down HMGA2 in CRC cells could inhibit HUVECs proliferation, metastasis and tumor formation.
Fig. 5: Conditional medium of HMGA2 knockdown CRC cells inhibits HUVECs proliferation and metastasis in vitro, (A and B): HMGA2 knockdown efficiency was examined in two colon cancer epithelia cells and cell proliferation was determined by culturing with CM at different time point and (C and D): HMGA2 knockdown conditional medium significantly inhibited metastasis of HUVECs Note: (A and B): ( ): NC and (
): NC and ( ): HMGA2 KD
): HMGA2 KD
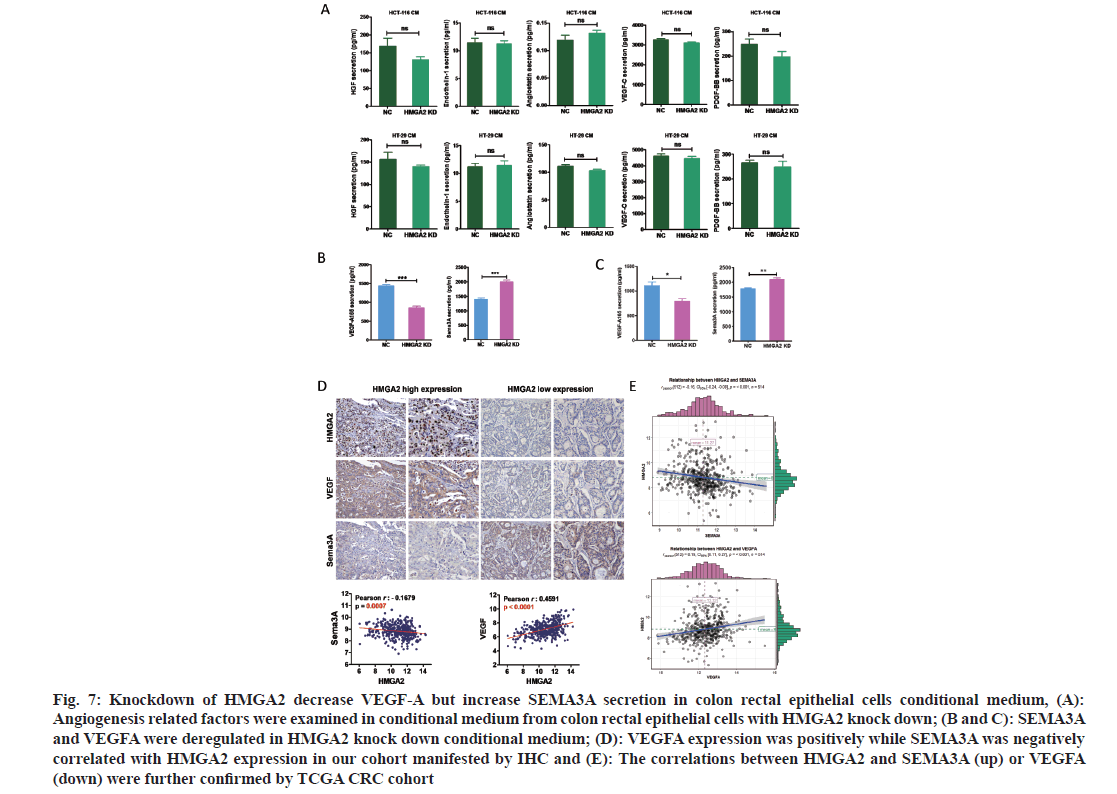
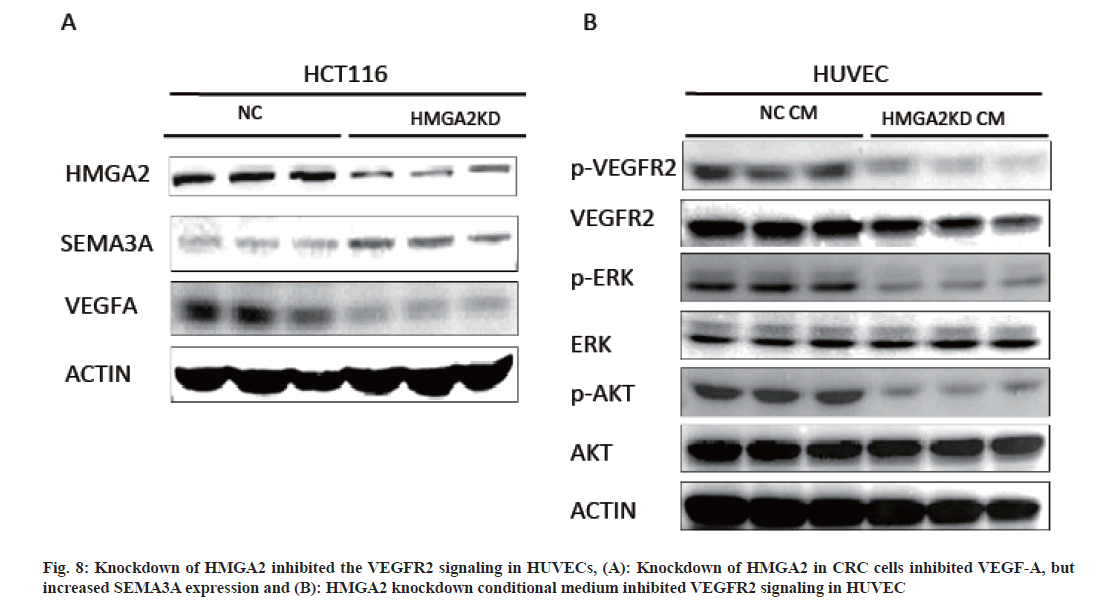
TME was important for angiogenesis was maintained by cancer cells and stromal cells via the secretion of various cytokines, growth factors and other effector molecules. Since the CM from HMGA2 KD could suppress the proliferation and metastasis of HUVECs (fig. 5), we speculated that overexpression of HMGA2 in colorectal epithelial cells might regulated vascular endothelial cells by secreting angiogenesis related molecules. To unveil the possible signal molecules involved in this process the levels of angiogenesis related secreted factors; HGF, endothelin-1, ANG, VEGF-A, VEGF-C, SEMA3A and PDGF-BB in the supernatant of HMGA2 KD and control cells were firstly detected. While HGF, endothelin-1, ANG, VEGF-C and PDGF-BB were not changed significantly (fig. 7A), VEGF-A level was lower while SEMA3A level was higher in the HMGA2 KD supernatant (fig. 7B and fig. 5C). Then we detected the expression of VEGFA and SEMA3A in CRC patient tissues. We found VEGF-A level was lower while SEMA3A was up-regulated in HMGA2 low expression patients vice versa (fig. 7D). We further found that HMGA2 was positively correlated with VEGF-A but negatively correlated with SEMA3A by analyzing TCGA COAD data sets (fig. 7E). To confirm the direct regulation of HMGA2 on VEGF-A and SMA3A, we analyzed the protein expression by knocking down HMGA2 in HCT116 cells and found that HMGA2-KD up regulated SEMA3A and inhibited VEGF-A (fig. 8A) which was in consistent with the observation that HMGA2 KD increased VEGF-A and reduced SEMA3A levels in the supernatant of HCT-116 and HT-29 colon cancer cells.
Fig. 7: Knockdown of HMGA2 decrease VEGF-A but increase SEMA3A secretion in colon rectal epithelial cells conditional medium, (A): Angiogenesis related factors were examined in conditional medium from colon rectal epithelial cells with HMGA2 knock down; (B and C): SEMA3A and VEGFA were deregulated in HMGA2 knock down conditional medium; (D): VEGFA expression was positively while SEMA3A was negatively correlated with HMGA2 expression in our cohort manifested by IHC and (E): The correlations between HMGA2 and SEMA3A (up) or VEGFA (down) were further confirmed by TCGA CRC cohort
Furthermore we evaluated the activation of VEGFR2 pathway in HUVECs treated by CM through detecting total and phosphorylation expression of VEGF-R2, ERK, and AKT in HUVECs. We found that HMGA2 KD CM from HCT-116 decreased the phosphorylation of VEGFR2, ERK and AKT indicating that VEGF-A-VEGFR2 signaling was inhibited in HUVECs whereby cell migration, endothelium-dependent relaxation and angiogenesis were compromised (fig. 8B). Taken together these data suggested that HMGA2 overexpression in CRC epithelial cells promoted angiogenesis by enhancing VEGFA and suppressing SEMA3A simultaneously, which is responsible for the activated VEGFR2 on the surface of HUVECs to promote angiogenesis in CRC.
Although the survival outcome of CRC patients has dramatically increased due to the improvement of diagnostic and therapeutic strategies, up to 20 % of CRC patients showed metastatic phenotype upon initial diagnosis and at least 30 % of them will die of metastatic cancer[2], therefore making it one of the major public health issues worldwide[1].
Angiogenesis is the fundamental step of tumors transition from dormant to malignant state and it is recognized as one of the hallmarks of cancer. Rooted in the belief that blocking vessel supply starves tumors to death[5,6], it has become increasingly accepted that blocking tumor angiogenesis would provide cancer patients with maximal survival benefit[10]. Clinically different strategies for angiogenesis intervention are based on modulating key steps of the angiogenic process, including endothelial cell proliferation cell-matrix adhesion and metastasis[11]. It is well known that VEGF-A promoted tumor angiogenesis via banding to VEGFR2 on the vascular endothelial cells[12]. Indeed the monoclonal anti-VEGF antibody bevacizumab[13,14] and the second-generation multi targeted RTKIs sunitinib[15] and sorafenib[16] have prolonged the life of numerous cancer patients. However, drug resistance would be inevitably established out of unknown mechanisms. Thus, uncover novel anti-tumor angiogenesis pathways in CRC would provide potential target for clinical interventions.
Through bioinformatics and IHC analysis, we found that HMGA2 was significantly overexpressed especially in patients at advanced stage and associated with undesired disease progression. Specifically HMGA2 expression was positively correlated with the density of CD31 (marker of angiogenesis) in CRC patient tissues. Our data for the first time indicated that CNV played a role in regulating HMGA2 expression in COAD manifested as gain of copy number denoted high expression of HMGA2. CRC is featured by high heterogeneous property due to the varied types of cells such as epithelial cell, fibroblast, immune cell etc., constituting the tumorous niche[26]. Therefore to figure out which type of cell in CRC contribute to the high expression of HMGA2 we analyzed a CRC single cell data set (including 9 CRC patient samples). After tSNE reduction and cell type annotation we found that colorectal epithelial cells showed high expression of HMGA2 among 7 types of cells. Consequently we established HMGA2- deficient epithelial origin human CRC cell line to investigate the role of HMGA2 in CRC. TME was important for angiogenesis and it was maintained by cancer cells and stromal cells via the secretion of various cytokines, growth factors and other effector molecules[23]. Therefore CM from the HMGA2 KD and scramble cells were applied to culture HUVECs and the results showed that CM from HMGA2 KD inhibited cell proliferation and metastasis. To unveil the signal molecules involved in this process the levels of angiogenesis related secreted factors; HGF, endothelin-1, ANG, VEGF-A, VEGF-C, SEMA3A and PDGF-BB in the supernatant of HMGA2 KD cells were firstly detected. While HGF, endothelin-1, ANG, VEGF-C and PDGFBB were not changed significantly, VEGF-A level was lower while SEMA3A level was higher in the HMGA2 KD supernatant and this observation was further confirmed in the HMGA2 KD, CRC cells. Previous studies showed that VEGF-A was a major factor that regulates angiogenesis by activating the tyrosine kinase receptor VEGFR-2[28] while SEMA3A could competitively bond to VEGFR2 to suppress tumor angiogenesis[29]. The VEGFR2 pathway was significantly inhibited in the HUVECs by the treatment of CRC HMGA2 KD CM manifest as the decreased level of phosphorylated VEGFR2, ERK and AKT[30,31]. Taken together our study for the first time showed that HMGA2 was up regulated in colorectal epithelial cells due to copy number amplification and knock down HMGA2 in CRC epithelial cells could inhibit the proliferation and metastasis of HUVECs via decreasing VEGF-A and increasing SEMA3A.
In the current study, we showed that HMGA2 was up regulated in CRC patients and its expression was associated with clinical values furthermore we demonstrated that copy number amplification contribute to the up regulation of HMGA2 in CRC patients. Single cell data analysis showed that HMGA2 was specifically up regulated in the colorectal epithelial cells. Mechanism study indicated that knocking down of HMGA2 suppresses angiogenesis via dual regulation of VEGF-A and SEMA3A in CRC whereby inactivating VEGFR2 pathway in HUVECs. Thus HMGA2 might be a promising prognostic marker and target for treating advanced CRC patients.
Author contributions’:
Cancan Wang, Di Hu, Gendou Zhou, Yanyan Li and Qi Zhou have conceive the idea; Xiaoqiao Hu, Ruyan Liu and Hongbo Ma collected the patient’s samples and performed the IHC and Wei Wang, Miaomiao Tao and Zexin Wang scored the IHC results; Cancan Wang, Di Hu, Gendou Zhou and Xiaoqiao Hu conducted the most of the cell and molecular experiments; Meiyu Zhou and Guofa Xu performed the bio informatics analysis; Guofa Xu, Yanyan Li, and Qi Zhou supervised the experiment. All authors contributed to subsequent and final drafts of this manuscript and approved the final manuscript before publication. All authors are in agreement with the content of the manuscript.
Cancan Wang and Di Hu have contributed equally to this work.
Ethics approval:
All experimental animal procedures were approved by the Institutional Animal Care and Use Committee of Third Military Medical University, China. Patients are consent to participate this study by providing the tumor sample excised during the surgery. The study protocol was approved by the Ethical Committee of Southwest Hospital Third Military Medical University.
Funding:
This study was founded by the Research foundation of Chongqing Health committee (ZY201902003); Fuling research project (FLKJ2022BAN2038) and Natural science foundation cultivation project (FLYYGZKPY2022003).
Conflict of interests:
The authors declared no conflict of interests.
References
- Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin 2021;71:7-33.
- Mody K, Baldeo C, Bekaii-Saab T. Antiangiogenic therapy in colorectal cancer. Cancer J 2018;24(4):165-70.
[Crossref] [Google Scholar] [PubMed]
- Deng F, Zhou R, Lin C, Yang S, Wang H, Li W, et al. Tumor-secreted dickkopf2 accelerates aerobic glycolysis and promotes angiogenesis in colorectal cancer. Theranostics 2019;9(4):1001.
[Crossref] [Google Scholar] [PubMed]
- Redman JM, Rhea LP, Brofferio A, Whelpley M, Gulley JL, Gatti-Mays ME, et al. Successful 5-fluorouracil (5-FU) infusion re-challenge in a metastatic colorectal cancer patient with coronary artery disease who experienced symptoms consistent with coronary vasospasm during first 5-FU infusion. J Gastrointes Oncol 2019;10(5):1010-4.
[Crossref] [Google Scholar] [PubMed]
- Poleszczuk J, Hahnfeldt P, Enderling H. Therapeutic implications from sensitivity analysis of tumor angiogenesis models. PLoS One 2015;10(3):e0120007.
[Crossref] [Google Scholar] [PubMed]
- Meirson T, Gil-Henn H, Samson AO. Invasion and metastasis: The elusive hallmark of cancer. Oncogene 2020;39(9):2024-6.
[Crossref] [Google Scholar] [PubMed]
- Ntellas P, Dadouli K, Perivoliotis K, Sogka E, Pentheroudakis G, Ioannou M, et al. Microvessel density and impact of angiogenesis on survival of resected pancreatic cancer patients: A systematic review and meta-analysis. Pancreas 2019;48(2):233-41.
[Crossref] [Google Scholar] [PubMed]
- Saravanan S, Vimalraj S, Pavani K, Nikarika R, Sumantran VN. Intussusceptive angiogenesis as a key therapeutic target for cancer therapy. Life Sci 2020;252:117670.
- Wang Z, Dabrosin C, Yin X, Fuster MM, Arreola A, Rathmell WK, et al. Broad targeting of angiogenesis for cancer prevention and therapy. Semin Cancer Biol 2015;35:S224-S3.
[Crossref] [Google Scholar] [PubMed]
- Paredes F, Williams HC, San Martin A. Metabolic adaptation in hypoxia and cancer. Cancer Lett 2021;502:133-42.
[Crossref] [Google Scholar] [PubMed]
- Li J, Gao A, Zhang F, Wang S, Wang J, Wang J, et al. ILT3 promotes tumor cell motility and angiogenesis in non-small cell lung cancer. Cancer Lett 2021;501:263-76.
[Crossref] [Google Scholar] [PubMed]
- Ferrara N, Hillan KJ, Gerber HP, Novotny W. Discovery and development of bevacizumab, an anti-VEGF antibody for treating cancer. Nat Rev Drug Discov 2004;3(5):391-400.
[Crossref] [Google Scholar] [PubMed]
- Berretta M, Lleshi A, Zanet E, Bearz A, Simonelli C, Fisichella R, et al. Bevacizumab plus irinotecan-, fluorouracil-, and leucovorin-based chemotherapy with concomitant HAART in an HIV-positive patient with metastatic colorectal cancer. Oncol Res Treatment 2008;31(7):394-7.
[Crossref] [Google Scholar] [PubMed]
- Miller K, Wang M, Gralow J, Dickler M, Cobleigh M, Perez EA, et al. Paclitaxel plus bevacizumab vs. paclitaxel alone for metastatic breast cancer. New Engl J Med 2007;357(26):2666-76.
[Crossref] [Google Scholar] [PubMed]
- Paz-Ares L, García del Muro X, Grande E, González P, Brosa M, Díaz S. Cost-effectiveness analysis of sunitinib in patients with metastatic and/or unresectable gastrointestinal stroma tumours (GIST) after progression or intolerance with imatinib. Clin Transl Oncol 2008;10:831-9.
[Crossref] [Google Scholar] [PubMed]
- Escudier B, Eisen T, Stadler WM, Szczylik C, Oudard S, Siebels M, et al. Sorafenib in advanced clear-cell renal-cell carcinoma. New Engl J Med 2007;356(2):125-34.
[Crossref] [Google Scholar] [PubMed]
- Ebos JM, Lee CR, Cruz-Munoz W, Bjarnason GA, Christensen JG, Kerbel RS. Accelerated metastasis after short-term treatment with a potent inhibitor of tumor angiogenesis. Cancer Cell 2009;15(3):232-9.
[Crossref] [Google Scholar] [PubMed]
- Loges S, Mazzone M, Hohensinner P, Carmeliet P. Silencing or fueling metastasis with VEGF inhibitors: Antiangiogenesis revisited. Cancer Cell 2009;15(3):167-70.
[Crossref] [Google Scholar] [PubMed]
- Cleynen I, Van de Ven WJ. The HMGA proteins: A myriad of functions (Review). Int J Oncol 2008;32(2):289-305.
[Google Scholar] [PubMed]
- Fedele M, Battista S, Kenyon L, Baldassarre G, Fidanza V, Klein-Szanto AJ, et al. Overexpression of the HMGA2 gene in transgenic mice leads to the onset of pituitary adenomas. Oncogene 2002;21(20):3190-8.
[Crossref] [Google Scholar] [PubMed]
- Wang X, Wang J, Zhao J, Wang H, Chen J, Wu J. HMGA2 facilitates colorectal cancer progression via STAT3-mediated tumor-associated macrophage recruitment. Theranostics 2022;12(2):963.
[Crossref] [Google Scholar] [PubMed]
- Mansoori B, Mohammadi A, Ditzel HJ, Duijf PH, Khaze V, Gjerstorff MF, et al. HMGA2 as a critical regulator in cancer development. Genes 2021;12(2):269.
[Crossref] [Google Scholar] [PubMed]
- Mao X, Xu J, Wang W, Liang C, Hua J, Liu J, et al. Crosstalk between cancer-associated fibroblasts and immune cells in the tumor microenvironment: New findings and future perspectives. Mol Cancer 2021;20(1):1-30.
[Crossref] [Google Scholar] [PubMed]
- Kobayashi A. Roles of NRF3 in the hallmarks of cancer: Proteasomal inactivation of tumor suppressors. Cancers 2020;12(9):2681.
[Crossref] [Google Scholar] [PubMed]
- Chen X, Zeng K, Xu M, Liu X, Hu X, Xu T, et al. P53-induced miR-1249 inhibits tumor growth, metastasis and angiogenesis by targeting VEGFA and HMGA2. Cell Death Dis 2019;10(2):131.
[Crossref] [Google Scholar] [PubMed]
- Fessler E, Medema JP. Colorectal cancer subtypes: Developmental origin and microenvironmental regulation. Trends Cancer 2016;2(9):505-18.
[Crossref] [Google Scholar] [PubMed]
- Gu Y, Chen Y, Wei L, Wu S, Shen K, Liu C, et al. ABHD5 inhibits YAP-induced c-Met overexpression and colon cancer cell stemness via suppressing YAP methylation. Nat Commun 2021;12(1):6711.
[Crossref] [Google Scholar] [PubMed]
- Liu H, Mei FC, Yang W, Wang H, Wong E, Cai J, et al. Epac1 inhibition ameliorates pathological angiogenesis through coordinated activation of Notch and suppression of VEGF signaling. Sci Adv 2020;6(1):eaay3566.
[Crossref] [Google Scholar] [PubMed]
- Acevedo LM, Barillas S, Weis SM, Göthert JR, Cheresh DA. Semaphorin 3A suppresses VEGF-mediated angiogenesis yet acts as a vascular permeability factor. Blood 2008;111(5):2674-80.
[Crossref] [Google Scholar] [PubMed]
- Xu X, Wu L, Zhou X, Zhou N, Zhuang Q, Yang J, et al. Cryptotanshinone inhibits VEGF-induced angiogenesis by targeting the VEGFR2 signaling pathway. Microvasc Res 2017;111:25-31.
[Crossref] [Google Scholar] [PubMed]
- Dellinger MT, Brekken RA. Phosphorylation of Akt and ERK1/2 is required for VEGF-A/VEGFR2-induced proliferation and migration of lymphatic endothelium. PloS One 2011;6(12):e28947.
[Crossref] [Google Scholar] [PubMed]
 ): NC and (
): NC and ( ): HMGA2 KD
): HMGA2 KD