- *Corresponding Author:
- Xiaofeng Cang
Department of Rheumatology and Immunology, Suzhou Ninth People's Hospital, Suzhou, Jiangsu 215000, China
E-mail: taotao10222023@163.com
| This article was originally published in a special issue,“Exploring the Role of Biomedicine in Pharmaceutical Sciences” |
| Indian J Pharm Sci 2024:86(1) Spl Issue “1-11” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The active ingredients in Tripterygium wilfordii have anti-cancer, anti-inflammatory and many other effects. The main objective of this study is to explore the therapeutic mechanism of Tripterygium wilfordii in the treatment of systemic lupus erythematosus using network pharmacology and molecular docking techniques. The active components and targets of Tripterygium wilfordii bark were screened by using traditional Chinese medicine system pharmacology database, high-throughput experiment- and reference-guided database, PubChem and SwissTargetPrediction, and systemic lupus erythematosus disease targets were screened by GeneCards website. Bioconcentration analysis of relevant targets was conducted to derive a possible mechanism of action of Tripterygium wilfordii active ingredients on systemic lupus erythematosus. The molecular docking results showed that the docking of the core active ingredients of Tripterygium wilfordii with the core targets was successfully validated and the docking activity was good. The therapeutic effects of Tripterygium wilfordii on systemic lupus erythematosus are not limited to a single target and a single pathway, but rather act synergistically on multiple pathways to alleviate the inflammatory response and maintain metabolic and immune homeostasis. The present study provides preliminary insights into the biological processes and signaling pathways involved in the therapy of SLE with Tripterygium wilfordii, which will be a theoretical reference and basis for further studies.
Keywords
Tripterygium wilfordii, systemic lupus erythematosus, network pharmacology, molecular docking, nanostructured lipid carriers
Systemic Lupus Erythematosus (SLE) is a multifaceted autoimmune disease characterized by chronic inflammation and abnormalities of the immune system. It affects multiple organs and has various clinical manifestations such as lymphopenia, kidney injury, pleurisy, meningitis, keratitis and gastrointestinal symptoms[1,2]. Glucocorticoids and immunosuppressant’s are commonly used in clinical treatment of Western medicine, but they often bring serious adverse reactions[3,4]. Modern Chinese medicine considers that SLE is a result of the accumulation of heat toxin in the body, or because the patient's body has been deficient for a long time. When it is invaded by external pathogens, it causes the imbalance of the five internal organs, and yin and yang, thus causing the disease[5]. At present, most Traditional Chinese Medicine (TCM) has the characteristics of low toxicity and high efficiency, and the adverse reactions are mild, which also brings new ideas for its clinical application in the therapy of SLE. Active ingredients extracted from the root, leaf and flower of Tripterygium wilfordii belongs to the Celastraceae family. It has antiinflammatory and detoxification effects. Studies have suggested that Tripterygium wilfordii may have a significant effect as an anti-inflammatory agent. Tripterygium wilfordii preparations also have immunosuppressive, anti-inflammatory, antitumor and other biological activities, and have great potential for the treatment of SLE[6-8]. Numerous studies have confirmed that either by the use of different solvents or by changing the concentration of celastrol for assembling them into nanoparticles with controllable size using anti-solvent method may efficiently target the delivery of triptolides and further improve the effect of active ingredients of Tripterygium wilfordii[9,10].
However, TCM and its compound preparations have the characteristics of complex components, numerous pathways and targets, and synergistic effects. Network Pharmacology (NP) is based on the 'disease-gene-target-drug' interaction network. Using omics, system biology, gene connectivity and redundancy, gene pleiotropy, computational biology and network biology analysis, we can explore the correlation between drugs and diseases, discover drug targets, and guide new drug development more systematically and comprehensively[11]. In this report, a NP approach was used to predict, analyze the active components and action targets associated with SLE in Tripterygium wilfordii, to explore the main mechanisms of action of Tripterygium wilfordii in the therapy of SLE and to provide theoretical support for its application in clinical practice.
Materials and Methods
Collection of active components and corresponding targets:
The chemistry of Tripterygium wilfordii was searched in the “herb name” entry using the Traditional Chinese Medicine System Pharmacology (TCMSP) database and analysis platform with “Tripterygium wilfordii” as the keyword. Following the pharmacokinetic analysis of Absorption, Distribution, Metabolism, and Excretion (ADME) parameters of the compound, combined with the later dose analysis, the Oral Bioavailability (OB)≥30 % and Drug-Likeness (DL)≥0.18 was set, and the main active ingredients of Tripterygium wilfordii were screened[12]. The chemical structure of the active ingredients was obtained by using PubChem database. The active components information was imported from the High-throughput Experiment- and Reference-guided database (HERB) platform[13] and SwissTargetPrediction was used for active ingredient target prediction[14]. The target with a score (probability) >0.1 was selected in the SwissTargetPrediction platform as the corresponding target of the active ingredient of Tripterygium wilfordii.
Triptolide treatment of SLE target collection:
The GeneCards database was searched using “systemic lupus erythematosus” as a keyword to screen for SLE disease targets. The respective targets of the compounds of Tripterygium wilfordii were intersected with the respective targets of SLE to obtain the effective compounds of Tripterygium wilfordii and their respective targets of SLE.
Construction of Protein-Protein Interaction (PPI) network:
PPI is a network of individual proteins linked by interactions that can be used to study the molecular machinery of disease more systematically and comprehensively, and identify new drug genes. The SLE-related targets which correspond to the compounds in Tripterygium wilfordii were entered into the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database. The 'Hide unconnected nodes in the network' option was selected. After deleting the non-interacting proteins, the PPI data source was downloaded and imported into Cytoscape 3.9.1 tools. The network analysis was performed using the Network Analysis plug-in. The degree of the nodes was obtained and the PPI network diagram was plotted, and the target protein was found where the standard was considered greater than the average degree of the nodes[15].
Drug-Active Ingredient-Target Network (DAIT-N):
The targets and compounds not related to the condition were eliminated, and then the remaining compounds from Tripterygium wilfordii and the SLE targets were imported into Cytoscape 3.9.1 tools to construct a DAIT-N to show the relationship between compounds and SLE genes. By using the Network Analysis plug-in for network analysis, the effective component node degree was obtained and the core active components were screened.
Target enrichment analysis:
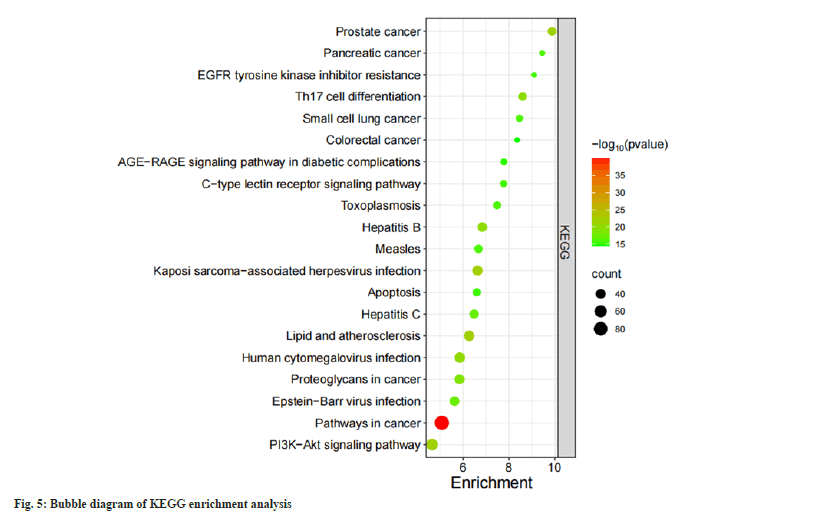
The Database for Annotation, Visualization and Integrated Discovery (DAVID) online analysis tool was used to perform Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis on SLE-related genes in the active components of Tripterygium wilfordii. GO analysis included Molecular Function (MF), Cellular Component (CC) and Biological Process (BP) and p<0.05 was limited for screening[15]. The top 10 of the three categories were drawn by the number of enriched genes. KEGG pathway enrichment was limited to p<0.05 for screening[15], and the top 20 data were imported into the Microbial Platform to draw the bubble diagram.
Molecular docking:
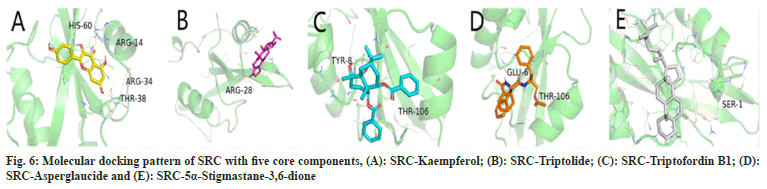
The resulting core compounds from Tripterygium wilfordii were molecularly docked to core functional targets. The molecule file in mol2 format was obtained from the TCMSP website. The core target genes structure was obtained from the Protein Data Bank (PDB) database in .pdb format file. Autodock Vina tools was used for molecular docking[16], and pictures of docking results processed by Proprietary Molecular visualization system (PyMOL) tools. In the molecular docking, the drug acts as the binding ligand and the protein transcribed and translated by the core target acts as the receptor. When the ligand-receptor binding energy is lower, the binding is more robust. It is generally believed that the lowest energy of binding is <0 kcal/mol with docking activity[17].
Results and Discussion
A total of 51 compounds were retrieved from the TCMSP database and a total of 1701 targets were obtained by HERB and Swiss Target Prediction. After summarizing and removing duplicates, a total of 660 targets were obtained, and then the compounds without targets were eliminated. A total of 44 compounds were obtained, mainly mairin, hederagenin, kaempferol, triptolide, triptinin B and isoxanthohumol, etc
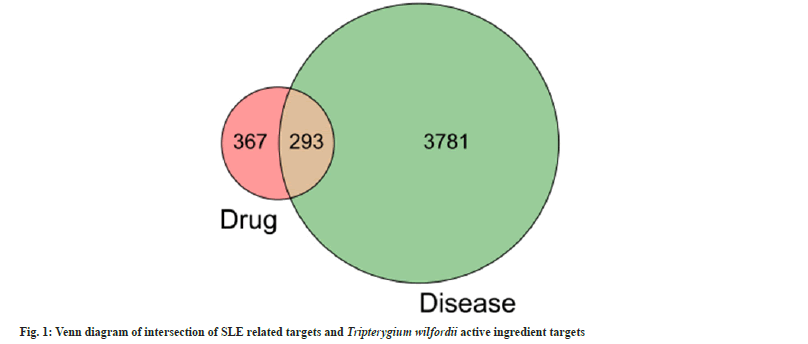
Using “systemic lupus erythematosus” as the key search string, 4074 SLE disease targets were retrieved from the GeneCards database. The targets corresponding to the compounds of Tripterygium wilfordii were intersected with SLE-related targets, and a grand total of 293 compounds of Tripterygium wilfordii and their respective SLErelated targets were obtained as shown in fig. 1. There are 36 active ingredients with therapeutic effect on SLE (Table 1).
| Molecular ID | Active ingredient | OB (%) | DL | Molecular formula | PubChem Id |
|---|---|---|---|---|---|
| MOL000211 | Mairin | 55.38 | 0.78 | C30H48O3 | 64971 |
| MOL000296 | Hederagenin | 36.91 | 0.75 | C30H48O4 | 73299 |
| MOL000358 | Beta-sitosterol | 36.91 | 0.75 | C29H50O | 222284 |
| MOL000422 | Kaempferol | 41.88 | 0.24 | C15H10O6 | 5280863 |
| MOL000449 | Stigmasterol | 43.83 | 0.76 | C29H48O | 5280794 |
| MOL002058 | 40957-99-1 | 57.2 | 0.62 | ||
| MOL003182 | (+)-Medioresinol di-O-beta-D-glucopyranoside_qt | 60.69 | 0.62 | C33H44O17 | 1.4E+07 |
| MOL003187 | Triptolide | 51.29 | 0.68 | C20H24O6 | 107985 |
| MOL003189 | Wilforlide A | 35.66 | 0.72 | C30H46O3 | 158477 |
| MOL003192 | Triptonide | 67.66 | 0.7 | C20H22O6 | 65411 |
| MOL003196 | Tryptophenolide | 48.5 | 0.44 | C20H24O3 | 173273 |
| MOL003198 | 5α-Benzoyl-4α-hydroxy-1 beta, 8α-dinicotinoyl-dihydro-agarofuran | 35.26 | 0.72 | C34H36N2O8 | 1.6E+08 |
| MOL003199 | 5,8-Dihydroxy-7-(4-hydroxy-5-methyl-coumarin-3)-Coumarin | 61.85 | 0.54 | C18H36O2 | 5281 |
| MOL003208 | Celafurine | 72.94 | 0.44 | C21H27N3O3 | 1.6E+08 |
| MOL003209 | Celallocinnine | 83.47 | 0.59 | C25H31N3O2 | 1.6E+08 |
| MOL003217 | Isoxanthohumol | 56.81 | 0.39 | C21H22O5 | 513197 |
| MOL003225 | Hypodiolide A | 76.13 | 0.49 | C20H30O3 | 72369 |
| MOL003229 | Triptinin B | 34.73 | 0.32 | C20H26O3 | 1E+07 |
| MOL003231 | Ttriptoditerpenic acid B | 40.02 | 0.36 | C21H28O3 | 192372 |
| MOL003232 | Triptofordin B1 | 39.55 | 0.84 | C29H34O6 | 1.2E+08 |
| MOL003238 | Triptofordin F1 | 33.91 | 0.6 | C37H42O13 | 1.6E+08 |
| MOL003239 | Triptofordin F2 | 33.62 | 0.67 | C35H40O13 | 1.4E+07 |
| MOL003241 | Triptofordin F4 | 31.37 | 0.67 | C35H40O12 | 1.6E+08 |
| MOL003242 | Triptofordinine A2 | 30.78 | 0.47 | C41H43NO12 | 1.6E+08 |
| MOL003244 | Triptonide | 68.45 | 0.68 | C20H22O6 | 65411 |
| MOL003245 | Triptonoditerpenic acid | 42.56 | 0.39 | C21H28O4 | 132263 |
| MOL003248 | Triptonoterpene | 48.57 | 0.28 | C20H28O2 | 1.6E+08 |
| MOL003266 | 21-Hydroxy-30-norhopan-22-one | 34.11 | 0.77 | C29H48O2 | 5318299 |
| MOL003278 | Salaspermic acid | 32.19 | 0.63 | C30H48O4 | 4.5E+07 |
| MOL003280 | Triptonolide | 49.51 | 0.49 | C20H22O4 | 4.6E+07 |
| MOL003283 | (2R,3R,4S)-4-(4-hydroxy-3-methoxy-phenyl)-7-methoxy-2,3-dimethylol-tetralin-6-ol | 66.51 | 0.39 | C20H24O6 | 160521 |
| MOL004443 | Zhebeiresinol | 58.72 | 0.19 | C14H16O6 | 192547 |
| MOL005828 | Nobiletin | 61.67 | 0.52 | C21H22O8 | 72344 |
| MOL007415 | Asperglaucide | 58.02 | 0.52 | C27H28N2O4 | 1E+07 |
| MOL007535 | 5α-Stigmastane-3,6-dione | 33.12 | 0.79 | C29H48O2 | 1.4E+07 |
| MOL011169 | Peroxyergosterol | 44.39 | 0.82 | C28H44O3 | 5351516 |
Table 1: 36 Active Components of Tripterygium Wilfordii in The Treatment of SLE
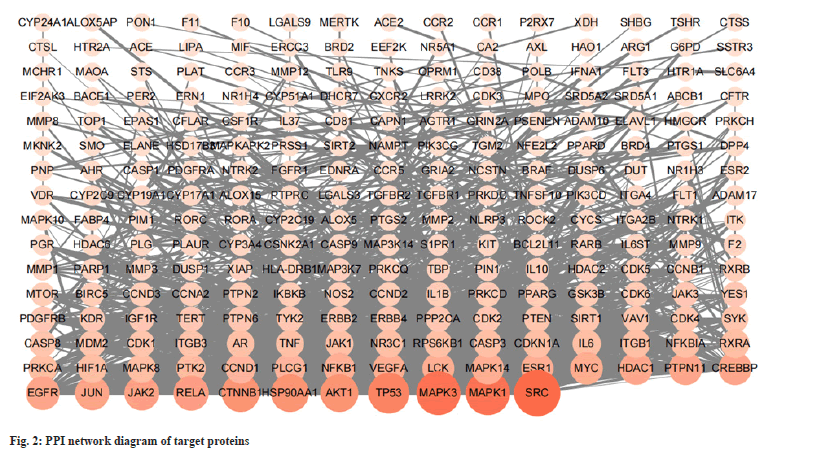
The target of Tripterygium wilfordii in the treatment of SLE was imported into the STRING website to obtain the target PPI network (fig. 2). The relevant PPI network source data was downloaded and loaded into the Cytoscape 3.9.1 software. Using the analysis network, the menu bar selected tools configures the node size and color to follow the node degree for continuous change. Then the edge thickness was configured to follow the combined score for continuous change, according to the node degree. The target gene is arranged in a rectangular arrangement. This network diagram includes 236 nodes and 1244 edges. As degrees increase, node shape increases and color deepens. According to the target PPI network diagram, the average degree of the relevant nodes was calculated to be 10.54. According to the principle that the initial screening was greater than the average degree of the nodes, 80 targets with a node degree >10 were selected as the key research which includes Proto-oncogene non-receptor tyrosine kinase (SRC), Mitogenactivated protein kinase 1 (MAPK1), MAPK3, Tumor Protein p53 (TP53) and Serine/threonine kinase 1 (AKT1), etc., and these targets were considered to be related to the treatment of SLE by Tripterygium wilfordii. The top 5 core targets of node degree and the corresponding compounds are listed in Table 2.
| Gene | Degree | Active ingredients of Tripterygium wilfordii |
|---|---|---|
| SRC | 63 | Kaempferol |
| Triptonoterpene | ||
| 5α-Stigmastane-3,6-dione | ||
| MAPK1 | 58 | Celafurine |
| Isoxanthohumol | ||
| (2R,3R,4S)-4-(4-hydroxy-3-methoxy-phenyl)-7-methoxy-2,3-dimethylol-tetralin-6-ol | ||
| Triptolide | ||
| MAPK3 | 58 | (2R,3R,4S)-4-(4-hydroxy-3-methoxy-phenyl)-7-methoxy-2,3-dimethylol-tetralin-6-ol |
| Triptolide | ||
| TP53 | 51 | Celallocinnine |
| 5α-Stigmastane-3,6-dione | ||
| AKT1 | 44 | Kaempferol |
| Triptolide |
Table 2: Top 5 core target Genes of Node Degree and the corresponding active compounds
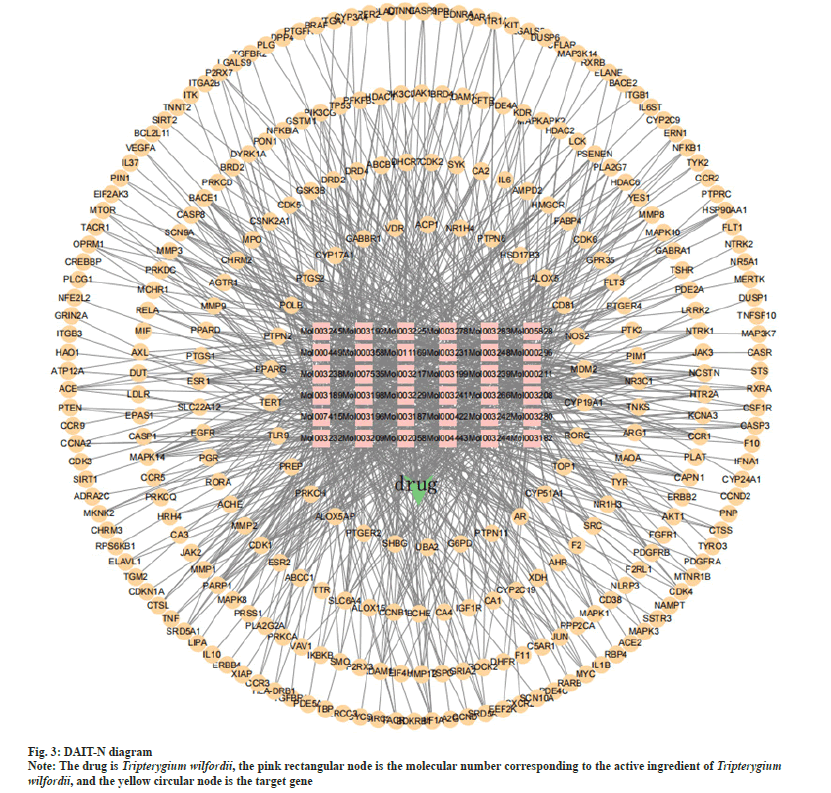
The drug components of Tripterygium wilfordii and the protein targets of SLE were imported into Cytoscape 3.9.1 to construct a DAIT-N. The network consists of 329 nodes and 908 edges. It can be seen from fig. 3 that Tripterygium wilfordii exerts its therapeutic effect on SLE mainly through multiple components corresponding to multiple target genes. According to the topological calculation results of the network graph, the degree of all nodes in the network was 5.45, and the average degree of the active components was 25.2. The top five components according to the degree of nodes were 5alpha (α)-Stigmastane-3,6- dione, asperglaucide, triptofordin B1, triptolide, kaempferol. The corresponding nodal degrees are 61, 59, 56, 55, 55, which are the main active constituents of Tripterygium wilfordii in the therapy of SLE.
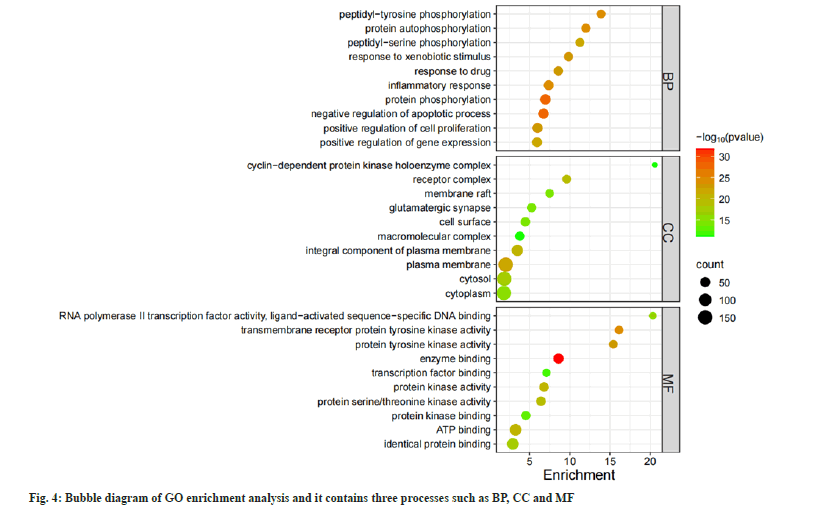
808 BPs were obtained in GO enrichment analysis. It is mainly manifested in protein phosphorylation responses, control of cell growth and apoptosis, and response to inflammation. There were 105 CCs, mainly involving in plasma membrane, receptor complex, protein kinase and other components. There are 174 MFs, mainly involving in enzyme binding, Adenosine Triphosphate (ATP) binding transmembrane receptor protein tyrosine kinase activity, protein tyrosine kinase activity and other functions (fig. 4). There are 168 KEGG enrichment pathways and the bubble diagram illustrates the main pathways (fig. 5). These pathways involve cell proliferation regulation, metabolic response, inflammatory response and other processes.
In this study, 5α-Stigmastane-3,6-dione, asperglaucide, triptofordin B1, triptolide and kaempferol were docked with SRC, MAPK1, MAPK3, TP53 and AKT1 respectively. The docking results indicate that the minimum binding energies of both the target protein and the corresponding component molecule are in the range of -4 kcal/ mol to -11 kcal/mol and indicate that there was good interaction between the target proteins and the component molecules (Table 3). Fig. 6 shows the molecular docking pattern of the protein target SRC with the highest-level node degree in the structure network as a receptor of the five core components.
| Active ingredient | SRC | MAPK1 | MAPK3 | TP53 | AKT1 |
|---|---|---|---|---|---|
| 5α-Stigmastane-3,6-dione | -7.7 | -9.32 | -10.05 | -9.71 | -10.8 |
| Asperglaucide | -5.13 | -6.28 | -8.02 | -7.93 | -7.22 |
| Triptofordin B1 | -7.78 | -7.79 | -9.22 | -9.43 | -8.13 |
| Triptolide | -6.11 | -6.97 | -9.28 | -7.52 | -8.8 |
| Kaempferol | -5.86 | -7.04 | -7.74 | -6.23 | -7.22 |
Table 3: Binding energy of core drug active components and core protein (Kcal/Mol)
The TCM-NP research method was used to explore and analyze the mechanism of action of Tripterygium wilfordii in SLE using relevant websites and tools. According to the results of the study, there were 51 potential activated ingredients of Tripterygium wilfordii in the therapy of SLE, and 36 compounds without targets were excluded among 293 potential targets. Starting from these targets, the BP and pathways of the therapeutic effect of Tripterygium wilfordii on SLE were further gathered. Molecular docking techniques were used to investigate the binding of proteins to small molecules in order to elucidate the therapeutic mechanism, which offer evidence for the therapy of SLE, and also provide a new way of thinking for the development of Tripterygium wilfordii drug formulations and delivery vehicles.
Based on the DAIT-N, it was found that the main ingredients of Tripterygium wilfordii against SLE were 5α-Stigmastane-3,6-dione, asperglaucide, triptofordin B1, triptolide, kaempferol. Triptolide is widely used to treat a wide range of inflammatory diseases such as SLE, Rheumatoid Arthritis (RA), psoriasis, etc. Its anti-inflammatory safety and efficacy have been confirmed by many studies. Zhao et al. found that triptolide can induce micro Ribonucleic acid-125a-5p (miR-125a-5p) by mediating Regulatory T (TReg) cells upregulation to improve lupus[18]. In addition, triptolide can upregulate the expression of anti-inflammatory factor Interluekin-37 (IL-37) by activating Extracellular signal-Regulated Kinase 1/2 (ERK1/2) and p38 MAPK pathways[19]. To better explain the mechanism of action of triptolide in SLE, Zhang et al. concluded that triptolide could decrease the amount of double standard-Deoxyribonucleic Acid (ds-DNA) and Immunglobulin G (IgG) in B cells of model mice, reduce the pathological damage of kidney tissue, upregulate the expression of miR- 146a, block the Toll-like receptor 7/Nuclear Factor kappa B (TLR7/NF-κB) signaling pathway, affect the activation of B cells, and play a significant role in the therapy of SLE[20]. Kaempferol is a flavonoid found naturally in many fruits and vegetables. Epidemiological studies have shown that taking kaempferol may reduce the chance of developing cancers of the lung, stomach, pancreas and ovaries. The use of kaempferol has a therapeutic effect in preventing and treating of inflammatory diseases such as SLE, RA and Ankylosing Spondylitis (AS) [21]. It has been documented that kaempferol can reduce the inflammatory activation of the TLR4/ NF-κB pathway, inhibit the overexpression of inflammatory cytokines such as Tumor Necrosis Factor-α (TNF-α), IL-1, IL-6 and monocyte chemoattractant protein-1, improve the integrity of the intestinal barrier, inhibit intestinal inflammation and maintain the intestinal microbial balance[22]. There are few studies on triptofordin B1 and 5α-Stigmastane-3,6-dione, asperglaucide in SLE, which may offer new ideas for drug therapy in SLE. In the study of drug utilization, the polyphenolic substance celastrol in Tripterygium wilfordii has anti-tumour and anti-inflammatory properties, but its clinical use is limited by its poor solubility and bioavailability. However, by constructing a highly soluble potential of Hydrogen (pH)- responsive nanoparticle drug for intra-articular injectable osteoarthritis treatment, the benefit to patients can be increased[23]. Therefore, the core active ingredient of Tripterygium wilfordii in the treatment of SLE is also expected to achieve better clinical therapeutic effect in the future combined with new materials.
The PPI network diagram and topological calculation results showed that the top 5 target proteins of node degree included SRC, MAPK1, MAPK3, TP53 and AKT1. SRC is a tyrosine kinase involved in a variety of cellular processes[24] and it has been reported to mediate inflammatory responses. Animal experiments have shown that inhibition of SRC expression can reduce microglia-induced neuroinflammation and has a protective effect on a mouse model of neuropathic pain[25]. At the same time, studies have shown that abnormal immune metabolism of Clusters of differentiation 4+ (CD4+) T cells and their subsets can affect the occurrence and development of SLE[26], and the expression of SRC is also related to cell metabolism regulation[27]. MAPK1 and MAPK3 are part of the MAPK family, which is widespread in mammals. They are related to cell proliferation, differentiation, cytokine synthesis and apoptosis. MAPK3 kinase abnormalities can cause peripheral blood T lymphocyte immunodeficiency, suggesting involvement in the pathogenesis of SLE[28,29]. Studies have shown that protein kinase is related to the immunopathological mechanism of SLE. The expression of MAPK1 in SLE patients is enhanced, which is related to the expression of inflammatory factor IL-10 indicating that MAPK1 is implicated in the control of the inflammatory response in patients[30]. MAPK1 is also a target gene regulated by many inflammatory diseases. For example, miR-320c suppresses the growth of articular chondrocytes and induction of apoptosis by targeting MAPK1, which is involved in the pathogenesis of osteoarthritis[31]; miR-129-5p can reduce the inflammatory response in the hippocampus of depressed mice by negatively regulating MAPK1[32]. TP53 protein is a transcription factor that inhibits tumor formation and plays an important biological role in promoting DNA damage repair, inducing apoptosis, senescence and cell cycle arrest[33]. Studies have found that SLE patients have a significantly higher risk of certain cancers than the general population, most commonly haematological malignancies, lung cancer and thyroid cancer[34]. AKT1 is involved in many BPs, including metabolism, proliferation, cell growth and angiogenesis. AKT1 activation depends on the Phosphoinositide 3-Kinase (PI3K) pathway, which can activate downstream inflammatory mediators from IL-17 to IL-4, inhibit TReg cells and be implicated in the pathogenesis of SLE[35].
The results of the GO analysis show that the target genes are involved in BPs such as protein phosphorylation, cell proliferation, differentiation, apoptosis cycle regulation and response to inflammation; CCs such as plasma membrane, receptor complex and protein kinases; MFs such as binding reactions of biological enzymes related to ATP energy and various protein kinase activities.
Among the 20 KEGG enrichment pathways of related targets, most of the targets were enriched in cancer related pathways like cancer pathways, prostate cancer related pathways, cancer proteoglycan, small cell lung cancer related pathways and colorectal cancer related pathways. Some targets were significantly enriched in viral infection-related pathways, such as hepatitis B pathway, hepatitis C pathway, human Cytomegalovirus (CMV) infection pathway, Epstein-Barr virus (EBV) infection pathway. Many scholars believe that viral infection is an essential factor in the pathogenesis and progress of SLE and increases the mortality of SLE patients, including Hepatitis B Virus, EBV, CMV, Measles virus, respiratory Enterovirus and so on[36,37]. In addition, some genes were also enriched in lipid and atherosclerosis-related pathways, PI3Kprotein kinase B (Akt) pathway and C-type Lectin Receptor (CLR) pathway. The abnormal inflammatory response of SLE itself is associated with the development of atherosclerosis. The study found that SLE and atherosclerosis share 246 common genes, including 189 upregulated and 57 downregulated genes, which are mainly enriched in TNF-α, IL-17 and NF-κB signaling pathway. The molecular mechanism underlying the association between SLE and atherosclerosis may be the above-mentioned pathways[38]. The PI3KAkt signaling pathway is a major signaling pathway in cells and it is closely related to mammalian Target of Rapamycin (mTOR). Akt and mTOR are both serine/threonine protein kinases. The interaction between the two can cause the increase of related inflammatory factors and the occurrence of pathological immune status and participate in the pathogenesis of SLE[39]. The use of mTOR inhibitor can stimulate the activity of T-cells, reduce the release of pro-inflammatory agents and increase the release of TReg, thereby alleviating the course of SLE and reducing relapses[40]. The CLR signaling pathway was implicated in inflammation, immunity and homeostasis regulation. Its dectin-1 receptor regulates the differentiation of T helper 17 (Th17) cells with the help of a series of factors. Th17 cells are a new type of helper T cells that can secrete specific effector molecule IL-17, which can mediate inflammatory reactions, tumors and autoimmune diseases (such as SLE), etc.[41].
In conclusion, this study applied the NP and molecular docking technology to theoretically explore the complex mechanism of the multiplecomponent, multiple-target, multiple-pathway active components of Tripterygium wilfordii in the therapy of SLE. The main constituents of Tripterygium wilfordii include asperglaucide, triptofordin B1, triptolide, 5α-stigmastane-3,6- dione and kaempferol, etc., which synergistically act on disease-related SRC, MAPK1, MAPK3, TP53, AKT1 and other target proteins to regulate cancer-related pathways, virus infection-related pathways, lipid and atherosclerosis-related pathways, PI3K-Akt signaling pathway and CLR signaling pathway. They play a part in reducing the inflammatory response and maintaining metabolic immune balance. In this way, the characteristics of the multicomponent, multi-target, multi-pathway therapy of diseases in TCM are fully exploited, and ideas and the basis for further research are provided. However, because it is a predictive discussion, the specific mechanism of Tripterygium wilfordii still needs further experiments to verify.
Conflict of interests:
The authors declared no conflict of interests.
References
- Liossis SN, Staveri C. What's new in the treatment of systemic lupus erythematosus. Front Med 2021;8:1-13.
[Crossref] [Google scholar] [PubMed]
- Barnett R. Systemic lupus erythematosus. Lancet 2016;387(10029):1711.
[Crossref] [Google scholar] [PubMed]
- Basta F, Fasola F, Triantafyllias K, Schwarting A. Systemic Lupus Erythematosus (SLE) therapy: The old and the new. Rheumatol Ther 2020;7(3):433-46.
[Crossref] [Google scholar] [PubMed]
- Popa R, Lautaru LA, Lucretiu R, Ruiu DC, Caragea D, Olteanu M, et al. Therapy side effects in systemic lupus erythematosus. Curr Health Sci J 2018;44(3):316-21.
[Crossref] [Google scholar] [PubMed]
- Ma YC, Lin CC, Li CI, Chiang JH, Li TC, Lin JG. Traditional Chinese medicine therapy improves the survival of systemic lupus erythematosus patients. Semin Arthritis Rheum 2016;45(5):596-603.
[Crossref] [Google scholar] [PubMed]
- Chen SR, Dai Y, Zhao J, Lin L, Wang Y, Wang Y. A mechanistic overview of triptolide and celastrol, natural products from Tripterygium wilfordii Hook F. Front Pharmacol 2018;9:1-13.
[Crossref] [Google scholar] [PubMed]
- Law SK, Simmons MP, Techen N, Khan IA, He MF, Shaw PC, et al. Molecular analyses of the Chinese herb Leigongteng (Tripterygium wilfordii Hook. f.). Phytochemistry 2011;72(1):21-6.
[Crossref] [Google scholar] [PubMed]
- Chen Y, Gong Z, Chen X, Tang L, Zhao X, Yuan Q, et al. Tripterygium wilfordii Hook F (a traditional Chinese medicine) for primary nephrotic syndrome. Cochrane Database Syst Rev 2013;11(8):1-45.
[Crossref] [Google scholar] [PubMed]
- Li JX, Zhang MJ, Shi JF, Wang SP, Zhong XM, Wu YH, et al. pH-sensitive nano-polyelectrolyte complexes with arthritic macrophage-targeting delivery of triptolide. Int J Pharm 2023;632:122572.
[Crossref] [Google scholar] [PubMed]
- Liu Y, Li J. Self-assembling nanoarchitectonics of size-controllable celastrol nanoparticles for efficient cancer chemotherapy with reduced systemic toxicity. J Colloid Interface Sci 2023;636:216-22.
[Crossref] [Google scholar] [PubMed]
- Li S, Fan TP, Jia W, Lu A, Zhang W. Network pharmacology in traditional Chinese medicine. Evid Based Complement Alternat Med 2014;2014:1-3.
[Crossref] [Google scholar] [PubMed]
- Xiao L, Xiao W, Zhan F. Targets of Tripterygium glycosides in systemic lupus erythematosus treatment: A network-pharmacology study. Lupus 2022;31(3):319-29.
[Crossref] [Google scholar] [PubMed]
- Fang S, Dong L, Liu L, Guo J, Zhao L, Zhang J, et al. HERB: A high-throughput experiment-and reference-guided database of traditional Chinese medicine. Nucleic Acids Res 2021;49(D1):D1197-206.
[Crossref] [Google scholar] [PubMed]
- Daina A, Michielin O, Zoete V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res 2019;47(W1):W357-64.
[Crossref] [Google scholar] [PubMed]
- Gao Y, Wang KX, Wang P, Li X, Chen JJ, Zhou BY, et al. A novel network pharmacology strategy to decode mechanism of Lang Chuang Wan in treating systemic lupus erythematosus. Front Pharmacol 2020;11:1-21.
[Crossref] [Google scholar] [PubMed]
- Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010;31(2):455-61.
[Crossref] [Google scholar] [PubMed]
- Guedes IA, de Magalhães CS, Dardenne LE. Receptor-ligand molecular docking. Biophys Rev 2014;6(1):75-87.
[Crossref] [Google scholar] [PubMed]
- Zhao X, Tang X, Yan Q, Song H, Li Z, Wang D, et al. Triptolide ameliorates lupus via the induction of miR-125a-5p mediating TReg upregulation. Int Immunopharmacol 2019;71:14-21.
[Crossref] [Google scholar] [PubMed]
- He L, Liang Z, Zhao F, Peng L, Chen Z. Modulation of IL-37 expression by triptolide and triptonide in THP-1 cells. Cell Mol Immunol 2015;12(4):515-8.
[Crossref] [Google scholar] [PubMed]
- Zhang Y, Zhang F, Gao Y, Wang M, Gao Y, Li H, et al. Triptolide in the treatment of systemic lupus erythematosus-regulatory effects on miR-146a in B cell TLR7 signaling pathway in mice. Front Pharmacol 2022;13:1-10.
[Crossref] [Google scholar] [PubMed]
- Ren J, Lu Y, Qian Y, Chen B, Wu T, Ji G. Recent progress regarding kaempferol for the treatment of various diseases. Exp Ther Med 2019;18(4):2759-76.
[Crossref] [Google scholar] [PubMed]
- Bian Y, Lei J, Zhong J, Wang B, Wan Y, Li J, et al. Kaempferol reduces obesity, prevents intestinal inflammation, and modulates gut microbiota in high-fat diet mice. J Nutr Biochem 2022;99:1-11.
[Crossref] [Google scholar] [PubMed]
- Jin T, Wu D, Liu XM, Xu JT, Ma BJ, Ji Y, et al. Intra-articular delivery of celastrol by hollow mesoporous silica nanoparticles for pH-sensitive anti-inflammatory therapy against knee osteoarthritis. J Nanobiotechnology 2020;18(1):1-5.
[Crossref] [Google scholar] [PubMed]
- Bagnato G, Leopizzi M, Urciuoli E, Peruzzi B. Nuclear functions of the tyrosine kinase Src. Int J Mol Sci 2020;21(8):1-14.
[Crossref] [Google scholar] [PubMed]
- Cai Y, Xu J, Cheng Q. Proto-oncogene tyrosine-protein kinase SRC (Src) inhibition in microglia relieves neuroinflammation in neuropathic pain mouse models. Bioengineered 2021;12(2):11390-8.
[Crossref] [Google scholar] [PubMed]
- Duarte-Delgado NP, Cala MP, Barreto A. Metabolites and metabolic pathways associated with rheumatoid arthritis and systemic lupus erythematosus. J Transl Autoimmun 2022;5:100150.
[Crossref] [Google scholar] [PubMed]
- Pelaz SG, Tabernero A. Src: Coordinating metabolism in cancer. Oncogene 2022;41(45):4917-28.
[Crossref] [Google scholar] [PubMed]
- Masselli E, Mecucci C, Gobbi G, Carubbi C, Pierini V, Sammarelli G, et al. Implication of MAPK 1/MAPK 3 signalling pathway in t (8; 9)(p22; 24)/PCM 1‐JAK 2 myelodysplastic/myeloproliferative neoplasms. Br J Haematol 2013;162(4):563-6.
[Crossref] [Google scholar] [PubMed]
- Bendix I, Pfueller CF, Leuenberger T, Glezeva N, Siffrin V, Müller Y, et al. MAPK3 deficiency drives autoimmunity via DC arming. Eur J Immunol 2010;40(5):1486-95.
[Crossref] [Google scholar] [PubMed]
- Garcia-Rodriguez S, Callejas-Rubio JL, Ortego-Centeno N, Zumaquero E, Ríos-Fernandez R, Arias-Santiago S, et al. Altered AKT1 and MAPK1 gene expression on peripheral blood mononuclear cells and correlation with T-helper-transcription factors in systemic lupus erythematosus patients. Mediators Inflamm 2012;2012:1-15.
[Crossref] [Google scholar] [PubMed]
- Zhao L, Zhou R, Wang Q, Cheng Y, Gao M, Huang C. MicroRNA‐320c inhibits articular chondrocytes proliferation and induces apoptosis by targeting Mitogen-Activated Protein Kinase 1 (MAPK1). Int J Rheum Dis 2021;24(3):402-10.
[Crossref] [Google scholar] [PubMed]
- Chang J, Zhang Y, Shen N, Zhou J, Zhang H. miR-129-5p prevents depressive-like behaviors by targeting MAPK1 to suppress inflammation. Exp Brain Res 2021;239(11):3359-70.
[Crossref] [Google scholar] [PubMed]
- Mijit M, Caracciolo V, Melillo A, Amicarelli F, Giordano A. Role of p53 in the regulation of cellular senescence. Biomolecules 2020;10(3):1-16.
[Crossref] [Google scholar] [PubMed]
- Hardenbergh D, Molina E, Naik R, Geetha D, Chaturvedi S, Timlin H. Factors mediating cancer risk in systemic lupus erythematosus. Lupus 2022;31(11):1285-95.
[Crossref] [Google scholar] [PubMed]
- Oaks Z, Winans T, Huang N, Banki K, Perl A. Activation of the mechanistic target of rapamycin in SLE: Explosion of evidence in the last five years. Curr Rheumatol Rep 2016;18(12):1-8.
[Crossref] [Google scholar] [PubMed]
- Iwata S, Tanaka Y. Association of viral infection with the development and pathogenesis of systemic lupus erythematosus. Front Med 2022;9:1-10.
[Crossref] [Google scholar] [PubMed]
- Krasselt M, Kippenhahn A, Baerwald C, Pietsch C, Seifert O. Relationship between cytomegalovirus prevalence and markers of disease activity in systemic lupus erythematosus. Rheumatology 2022;61(3):1288-90.
[Crossref] [Google scholar] [PubMed]
- Fan JL, Wu D, Zhu TT, Tian XL, Liu SJ, Zhang SL. The exploration of shared genes and molecular mechanisms of systemic lupus erythematosus and atherosclerosis. Lupus 2023;32(2):239-51.
[Crossref] [Google scholar] [PubMed]
- Tang H, Tan G, Guo Q, Pang R, Zeng F. Abnormal activation of the Akt-GSK3β signaling pathway in peripheral blood T cells from patients with systemic lupus erythematosus. Cell Cycle 2009;8(17):2789-93.
[Crossref] [Google scholar] [PubMed]
- Perl A. Activation of mTOR (mechanistic Target of Rapamycin) in rheumatic diseases. Nat Rev Rheumatol 2016;12(3):169-82.
[Crossref] [Google scholar] [PubMed]
- Vautier S, da Glória Sousa M, Brown GD. C-type lectins, fungi and Th17 responses. Cytokine Growth Factor Rev 2010;21(6):405-12.
[Crossref] [Google scholar] [PubMed]