- *Corresponding Author:
- M. X. Pan
Department of Hepatobiliary Surgery, Zhujiang Hospital, Southern Medical University/The Second School of Clinical Medicine, Southern Medical University, Guangzhou, Guangdong Province 510515, China
E-mail: pmxwxy@sohu.com
| This article was originally published in a special issue, “Recent Developments in Biomedical Research and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2022:84(4) Spl Issue “196-205” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
To investigate the effects of hepatocellular carcinoma cell-derived circ_0001649-mediated regulation of intercellular communication on the progression of hepatocellular carcinoma and their mechanisms is the main objective of the study. Immunohistochemical staining was used to detect the expression level of matrix metalloproteinase 9 in tumour and paracancer tissues. The expression levels of matrix metalloproteinase 9 messenger RNA, circ_0001649 and micro RNA-331-3p were measured by real-time fluorescence quantitative polymerase chain reaction. In vivo transfer models were established. Bagg Albino/c nude mice were injected subcutaneously with human hepatocellular carcinoma cell lines, Hep3B cell suspension, Hep3B cells stably shows overexpressing circ_0001649 and knockdown circ_0001649, respectively. The expression level of circ_0001649 in serum exosomes from hepatocellular carcinoma was significantly correlated with the expression level of matrix metalloproteinase 9 protein in tissues compared to serum exosomes from non-cancerous populations (p<0.05). Bioinformatics software was used to predict the target of circ_0001649 and micro RNA-331-3p and the 3’-untranslated region of micro RNA-331-3p binding site of matrix metalloproteinase 9 messenger RNA. There was no significant change in matrix metalloproteinase 9 protein expression when circ_0001649 and miR-331-3p were either up-regulated or down-regulated. In vivo experiments showed that the expression levels of circ_0001649 content and matrix metalloproteinase 9 proteins were reduced in the tumour of circ_0001649-knockdown group, while the expression levels of circ_0001649 content and matrix metalloproteinase 9 protein were significantly increased in the tumour of circ_0001649-over expression group. Circ_0001649 in hepatocellular carcinoma cell exosomes translocate to cancer-associated fibroblast and promoted matrix metalloproteinase 9 expressions and thus promote tumor metastasis through adsorption of micro RNA-331-3p.
Keywords
Circular RNA circ_0001649, micro ribonucleic acid-331-3p, exosome, hepatocellular carcinoma, matrix metalloproteinase 2, metastasis
Hepatocellular Carcinoma (HCC) is one of the leading causes of cancer deaths worldwide and the incidence of HCC tends to be younger[1]. A variety of risk factors (such as genetic factors, lifestyle, Hepatitis B Virus (HBV) infection and environmental influences) can contribute to the development of HCC[2]. Despite recent advances in HCC treatment (including chemotherapy and radiotherapy), the prognosis for HCC remains relatively poor[3], resulting in a relatively low survival rate for HCC patients, with an overall 5 y survival rate of 18 %[4]. Metastasis is the leading cause of death in patients with HCC, with portal vein and lung metastases accounting for approximately 60 % of these deaths[5], therefore the identification of molecular biomarkers associated with HCC metastasis is essential for the subsequent treatment of HCC and prolonging patient survival.
Exosomes are nanoscale (30-100 nm) extracellular vesicles that can be secreted by most types of cells and circulate in body fluids such as blood, urine, saliva and breast milk[6]. In recent years, studies have shown that exosomes have a broad role in the progression of HCC[7-9]. Exosomes have a wide range of contents and can encapsulate various growth factors, proteins, lipids, nucleic acids, as well as non-coding Ribonucleic Acids (RNAs) such as circular RNA (circRNA) and micro RNA (miRNA), and transport them to target cells. CircRNA is a novel endogenous RNA with a covalent closed-loop structure[10]. In cells, circRNA can not only bind to miRNAs or proteins to perform various functions, but can also be sorted into extracellular vesicles along with other nucleic acids, lipids and proteins. Exosomes are secreted from cells into body fluids, through which circRNAs begin their cycle and activate their biological functions.
Several studies have now found that circRNAs as miRNA molecular sponges are involved in the pathophysiological processes of proliferation, migration, invasion and apoptosis of HCC cells, and that certain circRNAs as miRNA molecular sponges are associated with radiotherapy sensitivity in HCC[11-13]. Circ_PRR34 Antisense RNA 1 (PRR34-AS1) activates the Translocase of Outer Mitochondrial Membrane 20 (TOMM20) and Integrin Subunit Alpha 6 (ITGA6) indirectly by adsorbing miR-498, thus promoting the progression of HCC[13]. Thus, the circRNA/miRNA/ target gene/target protein axis may be one of the important links in hepatocarcinogenesis. The aim of this study was to investigate the effect of miR-331-3p on the invasion and metastasis of HCC through the adsorption of miR-331-3p in exosome-encapsulated circ_0001649 transported to Cancer-Associated Fibroblast (CAF) by HCC cells and its possible mechanism of action, in order to provide a meaningful experimental basis for finding biomarkers of HCC invasion and metastasis.
Materials and Methods
Subjects:
Serum samples were randomly selected from 46 patients with HCC attending the Third Xiangya Hospital of Central South University from January 2020 to December 2021 and 46 serum samples from the non-cancer population (control group) admitted during the same period (Table 1). All patients were diagnosed with HCC by pathological examination and did not receive any cancer-related treatment prior to sampling.
| Clinicopathological features | Number of patients |
|---|---|
| Age/year | |
| <60 | 28 (60.9) |
| ≥60 | 18 (39.1) |
| Gender | |
| Male | 31 (67.4) |
| Female | 15 (32.6) |
| Tumor size | |
| <3 cm | 22 (47.8) |
| ≥3 cm | 24 (52.2) |
| Histopathological differentiation | |
| Low differentiation | 9 (19.6) |
| Medium differentiation | 32 (69.6) |
| High differentiation | 5 (10.8) |
| Lymph node status | |
| Negative | 34 (73.9) |
| Positive | 12 (26.1) |
| Grade | |
| I-II | 6 (13.0) |
| III-IV | 40 (87.0) |
Table 1: Clinicopathological Features of 46 HCC Patients [n (%)].
Experimental materials:
Peripheral serum specimens: All sera were collected in the morning under fasting conditions, stored in anticoagulation tubes at 4° and centrifuged at 1900×g for 15 min within 2 h. After centrifugation, the supernatant was collected in enzyme-removed 1.5 ml Eppendorf tubes and stored in a -80° refrigerator. All sera were collected with the informed consent of cancer patients and non-cancer subjects, in accordance with the ethical standards set by the Human Trials Committee and approved by the Ethics Committee of the Third Xiangya Hospital of Central South University.
Tissue specimens: A total of 40 pairs of tumour tissue specimens and paired paracancer specimens were collected from patients with surgically resected HCC, including 24 males and 16 females. Informed consent was obtained from patients and their families for the retention of all specimens, which complied with the ethical standards, set by the Human Trials Committee and were approved by the Ethics Committee of the Third Xiangya Hospital of Central South University.
Cell lines: Human HCC cell lines, Hep3B and Bel7402 were purchased from the Centre of Excellence in Molecular Cell Science, Chinese Academy of Sciences. HCC tissue-derived CAF and Normal Fibroblast (NF) cells were isolated from cancerous and paraneoplastic tissues of HCC patients.
Routine reagents and consumables: The luciferase reporter plasmid (p)-circ-miR-331-3p containing the miR-331-3p binding region was constructed by Kingsray Biosciences Ltd. The miR-331-3p mimics/ inhibitor and the corresponding controls were synthesized by Guangzhou Ribo Biotechnology Co. Ltd. TRIzol RNA extraction kit was purchased from Dalian Meilun Biotechnology Co. Ltd. and the Paul Karl Horan 26 (PKH26) membrane dye kit was purchased from Sigma, United States of America (USA).
Methods:
Cell culture: Hep3B and Bel7402 cell lines were cultured in Roswell Park Memorial Institute (RPMI)-1640 complete medium, CAF and NF cell lines were cultured in Dulbecco’s Modified Eagle Medium (DMEM) complete medium. Cells were cultured in a constant temperature and humidity cell culture chamber at 37° with a Carbon dioxide (CO2) volume fraction of 5 %.
Exosome extraction: The cell culture medium was drawn into centrifuge tubes and centrifuged in the following gradient: 1000×g for 10 min, 3000×g for 30 min, 10 000×g for 60 min and 100 000×g for 70 min at 4°. The exosomes were collected from the sediment and resuspended in Phosphate Buffered Saline (PBS). The extracted exosomes can be stored at 4° for 3 d and used as soon as possible.
Western blot: The total cellular protein was extracted using Radioimmunoprecipitation Assay (RIPA) lysates and the protein concentration of the experimental samples was determined by Bicinchoninic Acid (BCA) protein quantification kit. A total of 30 μg of protein was sampled on Sodium Dodecyl Sulphate-Polyacrylamide Gel Electrophoresis (SDS-PAGE) and transferred to nitrocellulose membrane. The sample was transferred to a nitrocellulose membrane and closed with 5 % skimmed milk powder at room temperature for 1 h. The sample was mixed with anti-Matrix Metalloproteinase 9 (MMP9) antibody (1:500), anti-Cluster of Differentiation 63 (CD63) antibody (1:2000), anti-Tumor Susceptibility Gene 101 (TSG101) antibody (1:1000), anti-Alix antibody (1:1000), anti-Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) antibody (1:1000) and anti-GAPDH antibody (1:3000) at 4° overnight. After overnight, the membranes were washed with Tris Buffered Saline with Tween 20 (TBST). The secondary antibodies were incubated for 1 h at room temperature, exposed to Electrochemiluminescence (ECL) and the results were collected.
Cell transfection: The cells were digested and resuspended in a 6-well plate at the appropriate density. The cells were transfected according to the instructions of Lipofectamine™ 2000 transfection reagent (Invitrogen, USA) when the cells were fully attached to the wall and the degree of fusion was 60 %-70 %, and incubated in the incubator for 6 h before changing to complete medium.
RNA extraction and Real-Time Fluorescence Quantitative Polymerase Chain Reaction (RTFQPCR): Total RNA was extracted from cultured cells and tissues using TRIzol reagent (Invitrogen, USA) according to the manufacturer’s instructions, and then the RNA was reverse transcribed to obtain complementary Deoxyribonucleic Acid (cDNA). SYBR Green Ⅰ RTFQ-PCR was used to detect the expression levels of miR-331-3p and MMP9 messenger RNA (mRNA) in each group of cells. The primers were designed as follows: The upstream primer for MMP9 was 5'-AAGGCGTTAGTTTTCTTCGGGG-3' and the downstream primer was 5'-CACCTTTTGCTCCACAGTGC-3'; circ_0001649 upstream primer is 5'-GCTGGGGTGTGTTAAGCTTT-3', downstream primer is 5'-ACCCACGTGTCCTTAGAGAA-3'; GAPDH upstream primer is 5'-AGAAGGCTGGGGCTCATTTG-3' and downstream primer is 5'-AGGGGCCATCCACAGTCTTC-3'. The reaction conditions were: Pre-denaturation at 94° for 3 min; denaturation at 94° for 30 s, annealing at 60° for 30 s, extension at 72° for 35 s for 1 cycle, 35 cycles in total; the last cycle was extended at 72° for 2 min, 3 replicate wells were set up for each group and repeated 3 times. The relative expression levels of miR-331-3p and MMP9 mRNA in each group were calculated according to the formula 2-ΔΔCt.
Dual luciferase reporter gene assay: Cells were inoculated in 24-well plates, mapped and ready for transfection. The luciferase reporter plasmids (1 μg/ well) were co-transfected with miR-331-3p mimics and miR-29b inhibitors (100 pmol/well), and a control group was set up for each group, totaling 6 groups. After incubation, the luciferase expression level of each group was measured with a luciferase assay kit (Promega, USA), according to the instructions.
Immunofluorescence experiments: The cell suspension was added to the 24-well plate and replaced with conditioned medium with 2 % Fetal Bovine Serum (FBS) when the wall was adhered and the density was suitable and 40 μl of exosomes were added to the medium to co-culture with the cells. The cells were fixed in 500 μl of 4 % paraformaldehyde solution for 15 min, washed with PBS; the plate was closed with 10 % Bovine Serum Albumin (BSA) for 2 h. The closure solution was removed and a primary antibody dilution of 1:250 was added to the 24-well plate and incubated overnight at 4° in a shaker. After washing with PBS, add Hochest dilution prepared with PBS at 1:1000 to the 24-well plate and incubate for 10 min at room temperature. After washing with PBS, seal the plate with anti-quenching agent and observe under confocal microscope and take photographs.
Immunohistochemical staining: Paraffin-embedded sections were dewaxed, hydrated, washed twice with PBS for 5 min each, closed with 3 % Hydrogen peroxide (H2O2) for 5-10 min at room temperature, washed three times with distilled water for 5 min each and then subjected to antigen repair, washed with PBS for 5 min and closed with BSA for 20 min at room temperature. The reagent Streptavidin and Biotinylated Horseradish Peroxidase Complex (SABC) was added dropwise for 20 min at 20°~37°, washed 3 times with PBS for 2 min each time, followed by 3, 3'-Diaminobenzidine (DAB) colour development kit for 5 min each time; washed 3 times with distilled water for 5 min each time, followed by hematoxylin re-staining for 2 min and ethanol fractionation with hydrochloric acid; dehydration, transparency, sealing and microscopic examination.
PKH26 staining: In this study, the PKH26 membrane dye kit was used to stain exosome membranes to observe and track the successful entry of exosomes into the recipient cells. The exosomal precipitate extracted by ultracentrifugation was fully resuspended with 100 μl of diluent C reagent (A solution); another 100 μl of diluent C reagent was added with 0.4 μl of PKH26 dye and mixed thoroughly (B solution); A solution was mixed with B solution using a pipette, co-warmed at room temperature for 15 min and 200 μl of serum was added to it. The exosomes were washed once with PBS and then resuspended with an appropriate amount of fresh PBS.
Establishment of a nude mouse tumour model: Bagg Albino (BALB)/c nude mice (6-8 w old) were randomly grouped and labeled, and Hep3B cells stably overexpressing circ_0001649 and Hep3B cells knocking down circ_0001649. Hep3B cells were fully expanded and single cell suspensions (1×107 cells per nude mouse) of the above cell lines were inoculated subcutaneously into nude mice and kept in a pathogen-free animal facility with a total of 4 groups of non-transplanted tumour-bearing nude mice, which were allowed to eat as they will. The nude mice were regularly observed and tumour growth was recorded subcutaneously. The nude mice were executed on d 48 after transplantation and the livers of the nude mice were taken and the number of liver metastases was counted.
Statistical treatment:
Statistical Package for the Social Sciences (SPSS) 20.0 statistical software was used for data analysis. The experimental data of each group was expressed by x̄ ±s and t-test was used, and p<0.05 was considered statistically significant difference.
Results and Discussion
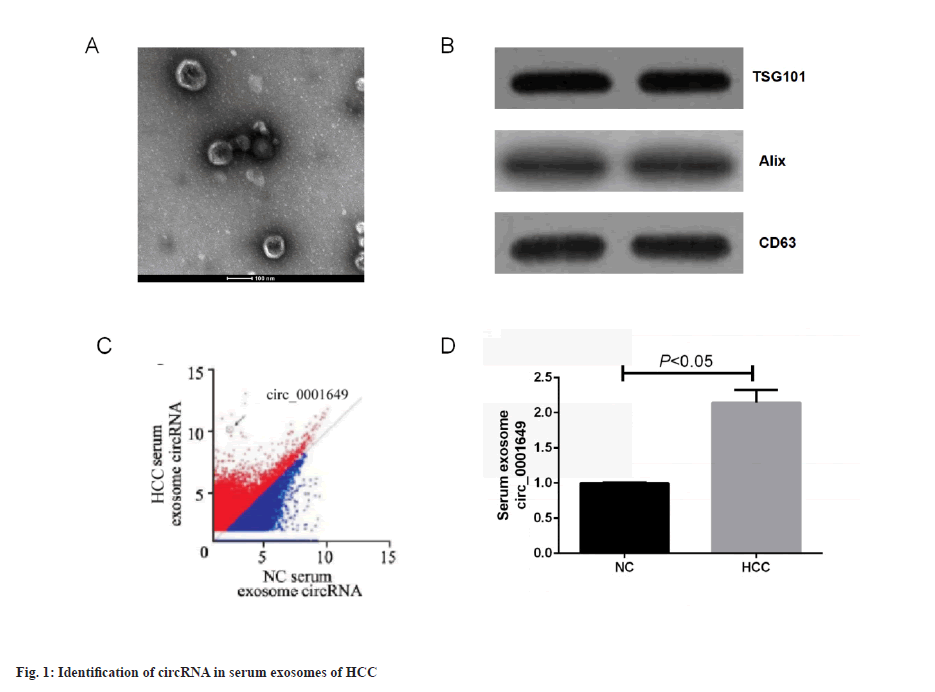
Identification of circRNA in HCC serum exosomes was shown in fig. 1. Serum exosomes were isolated from HCC patients and non-cancerous subjects using ultracentrifugation. The exosomes showed a membranous vesicle-like structure with a central depression and a diameter of approximately 100 nm under Transmission Electron Microscope (TEM). A TEM image of exosomes isolated from serum of HCC was shown in fig. 1A. The exosomes were next examined for the specific surface proteins, TSG101, Alix and CD63. Western blot of three representative exosome-specific markers: TSG101, Alix and CD63 were shown in fig. 1B. In summary, the pellets obtained from the cell cultures by ultracentrifugation matched the important characteristics reported for exosomes and the successful isolation of exosomes could be confirmed and subsequent experiments could be performed. Sequencing revealed that the expression of 1st group of exosomal circRNAs was highly variable in serum exosomes from HCC patients compared to non-cancer subjects. Preliminary screening of circRNA by an Agilent microarray scanner (n=46 and n=46, respectively) was shown in fig. 1C, with 95 circRNAs significantly up-regulated (fold change>4) and 94 circRNAs significantly down-regulated (fold change<0.25) in the HCC group, where hsa_circ_0001649 was 124-fold higher in the HCC group than in the non-cancer subject group, and subsequent studies focused on circ_0001649. RTFQ-PCR results showed that circ_0001649 was lowly expressed in normal human exosomes and highly expressed in serum exosomes of HCC patients (p<0.05, fig. 1D).
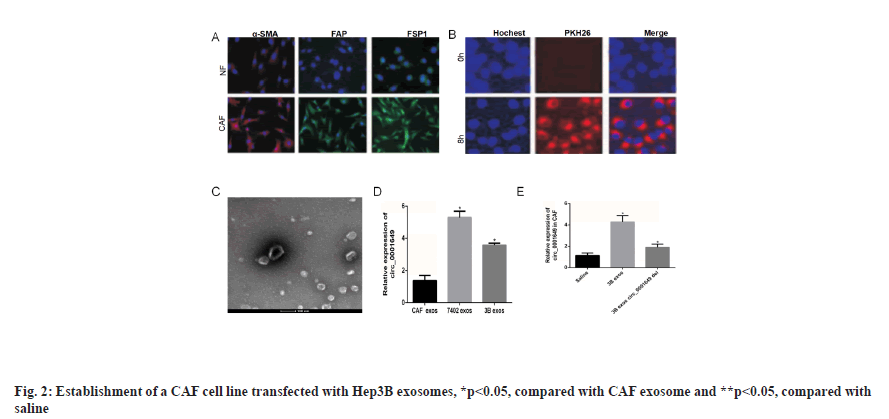
Establishment of CAF cell line transfected with Hep3B exosomes was shown in fig. 2. The tumour microenvironment consisted of a variety of cell types, among which CAF accounted for the largest proportion of mesenchymal cells. The CAF in tumour tissue and NF in paracancerous tissue were isolated, and immunofluorescence experiments showed that the expression of alpha-Smooth Muscle Actin (α-SMA), Fibroblast Activation Protein (FAP) and Fibroblast Specific Protein-1 (FSP1) were significantly higher in the CAF group than in the NF group. Immunofluorescence staining for α-SMA, FAP and FSP1 expressions of NFs and CAF were shown in fig. 2A. Hep3B exosomes were co-incubated with CAF cells and PKH26 was used to stain and trace the exosomes. The PKH26 labeled Hep3B exosomes successfully entered the recipient cells of CAF, confocal microscopy image of the internalization of fluorescently labeled 3B exosome (3B exos) in CAF, scale bars-50 μm (fig. 2B) and Hep3B cell exosomes transfected into CAF were observed under TEM (fig. 2C). The expression levels of circ_0001649 in exosomes from CAF, Hep3B and Bel7402 cell lines were examined. RTFQ-PCR results showed circ_0001649 was highly expressed in 3B exos and Bel7402 exosomes (7402 exos), while in CAF exosomes (CAF exos) circ_0001649 was significantly lower in CAF exos (p<0.05). Validation of circ_0001649 expression in exosome of CAF, Hep3B and Bel7402 using RTFQ-PCR (n=3) was shown in fig. 2D. The Hep3B cell line with the highest circ_0001649 expression was selected for subsequent experiments. After co-culturing CAF with 0.9 % Sodium chloride (NaCl) solution, 3B exos and 3B exos with circ_0001649 Knockdown (KD) (3B exos circ_0001649 del), RTFQ-PCR was performed and then subjected to RTFQ-PCR, which showed that the circ_0001649 content was lower in both the saline and 3B exos circ_0001649 del groups, whereas the circ_0001649 content was significantly higher in the 3B exos group (p<0.05, fig. 2E).
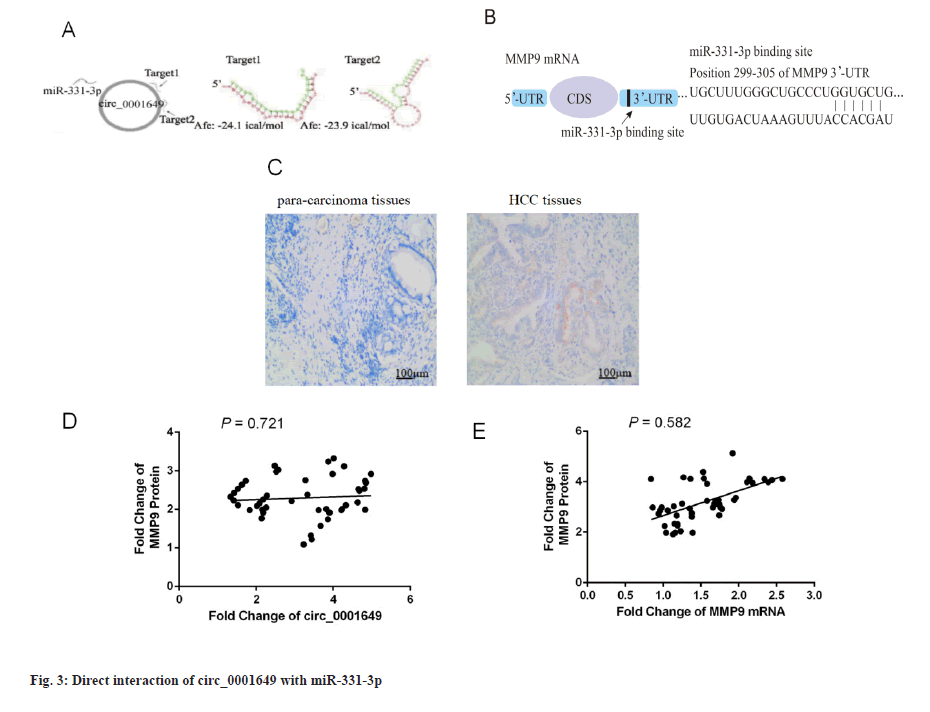
Circ_0001649 interacts directly with miR-331-3p (fig. 3). The presence of two targets between circ_0001649 and miR-331-3p was identified by raw signal analysis. Representative images of the potential interaction between circ_0001649 and miR-331-3p were shown in fig. 3A. The potential role of miR-331-3p in regulating MMP9 expression was cross-validated by TargetScan database (http://www.targetscan.org/), Database for Annotation, Visualization and Integrated Discovery (DAVID) database (https://david.ncifcrf.gov/), and predicted the 3’-Untranslated Region (UTR) binding region in MMP9 mRNA. Predicted binding region of miR-331-3p in the mRNA of MMP9 was shown in fig. 3B. The expression of MMP9 in para-carcinoma tissues and tumor tissues was detected by immunohistochemistry. Immunohistochemical analysis showed that MMP9 was highly expressed in HCC tissues and lowly expressed in paraneoplastic tissues (p<0.05, fig. 3C). Next, the clinical correlation between circ_0001649 and MMP9 protein in HCC cell exos was further identified: The expression level of circ_0001649 in exosomes was measured by RTFQ-PCR and MMP9 protein was quantified by a combination of Western blot and grey scale analysis. The relationship between exosome circ_0001649 and MMP9 (n=46, r=0.721) was shown in fig. 3D. Circ_0001649 expression level was positively correlated with the expression level of MMP9 protein (p<0.05, fig. 3D). The relationship between MMP9 mRNA and MMP9 (n=46, r=0.582) was shown in fig. 3E. The expression levels of MMP9 mRNA and MMP9 protein were measured by the same method and there was no significant correlation between the two (p>0.05, fig. 3E), therefore it was hypothesized that circ_0001649 regulates the expression of MMP9 through the post-transcriptional pathway.
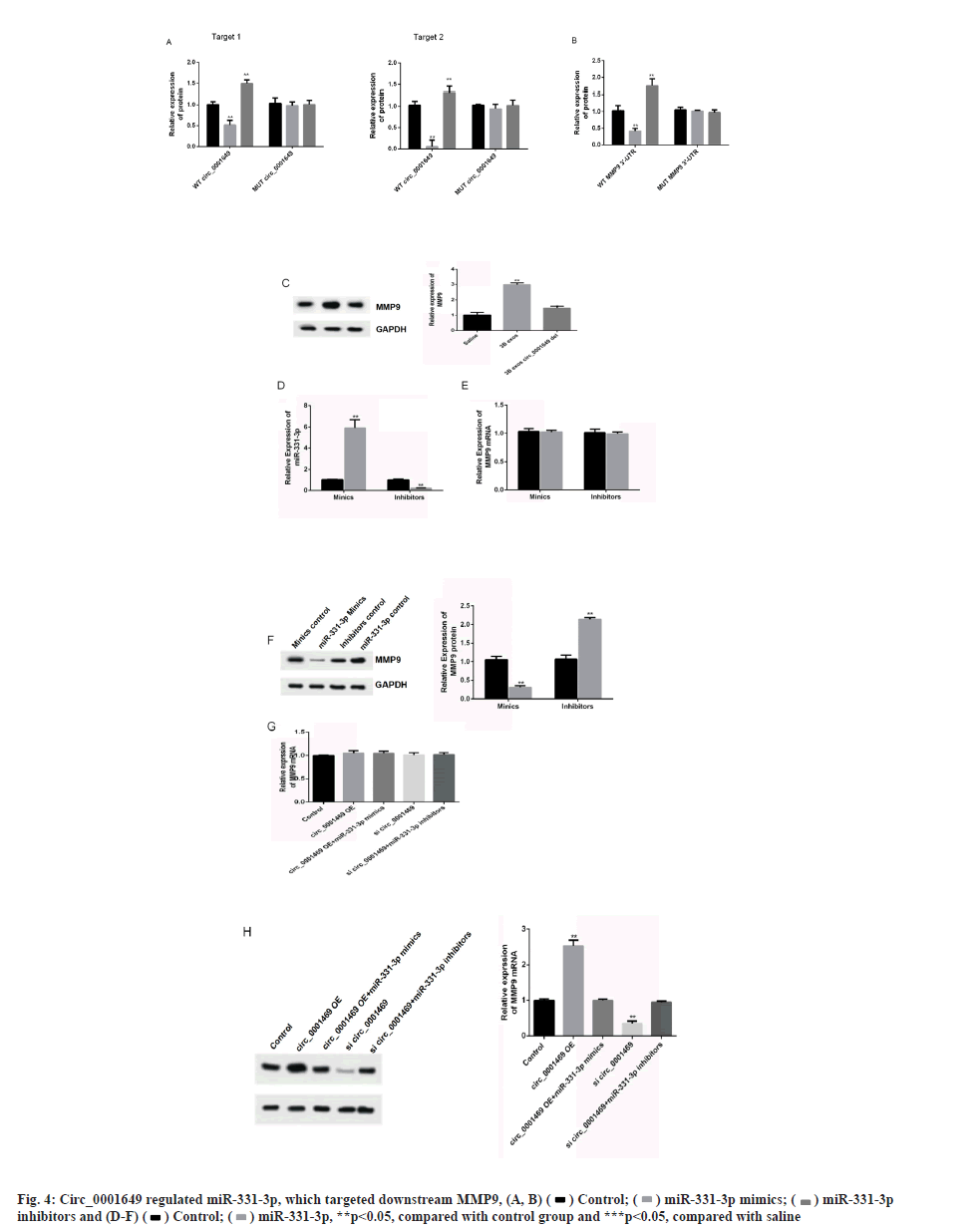
Circ_0001649 and MMP9 can target binding to miR-331-3p was shown in fig. 4A-fig. 4H. The relationship of circ_0001649 and miR-331-3p was measured by RTFQ-PCR (n=3) was shown in fig. 4A. The luciferase activity was detected by dual-luciferase reporter gene assay (n=3) which was shown in fig. 4B. The results of dual luciferase reporter gene assay showed that over expression of miR-331-3p significantly inhibited the fluorescence intensity of circ_0001649 and MMP9 mRNA 3'-UTR wild-type plasmids (p<0.05), while there was no obvious inhibitory effect on the fluorescence intensity of circ_0001649 and MMP9 mRNA 3'-UTR mutant plasmids (p>0.05, fig. 4A and fig. 4B). Inhibition of miR-331-3p significantly promoted the fluorescence intensity of circ_0001649 and MMP9 mRNA 3'-UTR wild-type plasmids (p<0.05), but no obvious effect on circ_0001649 and MMP9 3'-UTR mutant plasmids (p>0.05), demonstrating that circ_0001649 targets binding to miR-331-3p, which post-transcriptionally regulates the 3'-UTR region of MMP9 mRNA. Western blot results showed that the expression level of MMP9 protein was elevated in CAF cells co-cultured with 3B exos, whereas the expression level of MMP9 was decreased in CAF co-cultured with 3B exos circ_0001649 del (fig. 4C), demonstrating that 3B exos-mediated circ_0001649 could enhance the expression level of MMP9 protein in CAF. Transfection of miR-331-3p mimics into CAF cells inhibited the expression of MMP9 protein, while transfection of miR-331-3p inhibitors promoted the expression of MMP9 (fig. 4D and fig. 4F), but had no significant effect on the mRNA level of MMP9 (p>0.05, fig. 4E), demonstrating that miR-331-3p can inhibit through the post-transcriptional pathway the expression level of MMP9 protein. Next, cell function recovery experiments were performed in CAF cells and the results showed that the expression levels of MMP9 protein in the circ_0001649-Over Expression (OE) group were significantly increased and reversed by miR-331-3p mimics, and the expression levels of MMP9 protein in the circ_0001649-KD group were significantly decreased and reversed by miR-331-3p inhibitors (fig. 4H), but had no significant effect on the mRNA levels of MMP9 (p>0.05, fig. 4G).
In vivo experiments to verify circ_0001649 promotes tumour growth was shown in fig. 5. The contents in fig. 5A-fig. 5G were described as follows, (A) Implanted cells and analysis; (B) Changes of diameters of tumors; (C) Serum exosomes of mice under TEM; (D, E) Levels of circ _0001649 in serum exosomes and tumors measured by RTFQ-PCR (n=3); (F) Levels of MMP9 mRNA measured by RTFQ-PCR and (G) Expression of MMP9 measured by Western blot (n=3).
To further evaluate the effect of circ_0001649 on tumour growth in vivo, a nude mouse transplantation tumour model was established in this study, with a total of 4 groups (fig. 5A), and the diameter of each group of nude mice ruffles was measured every 8 d during incubation (fig. 5B). In addition, serum exosomes were extracted from each of the four groups of nude mice (fig. 5C) and the RTFQ-PCR results showed that the serum exosome circ_0001649 content was reduced in the circ_0001649-KD group and significantly increased in the circ_0001649-OE group (fig. 5D). Meanwhile, the circ_0001649 content and the expression level of MMP9 protein were reduced in the tumour of the circ_0001649-KD group, while the circ_0001649 content and the expression level of circ_0001649 protein were significantly increased in the tumour of the circ_0001649-OE group.
The expression level of MMP9 protein was significantly higher (fig. 5E and fig. 5G). There was no significant difference in the mRNA level of MMP9 in the two groups (fig. 5F).
CircRNAs are non-coding RNAs that are resistant to nucleic acid exonuclease-mediated degradation, which are stable cyclic structures and are commonly expressed in a variety of cell types[10]. These circRNAs are often used as competitive endogenous RNAs to influence the function of downstream miRNAs[14]. Studies have shown that circRNAs are enriched and stable in exosomes and can be detected in circulating blood and urine[15,16]. Exosomes can be accepted by many types of cells, including macrophages and can also act as intercellular messengers through which cells can transfer circRNAs.
With significant advances in microarray and RNA sequencing technologies, an increasing number of circRNAs have been identified as playing critical roles in numerous disease processes, particularly in cancers including HCC. Zhang et al. have demonstrated that the circ_Mothers against Decapentaplegic Homolog 2 (SMAD2) inhibits Epithelial-Mesenchymal Transition (EMT) by targeting miR-629 in HCC[17]. A study of esophageal squamous cell carcinoma showed that circ-Itchy E3 Ubiquitin Protein Ligase (ITCH) has an adsorptive effect on miRNA and that increasing circ-ITCH levels promotes tumour progression[18]. There is also growing evidence that exosomal circRNAs play a key role in tumour growth, immune escape, angiogenesis, metastasis and drug resistance formation in HCC[19-21].
In this study, we explored the role of circ_0001649 in HCC invasion and metastasis and demonstrated that the level of circ_0001649 was significantly higher in serum exosomes of HCC patients than in normal serum exosomes. It was confirmed by luciferase reporter gene assay that circ_0001649 targeted binding to miR-331-3p and miR-331-3p was significantly correlated with the expression level of MMP9 protein. In this study, we first found that exosome-mediated circ_0001649 could promote the migration of HCC cells and trans-cellular communication through exosomes significantly promoted the invasion and metastasis of HCC cells.
MMP9, found in the extracellular matrix, is a protein with type IV collagen degradation properties and contains three fibronectin type II, repeats in its catalytic site, which binds denatured type IV and type V collagen to elastin. Studies have shown that MMP9 expression is associated with the progression of various cancers including HCC, lung cancer, papillary thyroid cancer, adult neuroblastoma and bladder cancer[22-24].
In summary, this study shows that knocking down circ_0001649 in HCC cells is expected to inhibit HCC invasion and metastasis by regulating the miR-331-3p/ MMP9 axis, providing a meaningful experimental basis for exploring biomarkers for the diagnosis and treatment of HCC metastasis, as well as providing a new idea for regulating cross-cellular communication in the treatment of metastatic HCC, opening up a new research perspective, which is of great importance in the diagnosis of metastatic HCC, prognosis and prolonging patients survival and improving their quality of life from the perspective of metastasis inhibition.
Conflict of interests:
The authors declared no conflict of interest.
References
- Sempokuya T, Forlemu A, Azawi M, Silangcruz K, Khoury N, Ma J, et al. Survival characteristics of fibrolamellar hepatocellular carcinoma: A surveillance, epidemiology, and end results database study. World J Clin Oncol 2022;13(5):352-65.
- Taflin H, Hafström L, Holmberg E, Castedal M, Lindner P. The impact of increased immigration to Sweden on the incidence and treatment of patients with HCC and underlying liver disease. Scand J Gastroenterol 2019;54(6):746-52.
[Crossref] [Google scholar] [PubMed]
- Zeng Z, Cao Z, Tang Y. Increased E2F2 predicts poor prognosis in patients with HCC based on TCGA data. BMC Cancer 2020;20(1):1-5.
- Akamatsu N, Cillo U, Cucchetti A, Donadon M, Pinna AD, Torzilli G, et al. Surgery and hepatocellular carcinoma. Liver Cancer 2017;6(1):44-50.
[Crossref] [Google scholar] [PubMed]
- Song Y, Park IS, Kim J, Seo HR. Actinomycin D inhibits the expression of the cystine/glutamate transporter xCT via attenuation of CD133 synthesis in CD133+ HCC. Chem Biol Interact 2019;309:108713.
- Wang Y, Liu J, Ma J, Sun T, Zhou Q, Wang W, et al. Exosomal circRNAs: Biogenesis, effect and application in human diseases. Mol Cancer 2019;18(1):1-10.
[Crossref] [Google scholar] [PubMed]
- Tang Z, He J, Zou J, Yu S, Sun X, Qin L. Cisplatin-resistant HepG2 cell-derived exosomes transfer cisplatin resistance to cisplatin-sensitive cells in HCC. PeerJ 2021;9:e11200.
[Crossref] [Google scholar] [PubMed]
- Qu Z, Feng J, Pan H, Jiang Y, Duan Y, Fa Z. Exosomes derived from HCC cells with different invasion characteristics mediated EMT through TGF-β/Smad signaling pathway. Onco Targets Ther 2019;12:6897-905.
[Crossref] [Google scholar] [PubMed]
- Qu Z, Wu J, Wu J, Luo D, Jiang C, Ding Y. Exosomes derived from HCC cells induce sorafenib resistance in hepatocellular carcinoma both in vivo and in vitro. J Exp Clin Cancer Res 2016;35(1):1-2.
[Crossref] [Google scholar] [PubMed]
- Patop IL, Wüst S, Kadener S. Past, present, and future of circ RNAs. EMBO J 2019;38(16):e100836.
[Crossref] [Google scholar] [PubMed]
- Yang L, Jiang MN, Liu Y, Wu CQ, Liu H. Crosstalk between lncRNA DANCR and miR-125b-5p in HCC cell progression. Tumori J 2021;107(6):504-13.
[Crossref] [Google scholar] [PubMed]
- Liu YM, Cao Y, Zhao PS, Wu LY, Lu YM, Wang YL, et al. CircCCNB1 silencing acting as a miR-106b-5p sponge inhibited GPM6A expression to promote HCC progression by enhancing DYNC1I1 expression and activating the AKT/ERK signaling pathway. Int J Biol Sci 2022;18(2):637-51.
[Crossref] [Google scholar] [PubMed]
- Yang X, Song D, Zhang J, Feng H, Guo J. PRR34-AS1 sponges miR-498 to facilitate TOMM20 and ITGA6 mediated tumor progression in HCC. Exp Mol Pathol 2021;120:104620.
- Shang Q, Yang Z, Jia R, Ge S. The novel roles of circRNAs in human cancer. Mol Cancer 2019;18(1):1-10.
[Crossref] [Google scholar] [PubMed]
- Altesha MA, Ni T, Khan A, Liu K, Zheng X. Circular RNA in cardiovascular disease. J Cell Physiol 2019;234(5):5588-600.
[Crossref] [Google scholar] [PubMed]
- Lei B, Tian Z, Fan W, Ni B. Circular RNA: A novel biomarker and therapeutic target for human cancers. Int J Med Sci 2019;16(2):292-301.
[Crossref] [Google scholar] [PubMed]
- Zhang X, Luo P, Jing W, Zhou H, Liang C, Tu J. circSMAD2 inhibits the epithelial-mesenchymal transition by targeting miR-629 in hepatocellular carcinoma. Onco Targets Ther 2018;11:2853-63.
[Crossref] [Google scholar] [PubMed]
- Li F, Zhang L, Li W, Deng J, Zheng J, An M, et al. Circular RNA ITCH has inhibitory effect on ESCC by suppressing the Wnt/β-catenin pathway. Oncotarget 2015;6(8):6001-13.
- Huang XY, Huang ZL, Huang J, Xu B, Huang XY, Xu YH, et al. Exosomal circRNA-100338 promotes hepatocellular carcinoma metastasis via enhancing invasiveness and angiogenesis. J Exp Clin Cancer Res 2020;39(1):1-6.
[Crossref] [Google scholar] [PubMed]
- Liu D, Kang H, Gao M, Jin L, Zhang F, Chen D, et al. Exosome‐transmitted circ_MMP2 promotes hepatocellular carcinoma metastasis by upregulating MMP2. Mol Oncol 2020;14(6):1365-80.
[Crossref] [Google scholar] [PubMed]
- Chen W, Quan Y, Fan S, Wang H, Liang J, Huang L, et al. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett 2020;475:119-28.
[Crossref] [Google scholar] [PubMed]
- Ouyang X, Feng L, Yao L, Xiao Y, Hu X, Zhang G, et al. Testicular orphan receptor 4 (TR4) promotes papillary thyroid cancer invasion viaactivating circ-FNLA/miR-149-5p/MMP9 signaling. Mol Ther Nucleic Acids 2021;24:755-67.
- Wang JL, Wu DW, Cheng ZZ, Han WZ, Xu SW, Sun NN. Expression of high mobility group box-B1 (HMGB-1) and matrix metalloproteinase-9 (MMP-9) in non-small cell lung cancer (NSCLC). Asian Pac J Cancer Prev 2014;15(12):4865-9.
[Crossref] [Google scholar] [PubMed]
- Chen R, Cui J, Xu C, Xue T, Guo K, Gao D, et al. The significance of MMP-9 over MMP-2 in HCC invasiveness and recurrence of hepatocellular carcinoma after curative resection. Ann Surg Oncol 2012;19(3):375-84.
[Crossref] [Google scholar] [PubMed]
 **p<0.05, compared with control group and ***p<0.05, compared with saline.
**p<0.05, compared with control group and ***p<0.05, compared with saline.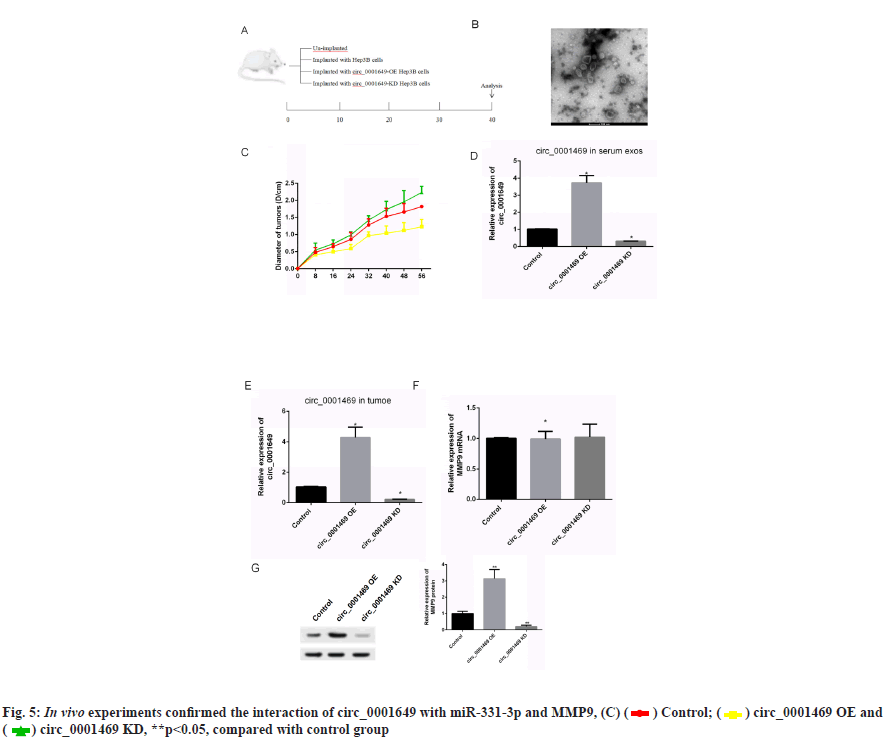
 **p<0.05, compared with control group.
**p<0.05, compared with control group.



