- *Corresponding Author:
- Kamal Ismail Bakr Al Otraqchi
Department of Basic Science, College of Medicine, Hawler Medical University, Erbil, Kurdistan, Iraq
E-mail: dr.kamalotraqchi@yahoo.com
| This article was originally published in a special issue, “Recent Developments in Biomedical Research and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2022:84(4) Spl Issue “279-289” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The main objective is to study the genotypic and characterization of plasmid deoxyribonucleic acid content of resistant isolates in Erbil city. A total of 39 Acinetobacter baumannii were isolated from 102 diabetic foot ulcer patients, whose ages ranged from 37 to 88 y, attending various medical wards of hospitals in Erbil city, Iraq, between May 2020 and March 2021. Isolates were tested for the presence of extended-spectrum beta-lactamases and metallo beta-lactamases. The presence and prevalence of multidrug-resistant isolates namely blaOXA-51- like, blaOXA-23-like, ISAba1 and outer membrane protein A expression of oxacillinase carbapenemases gene and biofilm formation respectively were screened on the molecular level. Finally, plasmid deoxyribonucleic acid profile was determined and the biofilm-forming capacity was measured. Most isolates showed high resistance to different antimicrobial agents tested, the most effective agents were colistin and polymyxin B. 12 (30.76 %) isolates were tested positive for extended-spectrum beta-lactamases, while all 39 (100 %) were metallo beta-lactamase producers. 27 (69.23 %) of isolates were biofilm producers, while 12 (30.76 %) were nonbiofilm producers. The blaOXA-51-like, blaOXA-23-like, ISAba1 and outer membrane protein A genes were detected in all 39 (100 %) isolates. The genes encoding resistance to certain antibiotics, in addition to genes that coding for extended-spectrum beta-lactamase and biofilm formation were located on plasmids. Analysis of plasmid deoxyribonucleic acid content revealed that 25 (64.10 %) isolates harbored from one to several plasmid bands ranging from 1 kilo base pairs to more than 10 kilo base pairs. No thermo-sensitive plasmids were detected. Due to the emergence of multidrug-resistance, Acinetobacter baumannii is evolving into a global health threat. Carbapenems are effective therapeutic agents for Acinetobacter baumannii nosocomial infections. Before deciding the course of treatment, it is important to consider the aforementioned interaction since the presence of multiple species may worsen the infection.
Keywords
Acinetobacter baumannii, plasmid profile, biofilm formation, genotypic characterization, carbapenems
Diabetes Mellitus (DM), usually known as diabetes, is a kind of metabolic diseases which characterized by a high blood sugar level during an extended time period as a result of insulin shortage, unusual insulin response or even both. It can make many problems, such as chronic kidney disease, cardiovascular disease and foot ulcers if not treated[1-3]. The diabetes prevalence is rising throughout the world, so that in 2010, about 200 million people were diabetic[4]. Based on the International Diabetes Federation (IDF) reports in 2019, almost 578.4 million and 700.2 million people have diabetes by the end of 2030 and 2045, respectively[5,6].
Acinetobacter baumannii (A. baumannii) as a Gramnegative pleomorphic bacterium which has a high occurrence among immunocompromised patients[7]. Most places have A. baumannii, an opportunistic pathogen that is particularly common in hospitals and other healthcare facilities, stays there for a long time and is easily transferred between patients[8]. Moreover, it is affecting severe illness like diabetes and prolonged hospital stay may have the risk of developing a hospital-acquired infection and prolonged exposure to antimicrobial agents may have the effect[9]. Nosocomial infections caused by A. baumannii are characterized by high morbidity and mortality, particularly in patients with severe medical conditions. These infections include skin and soft tissue infections, pneumonia, bacteremia and urinary tract infections[10]. Bacterial skin infections are a complicated problem[11]. According to recent data, 25 % of people with diabetes will experience a foot ulcer during their lifetime and the disease has developed into a very serious chronic condition with rising global prevalence[12]. Due to hyperglycemia’s impact on leukocyte function, diabetics are more likely to contract infections[13]. The infection in most cases makes an ulcer that has been open for a long time and not treated adequately; so that it is one of the most usual, critical and hard to treat[14,15].
Diabetic Foot Ulcers (DFUs) are a significant and growing public health concern. Multidrug-Resistant (MDR) A. baumannii is gradually becoming dominant among diabetes patients. The issue of plasmidmediated resistance to broad-spectrum-lactams is getting worse all over the world. A. baumannii is a frequently isolated pathogen in Diabetic Foot Ulcer (DFU), which can range from soft tissue to bone infections and are usually poly-microbial in nature[16,17]. A. baumannii has been identified as one of the microbes connected to a higher frequency of major amputation[18]. In the last two decades, the emergence and spread of bacterial infections resistant to beta (β)-lactams, particularly to third generations of cephalosporins and carbapenems, have grown to be an emerging problem on a global scale[19,20]. Infection risk factors are 10 times higher in DFU patients than in non-DFU patients and male patients are more likely to contact infections than female patients[21]. The second most common isolated micro-organism from non-fermented bacteria in foot ulcer infections is A. baumannii.
Although Acinetobacter wound infection showed an isolation rate of 11.1 %, it poses a problem to clinicians whether to treat it aggressively as a pathogen or as a colonizer[22]. When it comes to treating infections, brought by this organism, carbapenems, which were once used as the standard therapy, are no longer effective. In individuals with diabetic and non-DFU, a variety of variables contribute to the establishment and survival of MDR A. baumannii biofilm production which is the most common among them[23]. The rise in Metallo Beta- Lactamase (MBL) in A. baumannii is concerning development in the spread of β-lactam resistance, which affects many cephalosporins, carbapenems, penicillin’s and monobactams[24]. Age, length of hospital stay, underlying conditions like diabetes or tumors, or overcrowding in the hospital wards are additional risk factors for colonization or infection with MBL producers[25]. Infection outbreaks brought on by A. baumannii, that are resistant to various antibiotic classes, including carbapenems, are a severe concern in many specialized hospital units, such as Intensive Care Units (ICUs).
This resistance was proved by Ebrahem et al. They observed that A. baumannii was 100 % strong biofilm forming and it has capacity to extend the resistance to all antimicrobial substances used for treatment[26]. The main impact of a carbapenem-resistant A. baumannii infection is the requirement to use “lastline” antibiotics like Colistin (CL), Polymyxin B (PB) or Tigecycline (TIG)[27].
Beta-lactamase Oxacillinase (blaOXA) genes encoding carbapenem-hydrolysing β-lactamases (carbapenemases), belonging to molecular class-D (OXA enzymes), have emerged globally as the main mechanism responsible for this resistance. The blaOXA genes of Acinetobacter species can be divided into four phylogenic subgroups. The blaOXA51-like gene, which is intrinsic to A. baumannii[28], ordinarily expresses at a low level, but the Insertion Sequence A. baumannii (ISAba1) upstream of the gene can cause the gene to overexpress. In contrast, blaOXA23- like, blaOXA24-like and blaOXA58-like genes were consistently associated with resistance or at least, with reduced susceptibility[29]. For an epidemiological objective, phenotypic techniques like bio-typing and serotyping as well as molecular techniques like Polymerase Chain Reaction (PCR) and plasmid profile analysis were used[30]. This study is conducted according to the ethical committee at Hawler Medical University. The main aim is to study genotypic and characterization of plasmid Deoxyribonucleic Acid (DNA) content of MDR A. baumannii in Erbil city.
Materials and Methods
Bacterial species:
A total of 39 A. baumannii bacteria were observed in 102 DFU patients of all ages who were collected from various medical wards and ICU departments of hospitals in Erbil city for a period of 10 mo, from May 2020 to March 2021. Samples were inoculated on blood agar and MacConkey agar (BD BBLTM, United States of America (USA)), and isolates were recognized using standard microbiological techniques and biochemical tests. The VITEK 2® compact system was then used to identify species (bioMérieux, France). The antimicrobial susceptibility of A. baumannii isolates was tested by the modified Kirby-Bauer disc diffusion method as recommended by the Clinical and Laboratory Standard Institute (CLSL, 2015) and VITEK 2® compact guidelines[31]. MDR isolates underwent additional testing to determine whether they could produce MBL. Resistance to two or more drug classes with therapeutic relevance is referred to as MDR. While VITEK 2® compact identification and susceptibility testing confirmation were carried out by Gram-Negative (GN) card and Antimicrobial Susceptibility Testing (AST) N326 and A. baumannii American Type Culture Collection (ATCC) 19606 was used as quality control strains.
β-lactamase test:
All A. baumannii isolates were tested for β-lactamase using the Cefinase disc (BD BBLTM, USA). It is coated with nitrocefin, a chromogenic cephalosporin that changes color quickly from yellow to red when a β-lactamase hydrolyzes the amide bond in the β-lactum ring. The yellow-colored disc becomes red in the area where the isolate is spread on it when a bacterium produces this enzyme in substantial levels. As a positive control, Staphylococcus aureus (ATCC 29213) was used[32].
AST:
AST was carried out for 19 different therapeutically important antibiotics, such as Imipenem (IMP: 10 g), Meropenem (MEM: 10 μg), Ceftazidime (CAZ: 30 μg), Cefotaxime (CTX: 30 μg), Cefepime (CEP: 30 μg), Ciprofloxacin (CIP: 5 μg), Levofloxacin (LVX: 5 μg), Piperacillin (PIP: 100 μg), PIP-Tazobactum (TZM: 100/10 μg), Netilmicin (NET: 10 μg), Tobramycin (TN: 10 μg), Amikacin (AN: 30 μg), Gentamycin (GN: 10 μg), Tetracycline (TE: 30 μg), Trimethoprim (TMP)/ Sulfamethoxazole (STX: 1.25/23.75 μg), TIG: 15 μg, PB: 30 μg and CL: 10 μg by Kirby-Bauer disc diffusion method on Mueller-Hinton Agar (MHA) (BD BBLTM, USA) and incubated for 18-24 h at 35° according to Clinical Laboratory Standards Institute guidelines[31] and VITEK 2® compact (AST N326). The resistance to at least three categories of the drugs was considered as a MDR[33].
Extended-Spectrum Beta-Lactamase (ESBL) phenotypic detection of isolates:
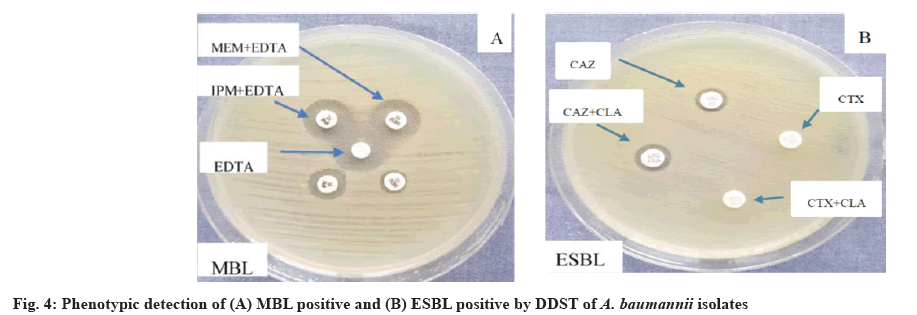
The Double-Disc Synergy Test (DDST) was used to identify ESBLs. The isolates were tested for the presence of an inhibition zone on MHA for CAZ (30 g), CTX (30 g) and CAZ (30 g)+clavulanic acid (10 g). On MHA, two pairs of disks, CAZ with CAZ/clavulanic acid and CTX with CTX/clavulanic were positioned with a 20 mm gap between them. If the inhibition zone diameter in the presence of clavulanic acid was ≥5 mm bigger than that without it, the ESBL test was considered positive[34].
Phenotypic detection of MBL:
Determination of MBL production among A. baumannii isolates, a recently prepared bacterial suspension adjusted to 0.5 McFarland was streaked for confluent growth on a MHA plate using a swab. 5 ml of Ethylenediaminetetraacetic Acid (0.5 M EDTA) solutions were added into a paper disk, 6 mm in diameter and dried without being overfull. The disks were placed at the center of the plate. 10 mg of MEM, MEM with EDTA, IMP and IMP with EDTA disks were placed at a distance of 10 mm from the center, and the plate was incubated at 37° for 16-18 h. Disks with EDTA alone were served as the negative control. The presence of a zone around the antibiotics containing EDTA disks would indicate an MBL producer. Zone of inhibition larger than 2 mm when EDTA was added to the MEM and IMP disks, the isolate was considered to be MBL positive. The test was repeated three times[35,36].
Biofilm production:
In vitro biofilm, the detection was done by the quantitative microtiter plate method described by Christensen in 1985 which was considered as the gold standard method for biofilm detection. Each isolate was grown overnight in Tryptic Soy Broth (TSB) with 0.5 % glucose. 200 ml of cell suspension was inoculated in sterile 96 well polystyrene microtiter plates. After 24 h of incubation, the wells were gently washed 3 times with 200 μl of Phosphate-Buffered Saline (PBS), then dried in an inverted position and stained with 1 % crystal violet for 15 min. The wells were rinsed again in 20 μl of ethanol 95 % to solubilize crystal violet. The Optical Density at 620 nm (OD620) was determined using a microplate reader. Each assay was performed in triplicate and the average OD was considered as Non-biofilm producer: OD<Optical Density cutoff value (ODc) negative, weak biofilm producer: ODc negative≤OD<2 ODc negative, medium biofilm producer: 2ODc negative≤OD<4 ODc negative and strong biofilm producer: 4 ODc negative≤OD[37].
Genotypic identification of A. baumannii isolates:
The molecular analysis of multidrug and biofilm formation associated genes blaOXA51-like, blaOXA23-like, ISAba1 and outer membrane protein A (ompA) in the A. baumannii isolates was employing standard multiplex PCRs. The defined and the sequences of the primers used in this study were listed in Table 1[38-40]. The primer mixing for carbapenemases and biofilm encoding genes were made in eppendorf tube and 20 μl of the reaction medium was employed for the PCR (2 μ DNA template+0.5 μM of each primer+4 μl hot start red load 5× Master Mix (3.5 mM Magnesium chloride (MgCl2) final concentration)+fill to 20 μl with distilled water). The PCR conditions are as follows-Initial incubation at 95° for 5 min followed by 30 cycles of reaction with the step of denaturation at 95° for 30 s, annealing at OXA51-like and OXA23-like 57° and ompA 56° for 30 s and elongation at 72° for 1 min, followed by 30 cycles and the step of final extension was at 72° for 7 min. The electrophoretically separated PCR amplicons with red safe dye on a 1 % agarose and observed under Ultraviolet (UV) light.
| Target | Primer | Sequence 5’→3’ | Amplicon size (bp) | Reference |
|---|---|---|---|---|
| blaOXA-51-like | OXA-51-like | F: 5-TAATGCTTTGATCGGCCTTG-3 | 353 | [38] |
| OXA-51-like | R: 5-TGGATTGCACTTCATCTTGG-3 | |||
| blaOXA-23-like | OXA-23-like | F: 5-GATCGGATTGGAGAACCAGA-3 | 501 | [38] |
| OXA-23-like | R: 5-ATTTCTGACCGCATTTCCAT-3 | |||
| ISAba1 | ISAba1 | F: 5’-CACGAATGCAGAAGTTG-3’ | 451 | [39] |
| ISAba1 | R: 5’-CGACGAATACTATGACAC-3’ | |||
| ompA | ompA | F: 5’-GTTAAAGGCGACGTAGACG-3’ | 578 | [40] |
| ompA | R: 5’-CCAGTGTTATCTGTGTGACC-3’ |
Table 1: Primes Used for Amplification of Genes of A. baumannii
Characterization of plasmid DNA content:
Isolation of plasmid DNA content from A. baumannii isolates: Preparation of plasmid miniprep kit had been optimized for the rapid high-yield plasmid DNA purification from 1-2 ml liquid. From a freshly streaked bacterial plate, one colony was taken and inoculated into Luria-Bertani (LB) broth with the appropriate antibiotic. The culture was shaken all night long during incubating. The extraction procedure was performed at room temperature according to the plasmid DNA miniprep kit guide.
Determination of plasmid DNA concentration: Plasmid DNA quantity was measured by Nano-Drop spectrophotometer to estimate the concentration and purity of extracted plasmid DNA through reading the absorbance at OD 260/280 nm and the ratio of reading should be between 1.9-2.2 in order to confirm the purity.
Detection of the thermo-sensitive plasmid DNA among A. baumannii isolates: A single colony from fresh bacterial isolate was inoculated onto sterile nutrient agar plates and cetrimide agar with and without antibiotics. The inoculated plates were incubated at 25°, 37° and 42° overnight respectively, then the results of bacterial growth in each condition were recorded[41].
Determination of the site of conferring resistance to antimicrobial agents and biofilm formation: This is carried out by isolation of the plasmid DNA from the A. baumannii isolates and then transforming into standard bacterial strain (Escherichia coli (E. coli) Douglas Hanahan alpha (DH5-α)) that acts as bacterial hosts for DNA uptake. Before 24 h of transformation, the Calcium chloride (CaCl2) treated DH5-α competent cells were prepared and the transformation process was included briefly. 200 μl of component cells were transferred to a sterile chilled 1.5 ml eppendorf tube, then 100 μl of the DNA was added, mixed well and stored in ice for 30 min. The tube was transferred to 42° and incubated for 90 s, then directly the tube was transferred to ice and allowed to chill for 1-2 min. 800 μl of LB broth was added to the mixture and incubated for 45 min in a 37° water bath. The mixture was cultured on LB-appropriate antimicrobial plates and incubated overnight at 37° and then the genetic transformation frequency was calculated[42].
Results and Discussion
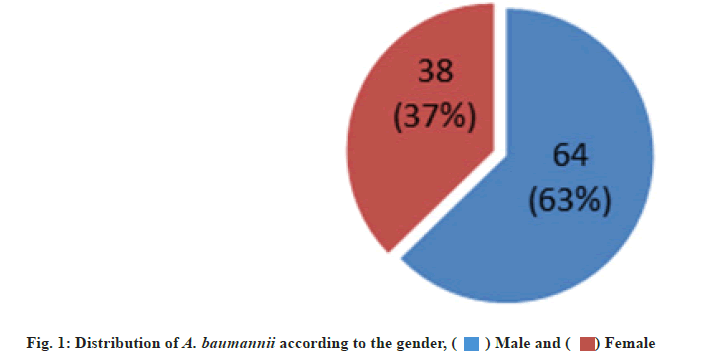
A total of 39 A. baumannii were collected from 102 DFU patients (64 males and 38 females), their ages ranged from 37-88 y, attending Rizgary emergency department and different private hospitals in Erbil city, Iraq for a period of 10 mo from May 2020 to March 2021.
A. baumannii isolates were identified based on the colonial morphology present on various culture media, specific biochemical tests and VITEK 2® compact confirmation. The distribution of A. baumannii was 38.23 % (39 out of 102), in which 27 (68.75 %) and 12 (30.76 %) of isolates recovered from males and females respectively (fig. 1). In many specialized hospital units as well as medical ICUs, outbreaks of infection caused by A. baumannii that are resistant to multiple antibiotic groups, including carbapenems, are a major concern. The main effect of having a carbapenem-resistant A. baumannii infection is that “last resort” antibiotics like CL, PB or TIG must be used[41]. The percentage of infections among males was higher than in females, thus males were more affected by DFUs than females. Our results are in agreement with those reported by other researchers in Iran and Iraq. They showed that the rate of infection in males were 67.4 % and 70 % respectively[43,44].
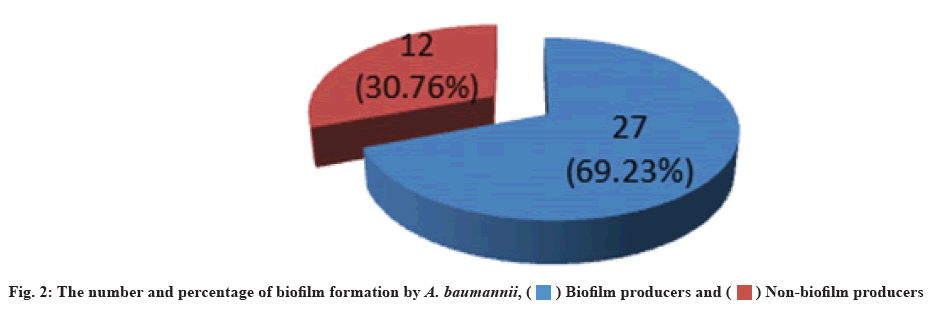
The biofilm forming capacity of isolates had been measured. A. baumannii can form biofilm on polystyrene surfaces. The microtiter plate method detected that 27 (69.23 %) of isolates were biofilm producers, while 12 (30.76 %) were non-biofilm producers (fig. 2). Similar results were recorded by Hassan et al.[45], they mentioned that 64.7 % of A. baumannii isolates from different clinical sources were biofilm producers. The capacity of A. baumannii to build biofilms is one of their most crucial virulencerelated attributes. Many recalcitrant infections were brought on by biofilm-producing bacteria, which are notoriously difficult to eradicate, so they show resistance to antibiotics by several methods such as decreased growth rate, limited penetration of antibiotic into biofilm and expression of resistant genes[46].
A. baumannii isolates from all 102 (100 %) DFU patients were found to contain beta-lactamase enzyme. There are four main molecular classes for betalactamases: A, B, C and D. Serine-beta-lactamases refer to the classes A, C and D of beta-lactamase, while MBL refers to group B beta-lactamases. Newer beta-lactamases that hydrolyze cephamycins, cephalosporins, monobactams and carbapenems are growing concern because they reduce the range of available treatments, which can result in treatment failures and poor prognoses[43]. Several mechanisms causing resistance include cabapenemase production, modification of Penicillin-Binding Proteins (PBPs), loss of porins, and/or altered efflux pump activity[47].
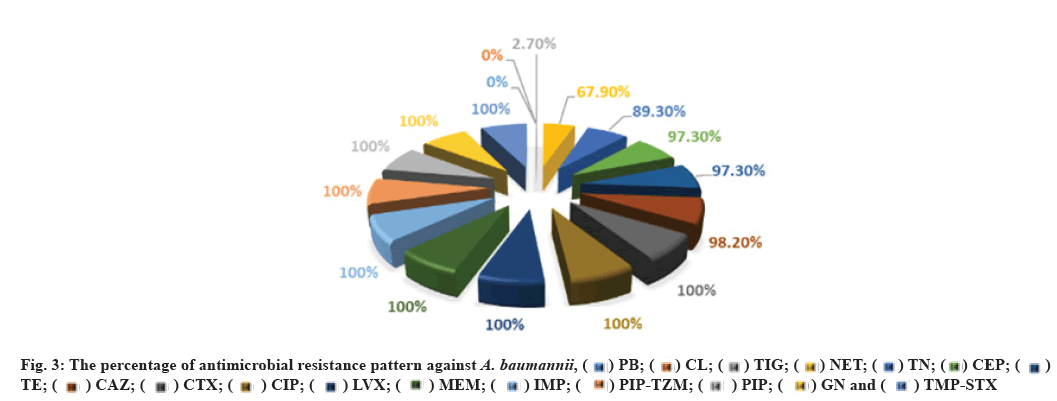
All 39 isolates in this case were resistant to all antimicrobials studied, including CTX, CIP, LVX, AN, GN, PIP, PIP-TZM, TMP-STX, IMP and MEM, with 38 isolates (97.43 %) being resistant to CAZ, while the number and rate of resistance to CEF and TE was 37 (94.87 %), TN 35 (89.74 %), NET 30 (76.92 %), TIG 1 (2.56 %) respectively. On the other hand, all 39 (100 %) of A. baumannii isolates were sensitive to CL and PB (fig. 3). These results agree with those mentioned by Hans et al. They found that all studied A. baumannii obtained from Indian hospitals were resistant to GN, erythromycin, TMP/ STX, PIP/TZM and CAZ, while they were sensitive to CL and TIG[43]. In addition, similar results were recorded in Iran among MDR A. baumannii in which 91.2 % were resistant to CAZ[48].
All of the examined A. baumannii isolates were discovered to produce MBL, while 12 isolates (30.76 %) were tested positive for ESBL (fig. 4). Similarly, ESBL production was detected in 42 (28 %) of isolates in India[49]. On the other hand, a study in Saudi Arabia only 7 (7 %) of A. baumannii isolates were positive for ESBL using DDST screening method[50]. Our results indicate the prevalence of A. baumannii producing MBL which may cause outbreak in immunocompromised patients, thus increase morbidity and mortality and limit therapeutic options due to the high degree of MDR. The higher rate of MBL-producing bacteria may be due to patients having predisposing factors such as duration of hospital stay, the severity of illness, compromised immunity, contamination of respiratory care unit tubes, unit hygiene and catheterization[51]. Another remarkable reason for the occurrence of a high rate of MBL-producing A. baumannii is the abuse, misuse and random use of antimicrobial agents, especially broad-spectrum antibiotics.
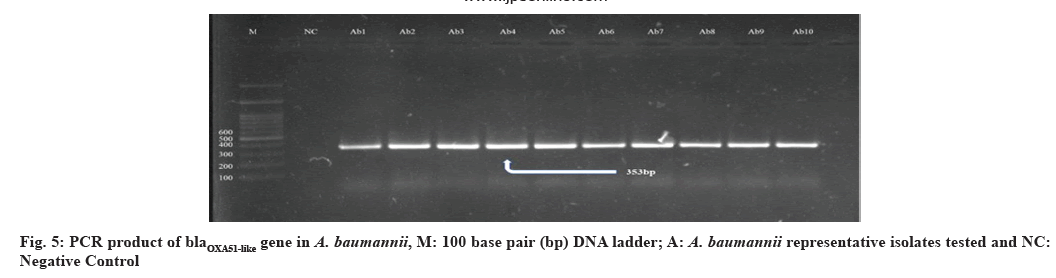
Presence and prevalence of certain important genes encoding MDR and biofilm formation among carbapenem-resistant A. baumannii were studied namely blaOXA51-like, blaOXA23-like, ISAba1 and ompA, which encode carbapenem-hydrolyzing oxacillinases, expression of OXA carbapenemase and biofilm formation respectively, were screened on the molecular level using conventional PCR technique. This is of valuable importance in predominant virulence genetic markers and their dissemination in local strains of this nosocomial pathogen circulating in the Kurdistan region. Analysis of the occurrence of OXA-type encoding genes revealed that the studied genes were detected in all 39 (100 %) of A. baumannii isolates. The genes encoding the blaOXA51-like β-lactamases are chromosomally located in all A. baumannii isolates[52]. Through the occasional detection of this gene in A. baumannii and Acinetobacter genomic species limits its use as a single identification method, on the other hand, the expression of intrinsic blaOXA51-like in A. baumannii due to ISAba1 insertion has been associated with carbapenem resistance. Our findings support those of other studies representing that all A. baumannii isolates were PCR positive for blaOXA51-like carbapenemase[38,53].
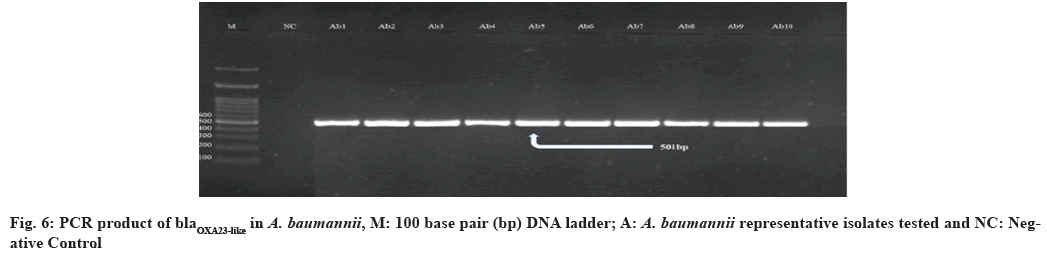
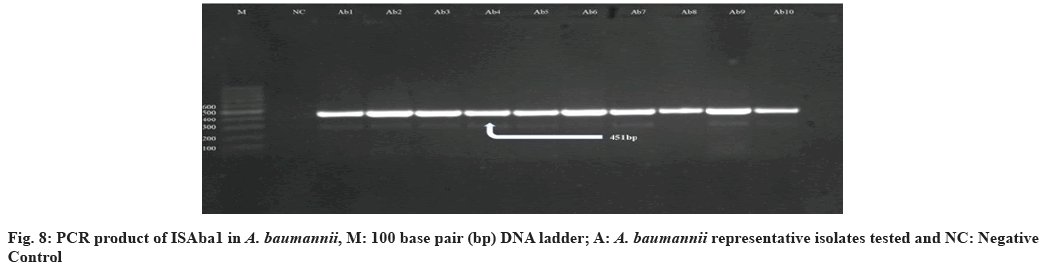
In this study, all 39 (100 %) A. baumannii isolates were confirmed as blaOXA23-like gene positive. Similar studies performed in Qatar and Italy mentioned that all tested isolates were positive for this gene[54,55]. In the findings, that among isolates with blaOXA51- like, as sole carbapenemase gene, IMP and MEM resistance was related only with isolates in which ISAba1 was upstream of blaOXA51-like suggests that ISAba1 providing the promoter for this gene, evidently suggesting that ISAba1 is important in the expression of this gene and the production of OXA- 23 was the major carbapenem resistance mechanism, with an upstream insertion of ISAba1, thus supporting international data about the worldwide emergence of this carbapenemase. Also the presence of ISAba1 in all 39 (100 %) isolates was detected positive for blaOXA51-like and blaOXA23-like gene and ISAba1 plays an important role in overexpression of these genes[51] (fig. 5-fig. 8). The incidence of coexistence with ISAba1 was 80 %, 66 % and 85 %, 80 % for blaOXA23-like and blaOXA51-like in Turkey and Egypt respectively[56,57]. The various results could be as a result of the various geographical locations, clinical samples and antibiotic patterns in various studies.
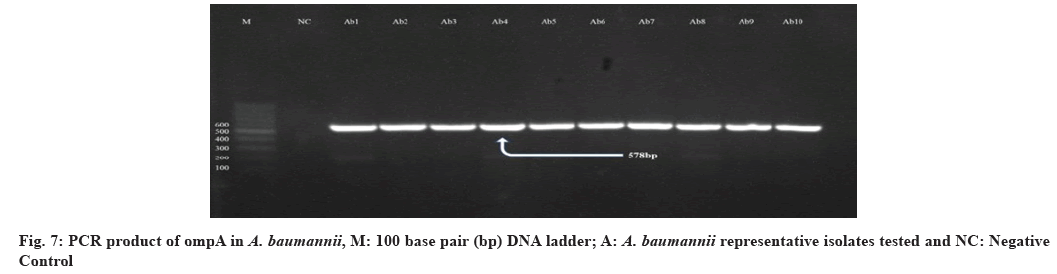
Meanwhile, the ompA was detected in all A. baumannii isolates. It has a significant role in several interactions during the host infection including initiation of apoptosis in host cells, adherence invasion to epithelial cells and differentiation of host immune cells. These findings clarify that ompA has a vital role in the pathogenesis of A. baumannii[58]. Our study agrees with that conducted in Iran by Azizi et al. They mentioned that all studied A. baumannii were PCR positive for ompA gene[59]. However, all the isolates are positive for ompA gene, but not all isolates were biofilm producers, this may be due to other genes like Biofilm-associated protein (Bap), Exopolysaccharide (EPS), chaperone-subunit usher E (csuE) and Biofilm-Based Fermentation Mode (BFM) which coding for biofilm formation also.
The results revealed that 25 (64.10 %) were harbored from one to four bands of plasmid ranged from 1 kbp to more than 10 kbp. Analysis of plasmid DNA content by agarose gel electrophoresis revealed variation in their size and number (fig. 9). Our findings are in accordance with that reported by Aris et al., who mentioned that 72.7 % of tested A. baumannii isolated from different clinical sources harbor plasmids, while Chen et al. mentioned that 90 % of A. baumannii contain plasmids[60,61]. The presence of more than one DNA band in the agarose gel is a good indicator for the existence of more than one plasmid DNA species. The presented findings clarify the dissemination of plasmids among A. baumannii isolates that may carry genes encoding resistance to broad spectrum of commonly used antibiotics which may explain the reason of developing antibioticresistant patterns in bacterial isolates under study, almost all our isolates harbored 1-2 high molecular weight plasmids more than 10 Kbp. According to the various researchers, the majority of A. baumannii isolates contain at least one large plasmid in addition to other smaller plasmids, which encode for several virulence factors such as hemolysin, toxins, biofilm formation, adherence ability, in addition to antibiotic-resistant markers. However, some of the isolates have no plasmids, yet they were resistant to a variety of antimicrobial agents commonly used. It’s possible that certain antibiotic resistance genes exist elsewhere, such as on transposable elements or the bacterial chromosome, rather than on the plasmid[61]. Since many species of bacteria contain plasmids, plasmid profiling or plasmid fingerprinting has been used to investigate outbreaks of numerous bacterial diseases and track the inter and intra-species spread of antibiotic resistance. Plasmids can also be used as marker of different bacterial strains[62].
Also, the ability of purified DH5-α transformant colonies to produce ESBL enzymes was tested and the results revealed that these transformant colonies were ESBL producers, indicating that the genes coding for ESBL production located on plasmid DNA and successfully transformed to the standard laboratory E. coli DH5-α strain. Furthermore, the ability of purified DH5-α transformant colony to produce biofilm was tested and the results demonstrated their ability to grow on Congo red agar and produce typical black colonies with a dry crystalline consistency. These findings indicate that the genes encoding biofilm formation are also located on plasmid DNA. Association of plasmid-mediated quinolone resistance and ESBL is well documented[49,61]. The presence or lack of plasmids is highly connected with resistance to different antimicrobial drugs and we observed a tendency for the resistance ratio to rise as the quantity of plasmids rose. Usually strains harboring multiple plasmids simultaneously show co-resistance to various antibiotic classes[62].
Furthermore, genetic transformation experiment was performed to determine the site of genes encoding antibiotic resistance in A. baumannii isolates A3, A11 and A28. The process of genetic transformation depends on the ability of laboratory E. coli DH5-α strain, which has known genotype to be transformed from sensitive strain to resistant one for certain antimicrobial agents after treating with CaCl2 and exposing to the prepared plasmid DNA from chosen bacterial isolates using heat shock technique. In this study, the genes that encode resistance to AN, TE, CAZ, CIP, GM and CXT were located on plasmid DNA studied A. baumannii isolates. DH5-α transformant colonies showed different levels of resistance to mentioned antibiotics, which might be explained by the antibiotic resistance genes present on several R-plasmid fragments[63]. The incorporation of a variety of DNA fragments during natural transformation with species-foreign DNA may affect the antimicrobial sensitivity profile and contribute to the development of antimicrobial resistance in bacteria[64]. A. baumannii genome is significantly formed by horizontal gene transfer events. This is especially true for genetic components that confer antibiotic resistance and that are thought to have been acquired from closely related species like Pseudomonas and Enterobacteriaceae. Recently, another possible horizontal gene transfer pathway was identified in A. baumannii showing that the outer membrane vesicles can mediate transfer of resistance genes[65]. No thermo-sensitive plasmids were found in this study.
The prevalence of DFU in diabetic patients is high. If left untreated, these infections, which are frequently poly-microbial in nature and have a higher level of morbidity, can result in amputations. Due to the emergence of MDR, A. baumannii is becoming global medical challenge and also the rate of infections with MDR especially, MBL and ESBL isolates of A. baumannii remarkably increased among patients admitted to ICU and different hospital wards in Erbil city. Carbapenems are known to be effective therapeutic agents for A. baumannii infections and their resistance limits the treatment options and restricts to use PB and CL. Increased expression of blaOXA51-like and blaOXA23-like genes is associated with presence of upstream insertion sequences of ISAba1 in all MDR studied isolates. A. baumannii develops antimicrobial resistance via horizontal gene transfer including plasmid acquisition in addition to other transposable elements and the genes that encode ESBL, biofilm formation and resistance to many commonly used antibiotics located on plasmid DNA. To distinguish cooperative or commensal behavior of uncommon species, more research is required to understand the interactions along with coexistence of the bacterial pathogens involved in DFU infections.
Conflict of interests:
The authors declared no conflict of interest.
References
- Yuniartika W, Nur Hidayati DA. Improving knowledge of diabetes mellitus patients using booklet. J Med Chem Sci 2021;4(3):238-45.
- Abba A, Nnenna UK, Nwaogu J. Antidiabetic, antioxidant and hypolipidemic potentials of Sterculia setigera methanol stem bark extract in alloxan-induced diabetic rats. Int J Adv Biol Biomed Res 2022;10(1):84-97.
- Zarei M, Ghafaryan H. Alfa-glucosidase inhibitory and antioxidant activity of hexane extract of flowers, leave and stems of Haplophyllum acutifolium DC. and Ferulahaussknechtii Wolff ex Rech. Int J Adv Biol Biomed Res 2020;8(2):153-64.
- Azizi S, Behzadi Andohjerdi R, Mohajerani H. Evaluation of two types of vitamin D receptor gene morphism in patients with type 2 diabetes and obesity. Int J Adv Biol Biomed Res 2020;8(1):86-91.
- Singh DK, Mondol S, Satpathy I, Patnaik BCM. Self-Care Practices (SCPs) among the type II diabetics affiliating to the self-help groups (SHGs) in Bangladesh. J Med Chem Sci 2022;5(6):1075-84.
[Crossref]
- Muhsen T, Risan M, Alqaysi N. Candida Berkh. (1923) species and their important secreted aspartyl proteinases (SAP) genes isolated from diabetic patients. Int J Adv Biol Biomed Res 2021;9(3):286-97.
- Alrifai SB, Mahmood WS, Jasem NH. Surveillance of multidrug-resistant Iraqibacter isolated from patients with urinary tract infection at a Baghdad urology center. J Med Chem Sci 2022;5(5):753-9.
[Crossref]
- Vafaei S, Mirnejad R, Amirmozafari N. Determining the patterns of antimicrobial susceptibility and the distribution of blaCTX-M genes in strains of Acinetobacterbaumannii isolated from clinical samples. J Isfahan Med Sch 2013;31(252):1443-51.
- Perez F, Hujer AM, Hujer KM, Decker BK, Rather PN, Bonomo RA. Global challenge of multidrug-resistant Acinetobacter baumannii. Antimicrob Agents Chemother 2007;51(10):3471-84.
[Crossref] [Google scholar] [PubMed]
- Karageorgopoulos DE, Kelesidis T, Kelesidis I, Falagas ME. Tigecycline for the treatment of multidrug-resistant (including carbapenem-resistant) Acinetobacter infections: A review of the scientific evidence. J Antimicrob Chemother 2008;62(1):45-55.
[Crossref] [Google scholar] [PubMed]
- Chisari LM, Antonin G, Arcidiacono G, Chisari EM. Human amniotic membrane in diabetic foot healing processes: A retrospective study. Acta Med Mediterr 2021;37(3):1399-403.
- Hurlow JJ, Humphreys GJ, Bowling FL, McBain AJ. Diabetic foot infection: A critical complication. Int Wound J 2018;15(5):814-21.
[Crossref] [Google scholar] [PubMed]
- Reiber GE, Smith DG, Carter J, Fotieo G, Deery HG, Sangeorzan JA, et al. A comparison of diabetic foot ulcer patients managed in VHA and non-VHA settings. J Rehabil Res Dev 2001;38(3):309-18.
[Google scholar] [PubMed]
- Antonin G, Maria CL, Giuseppe A, Santo DN, Margherita CE. Chronic stress in the healing processes of elderly subjects with chronic skin lesions (diabetic foot). Acta Med Mediterr 2021;37(6):2957-63.
- Gao R, Gu Y, Jing K, Zhang X, Wang C, Xiao M. Influence of empowerment theory plus graphic health education on self-care knowledge and behavior of high-risk diabetic foot patients. Acta Medica Mediterr 2022;38(4):2745-53.
[Crossref]
- Shettigar K, Jain S, Bhat DV, Acharya R, Ramachandra L, Satyamoorthy K, et al. Virulence determinants in clinical Staphylococcus aureus from monomicrobial and polymicrobial infections of diabetic foot ulcers. J Med Microbiol 2016;65(12):1392-404.
[Crossref] [Google scholar] [PubMed]
- Rahim F, Ullah F, Ishfaq M, Afridi AK, ur Rahman S, Rahman H. Frequency of common bacteria and their antibiotic sensitivity pattern in diabetics presenting with foot ulcer. J Ayub Med Coll Abbottabad 2016;28(3):528-33.
[Google scholar] [PubMed]
- Cardoso NA, Cisneiros LD, Machado CJ, Cenedezi JM, Procópio RJ, Navarro TP. Bacterial genus is a risk factor for major amputation in patients with diabetic foot. Rev Col Bras Cir 2017;44:147-53.
[Crossref] [Google scholar] [PubMed]
- Biyik I, Cayci YT, Birinci A. Investigation of KPC genes by PCR and evaluation of carbapenem inactivation method in carbapenem resistant Pseudomonas aeruginosa isolates. Acta Med Mediterr 2020;36(4):2331-5.
- Pfeifer Y, Cullik A, Witte W. Resistance to cephalosporins and carbapenems in Gram-negative bacterial pathogens. Int J Med Microbiol 2010;300(6):371-9.
[Crossref] [Google scholar] [PubMed]
- Shakil S, Khan AU. Infected foot ulcers in male and female diabetic patients: A clinico-bioinformative study. Ann Clin Microbiol Antimicrob 2010;9(1):1-10.
[Crossref] [Google scholar] [PubMed]
- Lahiri KK, Mani NS, Purai SS. Acinetobacter spp. as nosocomial pathogen: Clinical significance and antimicrobial sensitivity. Med J Armed Forces India 2004;60(1):7-10.
[Crossref] [Google scholar] [PubMed]
- Murali TS, Kavitha S, Spoorthi J, Bhat DV, Prasad AS, Upton Z, et al. Characteristics of microbial drug resistance and its correlates in chronic diabetic foot ulcer infections. J Med Microbiol 2014;63(10):1377-85.
[Crossref] [Google scholar] [PubMed]
- Rasmussen BA, Bush K. Carbapenem-hydrolyzing beta-lactamases. Antimicrob Agents Chemother 1997;41(2):223-32.
[Crossref] [Google scholar] [PubMed]
- Héritier C, Poirel L, Fournier PE, Claverie JM, Raoult D, Nordmann P. Characterization of the naturally occurring oxacillinase of Acinetobacter baumannii. Antimicrob Agents Chemother 2005;49(10):4174-9.
[Crossref] [Google scholar] [PubMed]
- Ebrahem AM, Jameel SK, Abbas AH. Investigation of biofilm formation and antibiotic resistant of bacteria isolated from septic neonates. J Med Chem Sci 2023;6(4):816-26.
[Crossref]
- Nath H, Barkataki D. Prevalence of ESBL and MBL producing Acinetobacter isolates in clinical specimens in tertiary care hospital, Assam, India. Int J Curr Microbiol App 2016;5(11):515-22.
- Brown S, Amyes S. OXA β-lactamases in Acinetobacter: The story so far. J Antimicrob Chemother 2006;57(1):1-3.
[Crossref] [Google scholar] [PubMed]
- Donnarumma F, Sergi S, Indorato C, Mastromei G, Monnanni R, Nicoletti P, et al. Molecular characterization of Acinetobacter isolates collected in intensive care units of six hospitals in Florence, Italy, during a 3-year surveillance program: A population structure analysis. J Clin Microbiol 2010;48(4):1297-304.
[Crossref] [Google scholar] [PubMed]
- Bakir SH, Ali FA. Comparison of different methods for detection of biofilm production in multi-drug resistance bacteria causing pharyngotonsillitis. Int J Res Pharm Biosci 2016;3(2):13-22.
- Wayne PA. Performance standards for antimicrobial susceptibility testing. 25th ed. National committee for clinical laboratory standards; 2015.
- Trivedi GR, Soni ST, Vegad MM, Yadav KS. Occurrence and detection of extended spectrum [Beta]-lactamase and ampc [beta]-lactamase in clinical isolates of Pseudomonasaeruginosa and Acinetobacter baumannii by inhibitor based method. Int J Microbiol Res 2012;4(8):299.
- Abbo S, Gopher A, Rubin B, Lev-Yadun S. On the origin of near Eastern founder crops and the ‘dump-heap hypothesis’. Genet Resour Crop Evol 2005;52(5):491-5.
- Moosavian M, Sirous M, Shams N. Phenotypic and genotypic detection of extended spectrum β-lactamase and carbapenemases production including blaTEM, blaPER and blaNDM-1 genes among Acinetobacter baumannii clinical isolates. Jundishapur J Microbiol 2017;10(12):1-6.
- Shahcheraghi F, Abbasalipour M, Feizabadi MM, Ebrahimipour GH, Akbari N. Isolation and genetic characterization of metallo-β-lactamase and carbapenamase producing strains of Acinetobacter baumannii from patients at Tehran hospitals. Iran J Microbiol 2011;3(2):68-74.
[Google scholar] [PubMed]
- Vijayakumar S, Rajenderan S, Laishram S, Anandan S, Balaji V, Biswas I. Biofilm formation and motility depend on the nature of the Acinetobacter baumannii clinical isolates. Front Public Health 2016;4:105.
[Crossref] [Google scholar] [PubMed]
- Badave GK, Kulkarni D. Biofilm producing multidrug resistant Acinetobacter baumannii: An emerging challenge. J Clin Diagn Res 2015;9(1):DC08.
[Crossref] [Google scholar] [PubMed]
- Turton JF, Ward ME, Woodford N, Kaufmann ME, Pike R, Livermore DM, et al. The role of ISAba1 in expression of OXA carbapenemase genes in Acinetobacter baumannii. FEMS Microbiol Lett 2006;258(1):72-7.
[Crossref] [Google scholar] [PubMed]
- Lopes BS, Amyes SG. Role of ISAba1 and ISAba125 in governing the expression of blaADC in clinically relevant Acinetobacter baumannii strains resistant to cephalosporins. J Med Microbiol 2012;61(8):1103-8.
[Crossref] [Google scholar] [PubMed]
- Smani Y, Fàbrega A, Roca I, Sánchez-Encinales V, Vila J, Pachón J. Role of OmpA in the multidrug resistance phenotype of Acinetobacter baumannii. Antimicrob Agents Chemother 2014;58(3):1806-8.
[Crossref] [Google scholar] [PubMed]
- Al-Otraqchi K. Molecular genetic studies of bacterial species isolated from different patients with diarrhea. Hawler Medical University, Iraq; 2006.
- Akingbade OA, Balogun SA, Ojo DA, Afolabi R, Motayo BO, Okerentugba PO, et al. Plasmid profile analysis of multidrug resistant Pseudomonas aeruginosa isolated from wound infections in South West, Nigeria. World Appl Sci J 2012;20(6):766-75.
- Hans R, Bisht D, Agarwal R, Irfan M. Phenotypic detection of MBL, Ampc beta-lactamase and carbapenemases in multi-drug resistant isolates of Acinetobacterbaumannii. Int J Med Res Health Sci 2015;4(2):311-6.
- Ganjo A. Molecular characterization of β-lactum resistant A. baumannii and P. aeruginosa isolated from patients in Erbil city-Kurdistan region. Hawler Medical University, Iraq; 2015.
- Hassan A, Usman J, Kaleem F, Omair M, Khalid A, Iqbal M. Evaluation of different detection methods of biofilm formation in the clinical isolates. Braz J Infect Dis 2011;15:305-11.
[Crossref] [Google scholar] [PubMed]
- Babu KV, Visweswaraiah DS, Kumar A. The influence of imipenem resistant metallo-beta-lactamase positive and negative Pseudomonas aeruginosa nosocomial infections on mortality and morbidity. J Nat Sci Biol Med 2014;5(2):345-51.
[Crossref] [Google scholar] [PubMed]
- Tsakris A, Ikonomidis A, Poulou A, Spanakis N, Vrizas D, Diomidous M, et al. Clusters of imipenem-resistant Acinetobacter baumannii clones producing different carbapenemases in an intensive care unit. Clin Microbiol Infect 2008;14(6):588-94.
[Crossref] [Google scholar] [PubMed]
- Farahani R, Moniri R, Farahani KD. Multi-drug resistant Acinetobacter-derived cephalosporinase and OXAsetC genes in clinical specimens of Acinetobacter spp. isolated from teaching hospital. Jundishapur J Microbiol 2013;6(2):181-5.
- Sinha M, Srinivasa H, Macaden R. Antibiotic resistance profile and extended spectrum beta-lactamase (ESBL) production in Acinetobacter species. Indian J Med Res 2007;126(1):63-7.
[Google scholar] [PubMed]
- Safari M, Nejad AS, Bahador A, Jafari R, Alikhani MY. Prevalence of ESBL and MBL encoding genes in Acinetobacter baumannii strains isolated from patients of intensive care units (ICU). Saudi J Biol Sci 2015;22(4):424-9.
[Crossref] [Google scholar] [PubMed]
- Varaiya A, Kulkarni N, Kulkarni M, Bhalekar P, Dogra J. Incidence of metallo beta lactamase producing Pseudomonas aeruginosa in ICU patients. Indian J Med Res 2008;127(4):398-402.
[Google scholar] [PubMed]
- Poirel L, Bonnin RA, Nordmann P. Genetic basis of antibiotic resistance in pathogenic Acinetobacter species. IUBMB Life 2011;63(12):1061-7.
[Crossref] [Google scholar] [PubMed]
- Tena D, Martínez NM, Oteo J, Sáez D, Vindel A, Azañedo ML, et al. Outbreak of multiresistant OXA-24-and OXA-51-producing Acinetobacter baumannii in an internal medicine ward. Jpn J Infect Dis 2013;66(4):323-6.
[Crossref] [Google scholar] [PubMed]
- Rafei R, Pailhoriès H, Hamze M, Eveillard M, Mallat H, Dabboussi F, et al. Molecular epidemiology of Acinetobacter baumannii in different hospitals in Tripoli, Lebanon using blaOXA-51-like sequence based typing. BMC Microbiol 2015;15(1):1-7.
[Crossref] [Google scholar] [PubMed]
- Rolain JM, Loucif L, Al-Maslamani M, Elmagboul E, Al-Ansari N, Taj-Aldeen S, et al. Emergence of multidrug-resistant Acinetobacter baumannii producing OXA-23 carbapenemase in Qatar. New Microbes New Infect 2016;11:47-51.
[Crossref] [Google scholar] [PubMed]
- Cicek AC, Saral A, Iraz M, Ceylan A, Duzgun AO, Peleg AY, et al. OXA-and GES-type β-lactamases predominate in extensively drug-resistant Acinetobacter baumannii isolates from a Turkish University Hospital. Clin Microbiol Infect 2014;20(5):410-5.
[Crossref] [Google scholar] [PubMed]
- Al-Agamy MH, Khalaf NG, Tawfick MM, Shibl AM, El Kholy A. Molecular characterization of carbapenem-insensitive Acinetobacter baumannii in Egypt. Int J Infect Dis 2014;22:49-54.
[Crossref] [Google scholar] [PubMed]
- Badmasti F, Siadat SD, Bouzari S, Ajdary S, Shahcheraghi F. Molecular detection of genes related to biofilm formation in multidrug-resistant Acinetobacter baumannii isolated from clinical settings. J Med Microbiol 2015;64(5):559-64.
[Crossref] [Google scholar] [PubMed]
- Azizi O, Shahcheraghi F, Salimizand H, Modarresi F, Shakibaie MR, Mansouri S, et al. Molecular analysis and expression of bap gene in biofilm-forming multi-drug-resistant Acinetobacter baumannii. Rep Biochem Mol Biol 2016;5(1):62-72.
[Google scholar] [PubMed]
- Aris P, Boroumand MA, Douraghi M. Amikacin resistance due to the aphA6 gene in multi-antibiotic resistant Acinetobacter baumannii isolates belonging to global clone 1 from Iran. BMC Microbiol 2019;19(1):1-6.
[Crossref] [Google scholar] [PubMed]
- Chen Y, Gao J, Zhang H, Ying C. Spread of the blaOXA–23-containing TN 2008 in carbapenem-resistant Acinetobacter baumannii isolates grouped in CC92 from China. Front Microbiol 2017;8:1-6.
[Crossref] [Google scholar] [PubMed]
- Jan NA, Meshram SU, Kulkarni A. Plasmid profile analysis of multidrug resistant E. coli isolated from UTI patients of Nagpur city, India. Rom Biotechnol Lett 2009;14(5):4635-40.
- Tortora GJ, Funke BR, Case CL. Microbiology: An introduction. Pearson; 2018.
- Domingues S, Harms K, Fricke WF, Johnsen PJ, da Silva GJ, Nielsen KM. Natural transformation facilitates transfer of transposons, integrons and gene cassettes between bacterial species. PLoS Pathog 2012;8(8):e1002837.
[Crossref] [Google scholar] [PubMed]
- Rumbo C, Fernández-Moreira E, Merino M, Poza M, Mendez JA, Soares NC, et al. Horizontal transfer of the OXA-24 carbapenemase gene via outer membrane vesicles: A new mechanism of dissemination of carbapenem resistance genes in Acinetobacterbaumannii. Antimicrob Agents Chemother 2011;55(7):3084-90.
[Crossref] [Google scholar] [PubMed]
 ) Male and (
) Male and ( ) Female
) Female