- *Corresponding Author:
- Jamal Sleman Sofi
Department of Biology, College of Education, Salahaddin University-Erbil, Kurdistan Region 44001, Iraq
E-mail: jamal.sofi@su.edu.krd
| This article was originally published in a special issue, “Modern Applications in Biomedical Research and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2022:84(3)Spl Issue “27-31” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The calreticulin gene has nine exons and most of the frequent mutations screened and observed in the 9th exon in myeloproliferative neoplasm patients, especially essential thrombocythemia and primary myelofibrosis. In the current study, an uncommon calreticulin mutation was observed in a 51 y old man with a polycythemia vera phenotype. Somatic novel homozygous mutations affect exon 9 of the calreticulin gene. These recent genetic variations were not reported in previous studies. Molecular screening of calreticulin may be important for polycythemia vera patients that are negative for the Janus kinase 2 V617F and exon 12 Janus kinase 2. A total of 11 negative Janus kinase 2 V617F polycythemia vera patients were previously diagnosed by the amplification refractory mutation system-polymerase chain reaction technique. Sanger sequencing was performed to screen exon 12 of the Janus kinase 2 gene and the last exon of the calreticulin gene. In addition, all 11 negative V617F patients were wild for exon 12 of the Janus kinase 2 gene. The calreticulin lesion in myeloproliferative neoplasms as a second biomarker, particularly in essential thromobocythemia and primary myelofibrosis, is more informative for diagnosis. Molecularly, in negative Janus kinase 2 V617F and exon 12 patients with polycythemia vera phenotypes, genotyping calreticulin may be useful.
Keywords
Myeloproliferative neoplasms, calreticulin, exon 12, Janus kinase 2
The Philadelphia-negative Myeloproliferative Neoplasms (MPNs) are defined by the clonal proliferation of an aberrant hematopoietic stem/ progenitor cell involving Polycythemia Vera (PV), Essential Thrombocythemia (ET) and Myelofibrosis (MF)[1]. In general, more than 95 % of PV cases carried with Janus Kinase 2 (JAK2) V617F mutation at nucleotide position c.1849G>T in 617th amino acid results substitution of a valine for a phenylalanine. While mutations in exon 12 of the JAK2 gene were heterogeneous and variable, including substitutions, deletions and duplications, they were approximately found in 3 % of PV patients without JAK2 V617F[2,3]. In addition, incidence of JAK2 V617F ranges from 51 %-55 % in ET and 50 %-65 % in Primary Myelofibrosis (PMF)[2,4]. Clinically, PV patients had a higher level of hemoglobin and hematocrit. In contrast, thrombocytosis was significantly increased in ET7 than in PV and PMF patients, which acted as a risk factor for the occurrence of thrombosis[5]. Annual incidence of PV and ET were relatively similar 1.0-2.0 per 100 000 people while, frequency of PMF was rare 0.3 per 100 000 people[6]. Moreover, life expectancy was low in PV as found in ET, 15 to 18 y, respectively[7]. The diagnosis of V617F mutation in JAK2 greatly advanced our understanding of the biology of these disorders[8]. Molecularly, mutations in the JAK2 gene are mostly associated with PV in all cases. In contrast, ET can be driven either by JAK2, Calreticulin (CALR) or Myeloproliferative Leukemia (MPL)-mutated, as well as triple negative[9]. In the 32 various types of CALR mutations, a 52 base pair (bp) deletion called “type 1” and a 5 bp insertion called type 2 mutation, were the most frequent variants identified in negative JAK2 V617F MPNs. All of them conferred frameshift and produced unique c-terminus[10]. In multi- gene screening among 27 relevant genes, CALR was considered the second-most prevalent target gene in ET, while in PV patients it was not identified[11]. Normally, wild-type CALR participates in hematopoiesis, including megakaryocytic/erythrocytic differentiation and Haematopoietic Stem Cells (HSCs) self-renewal[12].
Materials and Methods
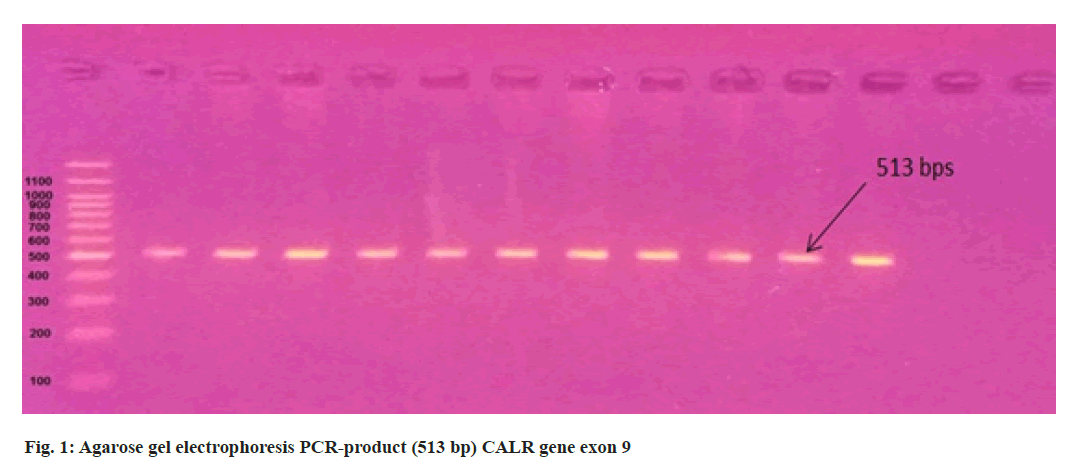
3 ml of blood from 11 suspected PV patients were collected in an Ethylenediamine Tetraacetic Acid (EDTA) tube from Hiwa Hemato Oncology Hospital in Sulaimani city. The total genomic Deoxyribonucleic Acid (DNA) was extracted from peripheralblood samples using the (AddbioPrep) Genomic DNA extraction kit according to the manufacturer’s protocols and the quality of DNA samples was assessed using NanoDropTM One (Thermo Scientific) and stored at -20° for later downstream applications. The negative samples for the V617F mutation were screened for mutations in exon 9 of the CALR gene. Primer for CALR exon 9 forward 5’-GCTATCGGGTATCACCTCTGAC-3’ and CALR reverse 5’-AGTTCTCGAGTCTCACAGAGAC-3’. Exon 12 JAK2 gene primer include forward 5’-CAAAGTTCAATGAGTTGACCCC-3’ and reverse 5’-CTAACATCTAACACAAGGTTGGC-3’. 40-80 ng of patient DNA were used for Polymerase Chain Reaction (PCR) amplification (513 bps) and (463 bps) of the CALR and JAK2 genes respectively. Using AddStart Taq DNA Polymerase kit (Addbio) according to the manufacturer’s protocol, 3 µl buffer, 2 µl Magnesium Chloride (MgCl2), 2 µl deoxynucleotide Triphosphate (dNTP), 1 µl of each primer, 0.4 µl Taq DNA polymerase, 13.6 µl PCR grade water and 2 µl template DNA were mixed in PCR tube and amplified in the following PCR conditions for CALR and JAK2 genes. Initial denaturation for 5 min at 94° followed by 35 cycles at 94° for 30 s, 61° for 30 s and 72° for 30 s, followed by final extension 72° for 7 min and PCR amplification products were separated using 1.5 % agarose gel electrophoresis at 90 V, 100 mA, 9 W for 1 h. The PCR products were purified and sequenced by the Sanger sequencing method using the 3130 Genetic Analyzer (Applied Biosystems, Hitachi High- Technologies, Tokyo, Japan) in Immunogene Center, Erbil, Iraq. The study was authorized by Salahaddin University’s medical ethics committee.
Results and Discussion
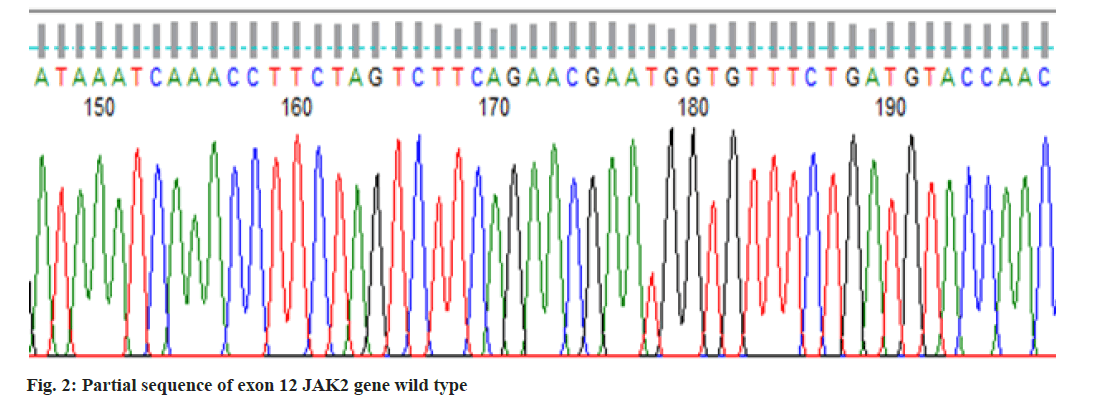
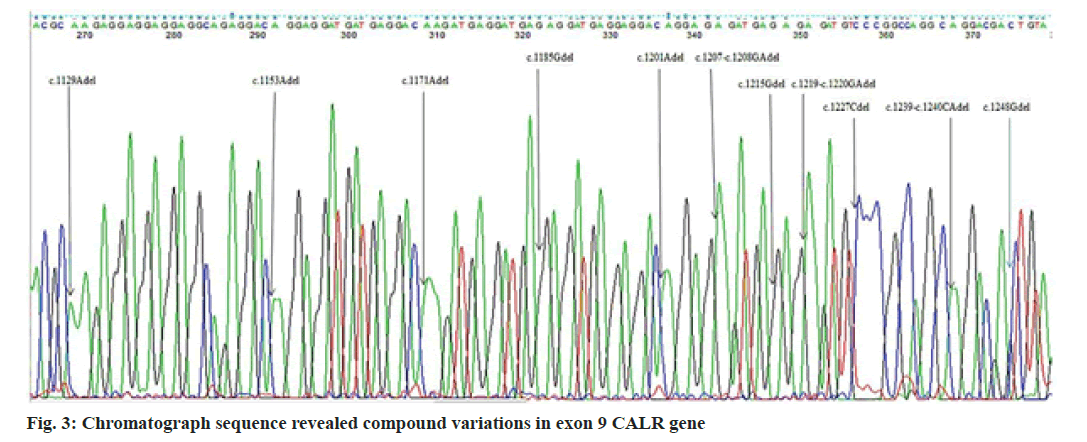
In the present study, we performed Sanger sequencing for the exon 12 of JAK2 gene and PCR product (513 bps) as outlined in fig. 1 for exon 9 of CALR gene. The results revealed only in a 51 y old male PV patient harboring compound mutations in the CALR gene. In contrast all 11 patients for exon 12 with wild type (fig. 2) presented normal sequence. 11 distinct unique homozygous variations observed and distributed at several positions that affected exon 9 is demonstrated in fig. 3. Among these mutations, the first two rare homozygous frameshift CALR lesions (c.1129Adel and c.1153Adel) were critical as shown in Table 1, which led to generate stop codons, producing truncated protein and eliminating almost last negative amino acids that modified the normal C-terminus and loss of the K-Lysine, D-Aspartic acid, E-Glutamic acid, L-Leucine (KDEL) signal. Hematologic parameters of a PV patient had a higher values for hemoglobin level, 16.5 g/dl and hematocrit, 52.5 % while red blood cells, white blood cells and platelet counts were normal at 5.82×106/µl, 10.5×103/µl, 177×103/µl respectively.
The unique compound mutations as shown in Table 1 were investigated in the current study that were not resembled by previous studies, or not previously described in PV, even in ET patients. CALR has three main domains: A globular N-domain, a proline-rich P-domain and an acidic C-domain. The C-terminal domain is enriched with a large number of negatively charged residues of acidic amino acids that are responsible for the calcium regulating function of the protein. It contains an ER retention signal (KDEL) that prevents the CALR protein from leaving the ER [13-15]. In our investigation, the complex instability of CALR was gain-of-function mutations. The outcome of the present mutations was similar to type 1 and type 2 CALR variations, which cause loss of KDEL signal. According to the majority of studies, the frequency of JAK2 V617F mutation in negative-Breakpoint Cluster Region protein-Proto-Oncogene ABL (BCR-ABL) Philadelphia MPNs represented 75 %-95 % in PV, 60 %-73 % in ET and 45 %-63 % in MPF[16-20], whereas the incidence of JAK2 exon 12 lesion is 4.5 %-5.76 % with wild JAK2 V617F PV of patients[17,21]. ET and PMF patients that lack JAK2 V617F harbor CALR mutations 7.3 %-21.9 %, 5.3 %-30 %, respectively, whereas in all cases of PV had nonmutated CALR aberrations[16-20]. Type 1, 52 bp deletions were of most prevalence in ET[16,18,19,21-23], in contrast others reported type 2, 5 bp insertions[17]. Other than two major types, minor CALR variations were found in ET but not in PMF[16,19] and not observed in ET and PMF[18] but incidence were similar[17]. The previous studies for PV individuals did not agree with our investigation. Furthermore, heterozygous CALR type 1, 52 bp deletions observed in two patients who had phenotypic PV, increased hemoglobin and hematocrit with unmutated JAK2 V617F and exon 12 relatively with thrombocytosis[24]. In a proteomic study, overexpression of endoplasmic reticulum proteins such as Calnexin (CANX) and CALR was presented on the red blood cell surface of PV patients[25]. The results may be supportive and comparable with the current findings. CALR lesion occurs in a heterozygous state[26-28]. However, a recent report showed that a PMF patient carried a rare homozygous mutation[29]. In recent large cohort study frequency of CALR mutations in negative JAK2 V716F MPNs with low 4.4 % is mostly seen in type 1 in ET and PMF, 5 bp insertion was not documented in PMF, type 1 and uncommon CALR mutations identified 1:2 in MPN-Unclassifiable (MPN-U) respectively[23]. In addition, a PV case, concurrent variations in JAK2 V617F and 3 bp deletion in exon 9 of the CALR gene were demonstrated, but frameshift was not generated[30]. While type 1 (52 bp deletions) or type 2 (5 bp insertions) restricted to exon 9 induced frameshift that changed C-terminal domain of the CALR protein[31]. Furthermore, mutant CALR generate the novel C-terminus contains multiple positively charged amino acids constitutively promote activation of Janus Kinase (JAK)/Signal Transducer and Activator of Transcription (STAT) signaling via phosphorylation (P) through binding, altered glycosylation which lead to the activation of thrombopoietin receptor MPL that induce megakaryocytopoiesis[32]. Mutant CALR exposed on cell surface which prevent the phagocytosis by Dendritic Cells (DC)[33]. Hematocrit and hemoglobin levels increased in PV patients due to an increase in red blood cell production[34]. PV patients harboring genetic lesions in exon 12 of JAK2 with a wild JAK2 V617F[35-37]. JAK2 V716F mutant PV was significantly older, had higher hemoglobin, white blood cell counts and lower platelet counts than those with the ET harboring CALR variants[21].
| Nucleotide change | C-terminal amino acids change |
|---|---|
| Wild type CALR | ….KRKEEEEAEDKEDDEDKDEDEEDEEDKEEDEEEDVPGQAKDEL |
| Mutant CALR | ....KRKRRRRQRTGG**GQDEDERMRRTGEMREMSRPGRTTV |
| c.1129Adel | |
| c.1153Adel | |
| c.1171Adel | |
| c.1185Gdel | |
| c.1201Adel | |
| c.1207-c.1208GAdel | |
| c.1215Gdel | |
| c.1219-c.1220GAdel | |
| c.1227Cdel | |
| c.1239-c.1240CAdel | |
| c.1248Gdel |
Table 1: CALR Gene Variations Identified in a PV Patient
It is a possible way for the molecular screening of CALR gene as important tool for identification rather than other gene especially for patients that lack mutations in JAK2 exon 14 and exon 12.
Author’s contributions:
Formal analysis was done by Jamal Sleman Sofi; project administration was taken care by Jamal Sleman Sofi and Hazha Jamal Hidayat; supervision was done by Hazha Jamal Hidayat; validation was done by Jamal Sleman Sofi and Hazha Jamal Hidayat; writing original draft was done by Jamal Sleman Sofi and review and editing was done by Hazha Jamal Hidayat.
Conflict of interests:
The authors declared no conflict of interest.
References
- Koschmieder S, Mughal TI, Hasselbalch HC, Barosi G, Valent P, Kiladjian JJ, et al. Myeloproliferative neoplasms and inflammation: Whether to target the malignant clone or the inflammatory process or both. Leukemia 2016;30(5):1018-24.
[Crossref] [Google Scholar] [PubMed]
- Gong JZ, Cook JR, Greiner TC, Hedvat C, Hill CE, Lim MS, et al. Laboratory practice guidelines for detecting and reporting JAK2 and MPL mutations in myeloproliferative neoplasms: A report of the Association for Molecular Pathology. J Mol Diagn 2013;15(6):733-44.
[Crossref] [Google Scholar] [PubMed]
- Pardanani A, Lasho TL, Finke C, Hanson CA, Tefferi A. Prevalence and clinicopathologic correlates of JAK2 exon 12 mutations in JAK2V617F-negative polycythemia vera. Leukemia 2007;21(9):1960-3.
[Crossref] [Google Scholar] [PubMed]
- Baxter EJ, Scott LM, Campbell PJ, East C, Fourouclas N, Swanton S, et al. Acquired mutation of the tyrosine kinase JAK2 in human myeloproliferative disorders. Lancet 2005;365(9464):1054-61.
[Crossref] [Google Scholar] [PubMed]
- Chadi S, Dhaouadi T, Sfar I, Baccouche H, Nabli R, Ben Romdhane N, et al. Analysis of JAK2 V617F mutation in Tunisian patients with myeloproliferative neoplasms. Eur J Inflamm 2021;19:1-8.
- Shallis RM, Wang R, Davidoff A, Ma X, Podoltsev NA, Zeidan AM. Epidemiology of the classical myeloproliferative neoplasms: The four corners of an expansive and complex map. Blood Rev 2020;42:100706.
[Crossref] [Google Scholar] [PubMed]
- Tefferi A, Barbui T. Polycythemia vera and essential thrombocythemia: 2021 update on diagnosis, risk?stratification and management. Am J Hematol 2020;95(12):1599-613.
[Crossref] [Google Scholar] [PubMed]
- James C, Ugo V, Le Couédic JP, Staerk J, Delhommeau F, Lacout C, et al. A unique clonal JAK2 mutation leading to constitutive signalling causes polycythaemia vera. Nature 2005;434(7037):1144-8.
[Crossref] [Google Scholar] [PubMed]
- Bose P, Verstovsek S. Updates in the management of polycythemia vera and essential thrombocythemia. Ther Adv Hematol 2019;10:1-13.
[Crossref] [Google Scholar] [PubMed]
- Pietra D, Rumi E, Ferretti VV, Di Buduo CA, Milanesi C, Cavalloni C, et al. Differential clinical effects of different mutation subtypes in CALR-mutant myeloproliferative neoplasms. Leukemia 2016;30(2):431-8.
[Crossref] [Google Scholar] [PubMed]
- Tefferi A, Lasho TL, Guglielmelli P, Finke CM, Rotunno G, Elala Y, et al. Targeted deep sequencing in polycythemia vera and essential thrombocythemia. Blood Adv 2016;1(1):21-30.
[Crossref] [Google Scholar] [PubMed]
- Salati S, Prudente Z, Genovese E, Pennucci V, Rontauroli S, Bartalucci N, et al. Calreticulin affects hematopoietic stem/progenitor cell fate by impacting erythroid and megakaryocytic differentiation. Stem Cells Dev 2018;27(4):225-36.
[Crossref] [Google Scholar] [PubMed]
- Michalak M, Groenendyk J, Szabo E, Gold LI, Opas M. Calreticulin, a multi-process calcium-buffering chaperone of the endoplasmic reticulum. Biochem J 2009;417(3):651-66.
[Crossref] [Google Scholar] [PubMed]
- Pocanschi CL, Kozlov G, Brockmeier U, Brockmeier A, Williams DB, Gehring K. Structural and functional relationships between the lectin and arm domains of calreticulin. J Biol Chem 2011;286(31):27266-77.
[Crossref] [Google Scholar] [PubMed]
- Wang WA, Groenendyk J, Michalak M. Calreticulin signaling in health and disease. Int J Biochem Cell Biol 2012;44(6):842-6.
[Crossref] [Google Scholar] [PubMed]
- Ojeda MJ, Bragós IM, Calvo KL, Williams GM, Carbonell MM, Pratti AF. CALR, JAK2 and MPL mutation status in Argentinean patients with BCR-ABL1-negative myeloproliferative neoplasms. Hematology 2018;23(4):208-11.
[Crossref] [Google Scholar] [PubMed]
- Rumi E, Harutyunyan AS, Pietra D, Milosevic JD, Casetti IC, Bellini M, et al. CALR exon 9 mutations are somatically acquired events in familial cases of essential thrombocythemia or primary myelofibrosis. Blood 2014;123(15):2416-9.
[Crossref] [Google Scholar] [PubMed]
- Zulkeflee RH, Zulkafli Z, Johan MF, Husin A, Islam MA, Hassan R. Clinical and laboratory features of JAK2 V617F, CALR, and MPL Mutations in Malaysian patients with classical myeloproliferative neoplasm (MPN). Int J Environ Res Public Health 2021;18(14):7582.
[Crossref] [Google Scholar] [PubMed]
- Guo H, Chen X, Tian R, Chang J, Li J, Tan Y, et al. Frequencies, laboratory features and granulocyte activation in Chinese patients with CALR-mutated myeloproliferative neoplasms. PLoS One 2015;10(9):e0138250.
[Crossref] [Google Scholar] [PubMed]
- Ha JS, Kim YK. Calreticulin exon 9 mutations in myeloproliferative neoplasms. Ann Lab Med 2015;35(1):22-7.
[Crossref] [Google Scholar] [PubMed]
- Rumi E, Pietra D, Ferretti V, Klampfl T, Harutyunyan AS, Milosevic JD, et al. JAK2 or CALR mutation status defines subtypes of essential thrombocythemia with substantially different clinical course and outcomes. Blood 2014;123(10):1544-51.
[Crossref] [Google Scholar] [PubMed]
- Sun C, Zhou X, Zou ZJ, Guo HF, Li JY, Qiao C. Clinical manifestation of calreticulin gene mutations in essential thrombocythemia without Janus kinase 2 and MPL mutations: A Chinese cohort clinical study. Chin Med J 2016;129(15):1778-83.
[Crossref] [Google Scholar] [PubMed]
- Belcic Mikic T, Pajic T, Sever M. CALR mutations in a cohort of JAK2 V617F negative patients with suspected myeloproliferative neoplasms. Sci Rep 2019;9(1):1-9.
[Crossref] [Google Scholar] [PubMed]
- Broséus J, Park JH, Carillo S, Hermouet S, Girodon F. Presence of calreticulin mutations in JAK2-negative polycythemia vera. Blood 2014;124(26):3964-6.
[Crossref] [Google Scholar] [PubMed]
- Brusson M, Cochet S, Leduc M, Guillonneau F, Mayeux P, Peyrard T, et al. Enhanced calreticulin expression in red cells of polycythemia vera patients harboring the JAK2 V617F mutation. Haematologica 2017;102(7):e241.
[Crossref] [Google Scholar] [PubMed]
- Nangalia J, Massie CE, Baxter EJ, Nice FL, Gundem G, Wedge DC, et al. Somatic CALR mutations in myeloproliferative neoplasms with nonmutated JAK2. N Engl J Med 2013;369(25):2391-405.
[Crossref] [Google Scholar] [PubMed]
- Klampfl T, Gisslinger H, Harutyunyan AS, Nivarthi H, Rumi E, Milosevic JD, et al. Somatic mutations of calreticulin in myeloproliferative neoplasms. N Engl J Med 2013;369(25):2379-90.
[Crossref] [Google Scholar] [PubMed]
- Lin Y, Liu E, Sun Q, Ma J, Li Q, Cao Z, et al. The prevalence of JAK2, MPL and CALR mutations in Chinese patients with BCR-ABL1–negative myeloproliferative neoplasms. Am J Clin Pathol 2015;144(1):165-71.
[Crossref] [Google Scholar] [PubMed]
- Rizvi Q, Zaidi U, Shahid S, Ahmed S, Shamsi T. Homozygous CALR mutation in primary myelofibrosis and its effect on disease phenotype: A case report and review of the literature. Case Rep Hematol 2019;2019.
[Crossref] [Google Scholar] [PubMed]
- Xu N, Ding L, Yin C, Zhou X, Li L, Li Y, et al. A report on the co-occurrence of JAK2V617F and CALR mutations in myeloproliferative neoplasm patients. Ann Hematol 2015;94(5):865-7.
[Crossref] [Google Scholar] [PubMed]
- Bel?i? Miki? T, Paji? T, Zver S, Sever M. The contemporary approach to CALR-positive myeloproliferative neoplasms. Int J Mol Sci 2021;22(7):3371.
[Crossref] [Google Scholar] [PubMed]
- Merlinsky TR, Levine RL, Pronier E. Unfolding the role of calreticulin in myeloproliferative neoplasm pathogenesis. Clin Cancer Res 2019;25(10):2956-62.
[Crossref] [Google Scholar] [PubMed]
- Liu P, Zhao L, Kroemer G, Kepp O. Secreted calreticulin mutants subvert anticancer immunosurveillance. Oncoimmunology 2020;9(1):1708126.
[Crossref] [Google Scholar] [PubMed]
- Bellucci S, Michiels JJ. The role of JAK2 V617F mutation, spontaneous erythropoiesis and megakaryocytopoiesis, hypersensitive platelets, activated leukocytes, and endothelial cells in the etiology of thrombotic manifestations in polycythemia vera and essential thrombocythemia. Semin Thromb Hemost 2006;32(4):381-98.
[Crossref] [Google Scholar] [PubMed]
- Li S, Kralovics R, de Libero G, Theocharides A, Gisslinger H, Skoda RC. Clonal heterogeneity in polycythemia vera patients with JAK2 exon12 and JAK2-V617F mutations. Blood 2008;111(7):3863-6.
[Crossref] [Google Scholar] [PubMed]
- Lakey MA, Pardanani A, Hoyer JD, Nguyen PL, Lasho TL, Tefferi A, et al. Bone marrow morphologic features in polycythemia vera with JAK2 exon 12 mutations. Am J Clin Pathol 2010;133(6):942-8.
[Crossref] [Google Scholar] [PubMed]
- Geay A, Aral B, Bourgeois V, Martin P, Airaud F, Garrec C, et al. Diagnosis of exon 12?positive polycythemia vera rescued by NGS. Clin Case Rep 2020;8(5):790-2.
[Crossref] [Google Scholar] [PubMed]