- *Corresponding Author:
- Li Jiang Ji
Department of Anorectal Surgery,
Changshu Hospital Affiliated to Nanjing University of Chinese Medicine,
Changshu,
Jiangsu Province 215500,
China
E-mail: ji512@163.com
| This article was originally published in a special issue, “Trending Topics in Biomedical Research and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2022:84(1) Spl Issue “69-80” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Ultra-processed food and psychological stress have led to an increase in the new incidence of irritable bowel syndrome. Xiaoyao powder is a classic traditional Chinese medicine formula for improving the secretion and transportation of the gastrointestinal tract as well as eliminating intestinal allergy in irritable bowel syndrome clinical treatment. To investigate the targets and mechanisms of action of Xiaoyao powder in the treatment of irritable bowel syndrome and in this study, we investigated the compound of Xiaoyao powder and the effects of it on irritable bowel syndrome by network pharmacology. Meanwhile, molecular docking method was used to verify the affinity of the top 20 active components to the targets (including master regulator of cell cycle entry and proliferative metabolism, adrenoceptor beta 2, Erb-B2 receptor tyrosine kinase 2). 146 active components and 628 drug targets of Xiaoyao powder were obtained from traditional Chinese medicine systems pharmacology database and analysis platform database. A total of 2498 irritable bowel syndrome differential genes were obtained from the Gene Expression Omnibus database. 737 items were obtained by gene ontology functional enrichment analysis. A total of 83 pathways were obtained by Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis. The results of molecular docking showed that the first 20 active components of Xiaoyao powder had strong affinity with their targets (master regulator of cell cycle entry and proliferative metabolism, adrenoceptor beta 2, Erb-B2 receptor tyrosine kinase 2). Our study showed that quercetin, polyporenic acid C and kaempferol in Xiaoyao powder were key potential active components treating irritable bowel syndrome. These compounds were strongly associated with core target proteins (such as master regulator of cell cycle entry and proliferative metabolism, adrenoceptor beta 2, Erb-B2 receptor tyrosine kinase 2 etc.). This study reveals that neuroactive ligand-receptor interaction, calcium signaling pathway and steroid hormone biosynthesis pathways mediate the therapeutic effects of Xiaoyao powder on irritable bowel syndrome.
Keywords
Network pharmacology, molecular docking, active components, Xiaoyao powder, irritable bowel syndrome
People easily access to diversity food or ultra-processed food and they also withstand a sharp increase in psychological pressure[1,2]. These factors bring newonset of incidence of Irritable Bowel Syndrome (IBS) and maintain approximately 1.5 %-2.5 % per year in recent decade[3]. IBS, a common clinically functional disorder of the gastrointestinal tract, is characterized by recurrent abdominal pain and changes in stool frequency[4]. IBS is mainly caused by alteration of visceral sensitivity, intestinal peristalsis disorder, infectious diseases of the intestine as well as brain-gut axis disorder and mood disorder[2,3]. People with IBS always have frailty and psychological burden, which are caused by restroom repetition and abdominal pain[5,6]. And the costs associated with a diagnostic workup are also an expensive burden to healthcare[7]. Chronic IBS impacts on patient’s quality of life, but few therapies are currently available for effectively treating IBS. The recommended treatment for IBS is medication. Loperamide, cimetropium is considered as the first-option and tricyclic antidepressants, selective serotonin receptor inhibitors, lubiprostone, rifaximin and pregabalin are used for secondary options[8]. Although those medicines show clear curative effect on IBS symptoms, limited and temporal effects as well as multiple side effects remain to be a problem for satisfying chronic IBS patients[9]. Therefore, more effective and safer therapies are still needed to treat IBS.
The curative effect of Traditional Chinese Medicine (TCM) has been more and more recognized internationally. Chinese herbal formula has certain curative effect in treating IBS clinically[10,11], among which Xiaoyao Powder (XYP) is the classic formula for the treatment of IBS. XYP was first recorded in Taiping Huimin Heji Jufang (an official revised record of Chinese patent medicine in ancient China) in Song Dynasty of China. It has been used in China for more than 700 y and most widely in digestive system disorders. XYP is composed of eight traditional Chinese herbs: Chaihu (Radix Bupleuri), Fuling (Poria), Baizhu (Rhizoma Atractylodis macrocephalae), Baishao (Radix Paeoniae Alba), Danggui (Radix Angelica sinensis), Bohe (Herba Menthae), Gancao (Radix Glycyrrhizae) and Shengjiang (Rhizoma Zingiberis Recens). Related clinical studies have shown that XYP or modified XYP has a therapeutic effect on Functional Gastrointestinal Disorders (FGIDs) and IBS symptoms obtained including abdominal pain, distension, constipation and defecation habit which can be relieved[12-15]. A metaanalysis of randomized controlled trials demonstrated that XYP is an effective and safe option for functional dyspepsia[16]. In addition, a meta-analysis[17] showed that the modified XYP was superior to conventional western medicine in terms of the total effective rate. An experimental study[18] on visceral hypersensitive IBS rats showed that XYP could improve the secretion and transportation of gastrointestinal tract, reduce the visceral sensitivity of IBS rats and reduce the content of serum cortisol. Study has shown that XYP could reduce visceral hypersensitivity via increasing 5-Hydroxytryptamine (5-HT) levels in the hypothalamus and colonic mucosa as well as regulating gastrointestinal motility and abnormal secretory function[19]. However, the mechanism of XYP in improving IBS is still not fully understood, which is also the main factor restricting its extensive clinical application. Network pharmacology[20] was first proposed by Hopkins. It could be used to investigate the complicated Chinese herbal formula through the cross-application of multi-disciplines and the overall biological network analysis and reintegration, to reveal the complex biological network relationships among drugs, components, targets and diseases.
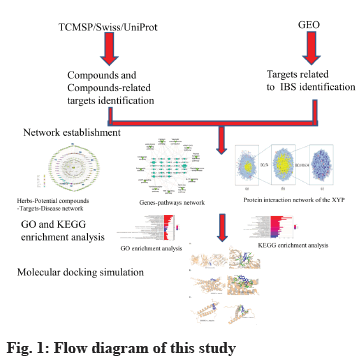
In this study, network pharmacology and molecular docking methods were used to preliminarily explore the potential mechanism of XYP in the treatment of IBS, to provide a reference for further pharmacological research and clinical application. The network flow chart is shown in fig. 1.
Materials and Methods
Screening of active components and prediction of drug targets in XYP:
In this study, the ingredients of XYP including Chaihu (Radix Bupleuri), Fuling (Poria), Baizhu (Rhizoma Atractylodis Macrocephalae), Baishao (Radix Paeoniae Alba), Danggui (Radix Angelica sinensis), Bohe (Herba Menthae), Gancao (Radix Glycyrrhizae) and Shengjiang (Rhizoma Zingiberis Recens) were captured via Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP)[21]. Oral Bioavailability (OB) and Drug-Likeness (DL) are two important indicators for bioinformatics evaluation of Chinese herbal medicine[22]. In this study, components with OB ≥30 % and DL ≥0.18 were selected as active components. OB is the main pharmacokinetic parameter of oral drugs and it is the proportion of oral drug dose in systemic circulation[23]. DL is used to qualitatively evaluate whether components have similar physical and chemical properties to existing drugs. Drug targets corresponding to active components were downloaded through TCMSP database[24]. For active components with no drug targets found on the TCMSP database, download their canonical smiles in the PubChem database[25], then, their drug targets were predicted by canonical smiles in the Swiss Target Prediction Online database[26] and all the obtained drug targets were input into the Uniprot database[27]. The protein species was set as "Homo sapiens" and the human gene abbreviations of each target were obtained.
Acquisition of IBS related targets:
The Differentially Expressed Genes (DEGs) of IBS were retrieved from the Gene Expression Omnibus (GEO) database (series: GSE36701; samples: There were 66 samples, 21 healthy volunteers and 45 patients with IBS) of National Center for Biotechnology Information (NCBI). The genes with a p value<0.05 and |log2 (fold change)|>0.5 were regarded as filter criteria for DEGs of IBS-related. To further screen the key DEGs for XYP-mediated alleviation of IBS, we intersected the targets regulated by active compounds in XYP with the disease target genes of IBS. After that, common targets were obtained via online tool Venny 2.1.
Network construction:
The intersection target genes of the active components in XYP and IBS and the Potential Active Components (PACs) acting on the genes were collected to construct TCM-potential components-targets-disease network. In addition, we identified the key nodes according to the degree of each node in the network and evaluated the effectiveness of XYP in treating IBS. Protein- Protein Interaction (PPI) networks were constructed by Cytoscape 3.8.0 software and its plug-in BisoGenet and then topology analysis was carried out by CytoNCA plug-in. Degree Centrality (DC) and Betweenness Centrality (BC) for each node[28] were also calculated.
Gene Ontology (GO) and pathway enrichment analysis:
R 4.2.0 and its installation package, bioconductor and cluster profiler were used to carry out GO function enrichment analysis on the potential target proteins associated with IBS. Significant items were selected with p<0.05 as the screening criteria. Biological Process (BP), Cell Component (CC) and Molecular Function (MF) of the top 10 functional categories were selected according to the ranking of p values to draw a histogram. Then the potential target protein was predicted by Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis and the significant signaling pathways were screened with p<0.05. According to the ranking of p values, the top 30 KEGG pathways were selected to draw a histogram. Potential target proteins were involved in the regulation of multiple pathways and signaling pathways closely related to IBS were selected. Genes-pathways network was mapped using Cytoscape 3.8.0 software to analyze their potential roles in the XYP network.
Molecular docking between the main active components of XYP and core proteins: By searching PubMed and Cochrane Library, the important target proteins Master Regulator of Cell Cycle Entry and Proliferative Metabolism (MYC), Adrenoceptor Beta 2 (ADRB2) and Erb-B2 Receptor Tyrosine Kinase 2 (ERBB2), related to IBS as reported in literatures were obtained. The crystal structures of MYC (Protein Data Bank (PDB) ID: 2OR9), ADRB2 (PDB ID: 6KR8) and ERBB2 (PDB ID: 5OB4) from the PDB (www.rcsb.org) were also downloaded. Subsequently, the structures of the active components in the XYP were downloaded from PubChem database (ranking among the top 20 in degree values) and the 20 compound structures were transformed into Three-Dimensional (3D) structures by Chem3D Ultra 14.0 software. The energy minimization process was performed using the Mining-Minima (MM2) algorithm and the minimum Root Mean Square (RMS) convergence parameter was set to 0.01. Then, the three target protein receptors dehydration and organic matter removal were performed by PyMOL software. The target protein receptor molecules were hydrogenated and charged by AutoDockTools 1.5.6, and the ligand and target protein receptor were converted into Protein Data Bank, Partial Charge (Q) and Atom Type (T) (PDBQT) parameter files, and the appropriate box center and box grid point parameters were set. Finally, molecular docking evaluation was carried out by Vina 1.5.6.
Results and Discussion
Active components and drug targets of XYP is explained here. By searching TCMSP database, 161 active components in XYP were obtained with threshold of OB≥30 % and DL≥0.18. Among them, 17 active components were from Chaihu (Radix Bupleuri), 15 from Fuling (Poria), 7 from Baizhu (Rhizoma Atractylodis Macrocephalae), 13 from Baishao (Radix Paeoniae Alba), 2 from Danggui (Radix Angelica sinensis), 10 from Bohe (Herba Menthae), 92 from Gancao (Radix Glycyrrhizae) and 5 from Shengjiang (Rhizoma Zingiberis Recens). After eliminating the duplicated components, there were 146 active components in XYP. 628 abbreviations of drug target genes were obtained in TCMSP database, the Swiss Target Prediction online database and Uniprot database. The top 30 active components of XYP and their corresponding drug targets are shown in Table 1.
| Related herbs | Molecule ID | Name | OB | DL | Targets |
|---|---|---|---|---|---|
| CH, GC | MOL000098 | Quercetin | 46.43 | 0.28 | 138 |
| SJ | MOL008698 | Dihydrocapsaicin | 47.07 | 0.19 | 100 |
| GC | MOL004905 | Glyuranolide | 34.32 | 0.55 | 100 |
| BS | MOL001910 | 11α,12α-Epoxy-3β,23-dihydroxy-30-norolean-20(29)-en-28,13β-olide | 64.77 | 0.38 | 100 |
| FL | MOL000285 | Polyporenic acid C | 38.26 | 0.82 | 100 |
| GC | MOL005013 | 18α-hydroxyglycyrrhetic acid | 41.16 | 0.71 | 98 |
| FL | MOL000287 | Eburicoic acid | 38.7 | 0.81 | 92 |
| FL | MOL000292 | Poricoic acid C | 38.15 | 0.75 | 78 |
| BS | MOL001928 | Albiflorin_qt | 66.64 | 0.33 | 68 |
| FL | MOL000300 | Dehydroeburicoic acid | 44.17 | 0.83 | 65 |
| BS | MOL001925 | Paeoniflorin_qt | 68.18 | 0.4 | 60 |
| BZ | MOL000028 | α-Amyrin | 39.51 | 0.76 | 56 |
| BS, CH, GC | MOL000422 | Kaempferol | 41.88 | 0.24 | 55 |
| BH | MOL000006 | Luteolin | 36.16 | 0.25 | 54 |
| FL | MOL000289 | Pachymic acid | 33.63 | 0.81 | 53 |
| FL | MOL000290 | Poricoic acid A | 30.61 | 0.76 | 39 |
| CH | MOL004628 | Octalupine | 47.82 | 0.28 | 39 |
| GC | MOL003896 | 7-Methoxy-2-methylisoflavone | 42.56 | 0.20 | 37 |
| BH, GC | MOL004328 | Naringenin | 59.29 | 0.21 | 35 |
| FL | MOL000291 | Poricoic acid B | 30.52 | 0.75 | 34 |
| FL | MOL000276 | 7,9(11)-dehydropachymic acid | 35.11 | 0.81 | 34 |
| BS | MOL001921 | Lactiflorin | 49.12 | 0.8 | 32 |
| GC | MOL000392 | Formononetin | 69.67 | 0.21 | 32 |
| GC | MOL000497 | Licochalcone A | 40.79 | 0.29 | 30 |
| CH, GC | MOL000354 | Isorhamnetin | 49.6 | 0.31 | 29 |
| GC | MOL002565 | Medicarpin | 49.22 | 0.34 | 28 |
| BH | MOL001790 | Linarin | 39.84 | 0.71 | 28 |
| BH | MOL011616 | Fortunellin | 35.65 | 0.74 | 28 |
| BS, DG, SJ | MOL000358 | β-sitosterol | 36.91 | 0.75 | 28 |
| CH, DG, SJ | MOL000449 | Stigmasterol | 43.83 | 0.76 | 27 |
Note: OB-Oral Bioavailability; DL-Drug-Likeness. CH stands for Chaihu (Radix Bupleuri); FL for Fuling (Poria); BZ for Baizhu (Rhizoma Atractylodis Macrocephalae); BS for Baishao (Radix Paeoniae Alba); DG for Danggui (Radix Angelica sinensis); BH for Bohe (Herba Menthae); GC for Gancao (Radix Glycyrrhizae); SJ for Shengjiang (Rhizoma Zingiberis Recens)
Table 1: Top 30 Active Components and Their Number of Targets of XYP
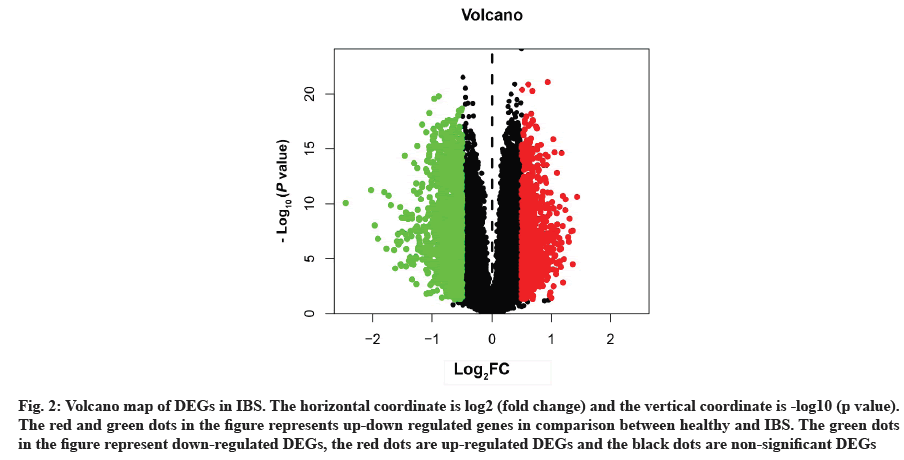
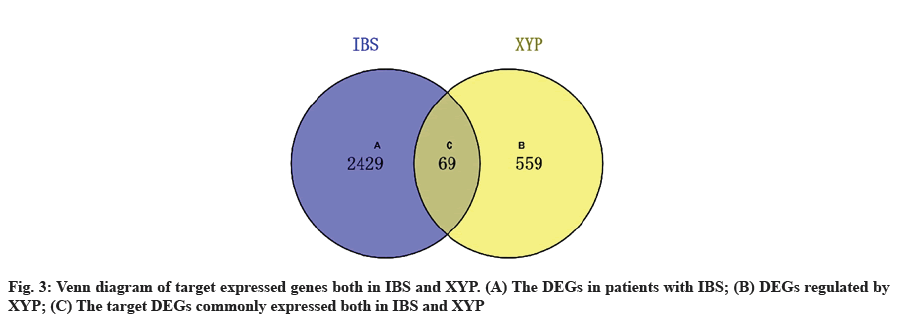
Acquisition of IBS related target genes is shown here. A total of 2498 DEGs in IBS were collected from the GEO database, including 963 up-regulated genes and 1535 down-regulated genes. A volcano map was plotted to show the distribution of DEGs (fig. 2). The red and green dots represented DEGs between IBS and the healthy. The key target genes of XYP treating IBS were captured by intersecting 628 drug targets with 2498 disease targets using online tool VENNY 2.1 (fig. 3). Notably, 107 crucial active components of XYP regulated 69 key target genes in treating IBS.
Fig. 2: Volcano map of DEGs in IBS. The horizontal coordinate is log2 (fold change) and the vertical coordinate is -log10 (p value). The red and green dots in the figure represents up-down regulated genes in comparison between healthy and IBS. The green dots in the figure represent down-regulated DEGs, the red dots are up-regulated DEGs and the black dots are non-significant DEGs
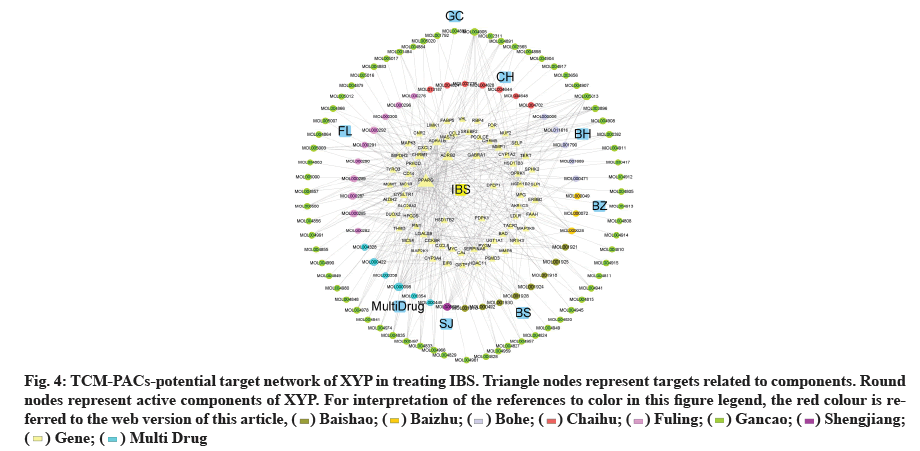
TCM-PACs-potential target network of XYP treating IBS was constructed with 107 PACs and 69 target genes (fig. 4). The network included 176 nodes (107 components, 69 intersection target genes) and 342 edges, elucidating the interaction of PACs-potential target genes. The top three active compounds included: MOL000098 (quercetin), MOL000285 (polyporenic acid C) and MOL008698 (dihydrocapsaicin) and they may influence gene expression related IBS. The gene counts that correspond to each of them are 18, 12 and 11.
Fig. 4: TCM-PACs-potential target network of XYP in treating IBS. Triangle nodes represent targets related to components. Round
nodes represent active components of XYP. For interpretation of the references to color in this figure legend, the red colour is referred
to the web version of this article, ( ) Baishao; (
) Baishao; ( ) Baizhu; (
) Baizhu; ( ) Bohe; (
) Bohe; ( ) Chaihu; (
) Chaihu; ( ) Fuling; (
) Fuling; ( ) Gancao; (
) Gancao; ( ) Shengjiang;
(
) Shengjiang;
( ) Gene; (
) Gene; ( ) Multi Drug
) Multi Drug
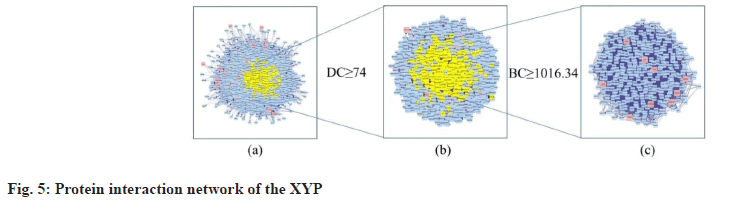
Construction of PPI network, topological analysis and determination of core targets is explained here. Based on one protein corresponding to one gene, proteins are main executors in various BPs. To clarify the mechanism of XYP in treating IBS, the network of intersection target genes was merged with the target network related to IBS and the nodes belonging to the intersection targets in the network were marked red to form a PPI network with 3289 nodes and 112 198 edges (fig. 5a). Studies have shown that the DC value of important nodes exceeds 74[29]. Therefore, the first screening was carried out with DC≥74 and a target network with 1050 nodes and 41 449 edges was obtained (fig. 5b). The second screening was performed using BC≥1016.34 (BC mean) and 330 nodes and 9804 edges were obtained (fig. 5c). Finally, 9 core targets for XYP treating IBS were screened out. The specific information of DC and BC values of the 9 intersection target proteins is shown in fig. 5c and Table 2.
| Proteins | Betweenness | Degree |
|---|---|---|
| MYC | 6380.12 | 222 |
| VHL | 4347.11 | 202 |
| EIF6 | 2562.09 | 117 |
| ADRB2 | 2139.02 | 112 |
| PSMD3 | 2134.03 | 106 |
| PIN1 | 2107.80 | 112 |
| MAPK3 | 1832.03 | 107 |
| PRKCD | 1784.76 | 95 |
| ERBB2 | 1617.87 | 86 |
Table 2: Information Of 9 Key Target Proteins For XYP In Treating IBS
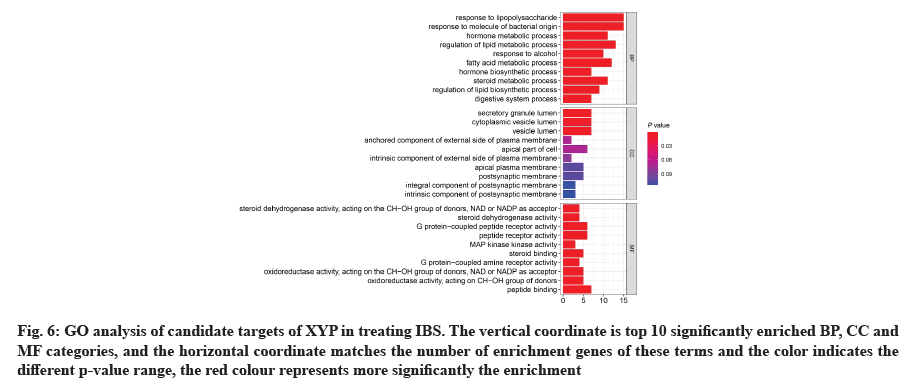
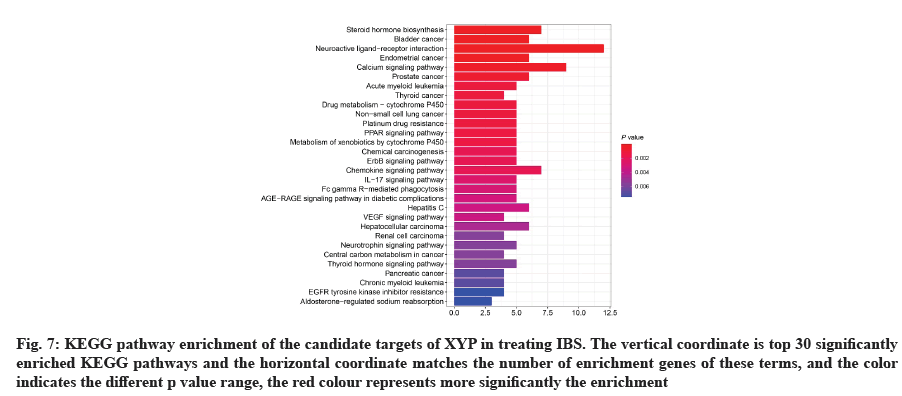
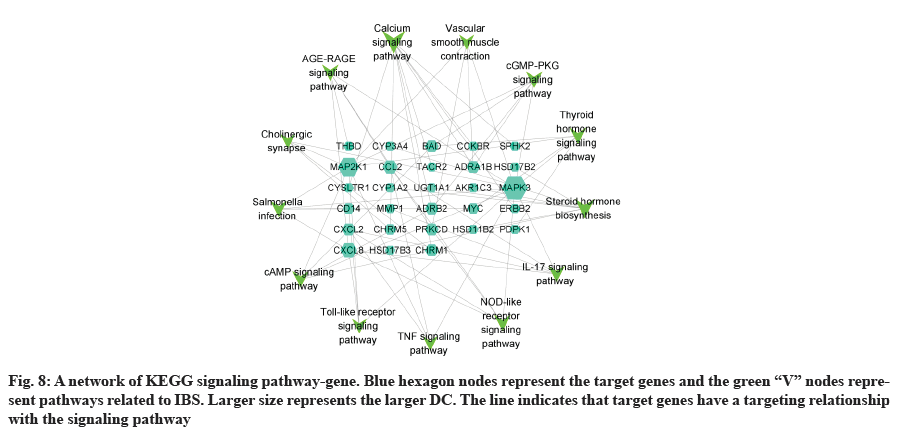
GO analysis was performed on 69 potential IBS related target proteins based on BP, CC and MF to explain potential mechanism in XYP treating IBS. The key targets of treating IBS signaling pathway were studied by KEGG enrichment analysis. GO enrichment analysis showed that there were 737 items with p<0.05. In the BP enrichment analysis, there were 684 items, including response to lipopolysaccharide, response to molecule of bacterial origin, hormone metabolic process and other items. In the CC enrichment analysis, there were 3 items, including secretory granule lumen, cytoplasmic vesicle lumen and vesicle lumen. In the MF enrichment analysis, there were 50 items, including steroid dehydrogenase activity, acting on the Hydroxyl (CH-OH) group of donors, Nicotinamide Adenine Dinucleotide (NAD) or Nicotinamide Adenine Dinucleotide Phosphate (NADP) as acceptor, steroid dehydrogenase activity, G protein-coupled peptide receptor activity and other items. According to the ranking of p value, the top 10 function categories were selected to draw a bar chart, as shown in fig. 6. To further show the BP of potential target proteins, KEGG enrichment analysis was performed and the results showed 83 signaling pathways with p<0.05. According to the ranking of p value, the top 30 signal pathways were selected to draw a histogram, as shown in fig. 7. In these signaling pathways, according to the number of target genes corresponding to the signaling pathways, the top 3 were Neuroactive ligand-receptor interaction, Calcium signaling pathway and steroid hormone biosynthesis, corresponding to 12, 9 and 7 potential target proteins, respectively. In addition, the signal pathways and target proteins associated with IBS were shown in fig. 8.
Fig. 6: GO analysis of candidate targets of XYP in treating IBS. The vertical coordinate is top 10 significantly enriched BP, CC and MF categories, and the horizontal coordinate matches the number of enrichment genes of these terms and the color indicates the different p-value range, the red colour represents more significantly the enrichment
Fig. 7: KEGG pathway enrichment of the candidate targets of XYP in treating IBS. The vertical coordinate is top 30 significantly enriched KEGG pathways and the horizontal coordinate matches the number of enrichment genes of these terms, and the color indicates the different p value range, the red colour represents more significantly the enrichment
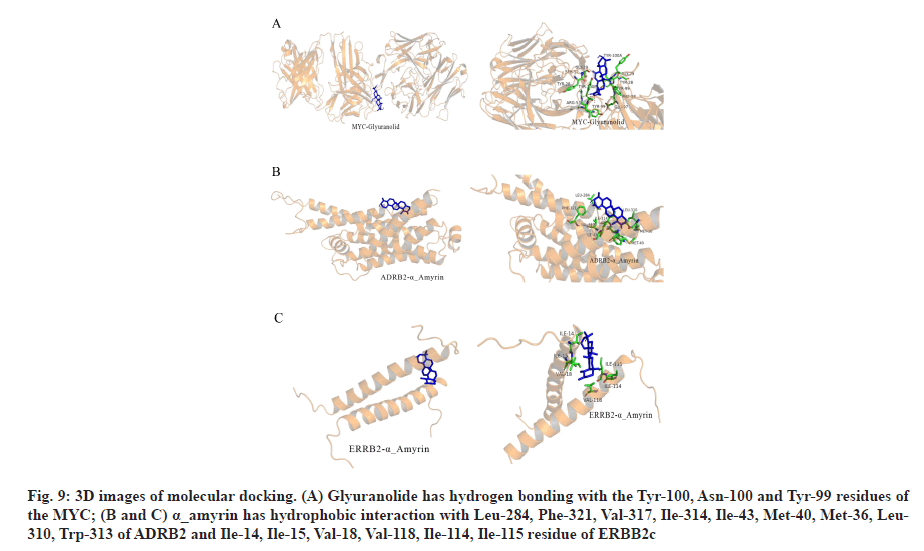
In the process of molecular docking, due to the different conformation of various components and target proteins, the affinity will be different. Affinity<-4.25 kcal/mol means that ligands and receptors have possibility of combination, affinity<-5.00 kcal/mol indicates good binding strength and affinity<-7.00 kcal/mol suggests satisfactory binding strength[30]. A total of 60 pairs of receptor-ligand combinations were obtained from 20 potential core components and 3 core targets of which 40 pairs (66.66 %) were <-7.00. The three combinations with the highest affinity were ADRB2 alpha (α)_amyrin (-9.6 kcal/mol), ADRB2-quercetin (-9.5 kcal/mol) and MYC-glyuranolide (-9.4 kcal/mol) (Table 3). The conformational 3D images of the components with the best binding activities corresponding to each target protein were drawn by PyMOL software for visual display, as shown in fig. 9. The results of molecular docking were consistent with the relationship between the target and the active component predicted in this study. Fig. 9A shows that glyuranolide has hydrogen bonding with the Tyr-100, Asn-100 and Tyr-99 residues of the MYC. Fig. 9B and fig. 9C also indicate that α_ amyrin has hydrophobic interaction with Leu-284, Phe- 321, Val-317, Ile-314, Ile-43, Met-40, Met-36, Leu-310, Trp-313 of ADRB2 and Ile-14, Ile-15, Val-18, Val-118, Ile-114, Ile-115 residue of ERBB2. It could be seen that hydrogen bonding and hydrophobic interaction were the main binding forces between components and target proteins. In short, molecular docking verified the interaction between the important components and the core target genes of XYP. In addition, the high affinity between other components and target genes suggested that XYP might play a therapeutic role by regulating those related components.
Fig. 9: 3D images of molecular docking. (A) Glyuranolide has hydrogen bonding with the Tyr-100, Asn-100 and Tyr-99 residues of the MYC; (B and C) α_amyrin has hydrophobic interaction with Leu-284, Phe-321, Val-317, Ile-314, Ile-43, Met-40, Met-36, Leu- 310, Trp-313 of ADRB2 and Ile-14, Ile-15, Val-18, Val-118, Ile-114, Ile-115 residue of ERBB2c
| Name | MYC (kcal/mol) | ADRB2 (kcal/mol) | ERBB2 (kcal/mol) |
|---|---|---|---|
| α-Amyrin | -8.5 | -9.6 | -9.0 |
| 7-Methoxy-2-methyl isoflavone | -6.8 | -7.3 | -7.3 |
| Licoagrocarpin | -7.4 | -8.0 | -7.2 |
| Glyuranolide | -9.4 | -8.8 | -7.1 |
| 11α,12α-Epoxy-3β,23-dihydroxy-30-norolean-20(29)-en-28,13β-olide | -9.3 | -8.6 | -7.0 |
| Polyporenic acid C | -8.3 | -8.7 | -7.0 |
| 18α-Hydroxyglycyrrhetic acid | -9.0 | -8.1 | -6.9 |
| Dehydroeburicoic acid | -7.0 | -8.1 | -6.7 |
| Eburicoic acid | -8.5 | -8.1 | -6.7 |
| Pachymic acid | -7.3 | -8.9 | -6.6 |
| b-sitosterol | -7.7 | -8.2 | -6.5 |
| Kaempferol | -7.9 | -8.1 | -6.3 |
| Licochalcone A | -5.6 | -7.8 | -6.3 |
| Naringenin | -7.8 | -8.6 | -6.2 |
| Quercetin | -7.7 | -9.5 | -6.2 |
| Paeoniflorin_qt | -7.6 | -8.0 | -6.0 |
| Albiflorin_qt | -7.1 | -8.3 | -5.9 |
| Luteolin | -8.0 | -8.2 | -5.9 |
| Poricoic acid C | -6.6 | -6.7 | -5.9 |
| Dihydrocapsaicin | -6.3 | -6.5 | -5.3 |
Table 3: Affinity Between Main PACs and Core Target Proteins of XYP
In this study, 107 active components and 69 potential target proteins were used to construct a TCM-PACspotential target gene network of XYP in treating IBS, in which most of XYP components regulate IBS via multiple targets. For example, quercetin, polyporenic acid C and kaempferol ranked higher in degree value in the network, which regulate 18, 12 and 11 target proteins, respectively. Quercetin is the common active component in Chaihu (Radix Bupleuri) and Gancao (Radix Glycyrrhizae). Polyporenic acid C is one of active components in Fuling (Poria) and kaempferol is a common active component in Baishao (Radix Paeoniae Alba), Chaihu (Radix Bupleuri) and Gancao (Radix Glycyrrhizae). Those could be indirectly elucidated that there exist common components in traditional Chinese herbs and compatibility use could function in synergistic effect.
Studies found that quercetin could alleviate visceral hypersensitivity in Post-Infectious IBS (PIIBS) rat model by reducing the effectiveness of intestinal enterochromaffin cell and 5-HT, providing preliminary data for the analgesic effect of quercetin in IBS[31]. Quercetin could improve intestinal epithelial cell permeability and alleviate IBS internal hypersensitivity[32]. In addition, one study has shown that quercetin could inhibit small intestinal peristalsis and delay intestinal transport in guinea pigs[33]. Polyporenic acid C is a triterpenoid in Poria, which could reduce NO level by inhibiting the expression of inducible Nitric Oxide Synthase (iNOS) enzymes to achieve the antiinflammatory effect[34]. Kaempferol can significantly inhibit the expression of Mitogen-Activated Protein Kinase (MAPK) pathway of Lipopolysaccharide (LPS)-induced human monocyte THP-1 (Human Monocytic Cell Line) and decrease the production of Macrophage Derived Chemokine (MDC), Interferon Induced Protein-10 (IP-10), Interleukin-8 (IL-8) and other inflammatory factors to effectively inhibit the occurrence of inflammation[35]. Studies have shown that IBS is closely related to psychological factors, especially depression and anxiety[36]. We also noticed that among the 107 key active components, Radix Glycyrrhizae had a larger proportion. Glycyrrhizic acid, one of the main components of Radix Glycyrrhizae extract, could regulate visceral hypersensitivity of IBS by influencing the release of local allergic mediators and spinal dorsal horn to function in the gastrointestinal nervous system to improve the symptoms of IBS[37].
By constructing the PPI network of IBS related targets, the important targets of IBS were obtained, such as MYC, Von Hippel-Lindau (VHL) syndrome, Eukaryotic Translation Initiation Factor 6 (EIF6), ADRB2, Proteasome 26S Subunit, Non-ATPase 3 (PSMD3), Peptidylprolyl Cis/Trans Isomerase, NIMA Interacting 1 (PIN1), MAPK3, Protein Kinase C Delta (PRKCD) and ERBB2. By searching in PubMed and Cochrane Library, only 3 of the above 9 targets were found to be related to IBS, namely MYC, ADRB2 and ERBB2. MYC plays an important role in the production of metabolic enzymes[38]. Activation of MYC contributes to the conversion of glucose to pyruvate[39]. MYC also blocks the entry of pyruvate into the Tricarboxylic Acid (TCA) cycle by regulating Pyruvate Dehydrogenase Kinase (PDK), prevents Lactate Dehydrogenase A (LDHA) and lactate transporter, Monocarboxylate Transporter 1 (MCT1) from transferring pyruvate and lactic acid out of the cell[40]. Glucose and amino acids (especially glutamine) can be directly absorbed in the colonic mucosa and then utilized for respiration, growth and repair[41]. At present, many studies have found the overgrown intestinal flora, disordered bacterial flora and disturbed energy supply of the colon in patients with IBS[42]. ADRB2 is the main target of catecholamine epinephrine and a major mediator of stress response. Blocking ADRB2 reduces pain sensitivity in humans and animals[43,44]. ADRB2 is widely expressed in gastrointestinal tract and central nervous system[45]. Some studies showed that, ADRB2 polymorphisms, in particular rs1042714, might play a role in the pathogenesis of FGIDs[46]. Clinically, this polymorphism was associated with higher Functional Gastrointestinal Disorder (FGID) and Extraintestinal Functional Disorder (EIFD) burden, especially in IBS patients. ERBB2 is a transmembrane glycoprotein with tyrosine kinase activity and belongs to the type I growth factor receptor family[47]. Studies have shown that probiotics isolated from traditional dairy products played an anti-cancer role in colon cancer cells by down-regulating the expression of ERBB-2 and ERBB- 3 genes[48].
83 signaling pathways were obtained by KEGG analysis. In this study, IBS related pathways were mainly concentrated in inflammatory response, endocrine, intestinal function, nerve sensitization and bacterial infection, and other related signaling pathways. Among them, IL-17 signaling pathway, Nucleotide-Binding Oligomerization Domain (NOD)- like receptor signaling pathway, cyclic Adenosine 3',5'-Monophosphate (cAMP) signaling pathway, tolllike receptor signaling pathway and Tumor Necrosis Factor (TNF) signaling pathway were associated with inflammation. More and more literatures have reported the close relationship between intestinal inflammatory response and IBS in recent years. Colonic mucosa in patients with IBS is often associated with mild inflammation and inflammatory response may increase colon permeability[49]. TNF-α has increased specificity in patients with IBS and is associated with symptoms of pain intensity and pain frequency, as well as sensitizing the colon afferent nerve to mechanical stimulation[50]. The thyroid hormone signaling pathway and steroid hormone biosynthesis are related to endocrine. Steroid hormone biosynthesis pathways significantly interfere with IBS based pathway analysis, and clinical and experimental studies[51] have shown that the decreased sex hormone levels might lead to the occurrence or aggravation of intestinal symptoms. The cyclic Guanosine 3',5'-Monophosphate (cGMP)-Protein Kinase G (PKG) signaling pathway and vascular smooth muscle contraction are related to intestinal function. cGMP, as a second messenger, can act on many biological effects such as PKG, participating in biological signaling, involving cell proliferation and apoptosis, inhibiting intestinal peristalsis and so on. PKG can play a role in relaxing smooth muscle by reducing intracellular calcium level and calcium sensitivity[52]. Calcium signaling pathway and cholinergic synapse are associated with neurosensitization. The activation of the cholinergic receptor causes smooth muscle contraction, leading to abnormal gastrointestinal dynamics, aggravating symptoms in patients with IBS[53]. Study showed that smalldose anticholinergic drug therapy was considered as one of the effective treatments for IBS by regulating gastrointestinal dynamics, reducing visceral high sensitivity and improving anxiety status[54]. Salmonella infection is associated with bacterial infections of the gastrointestinal tract. In recent years, more and more epidemiological and clinical data indicated the close relationship between early gastrointestinal infection and IBS. Two meta-analyses[55,56] suggested that patients with early gastrointestinal infection had a 6-7 fold increase in the risk of IBS. In conclusion, XYP may reduce intestinal permeability by alleviating inflammatory response, relaxing smooth muscle to regulate gastrointestinal motility, reducing visceral hypersensitivity and improving anxiety to treat IBS.
The present study systematically expounds on the core compounds and molecular mechanism of XYP in the treatment of IBS. The key active components of XYP in treating IBS are quercetin, polyporenic acid C and kaempferol via network pharmacology and molecular docking. The key target genes in XYP treating IBS pathway network were MYC, VHL, EIF6, ADRB2, PSMD3, PIN1, MAPK3, PRKCD and ERBB2. In addition, through molecular docking analysis, important components such as quercetin, polyporenic acid C, kaempferol, glyuranolide, α_amyrin had good affinity to the core proteins MYC, ADRB2 and ERBB2. Also, XYP can regulate signal pathways related to IBS disease, for example, neuroactive ligand-receptor interaction, calcium signaling pathway and steroid hormone biosynthesis. These findings suggest the potential synergistic role of XYP in treating IBS via multiple components, multi-targets, multiple biological functions and multiple signal pathways. Nevertheless, further studies are needed to verify these results.
Acknowledgements:
This study was supported by the Natural Science Foundation of Nanjing University of Chinese Medicine (No. XZR2020062), Changshu Municipal Science and Technology Bureau Supporting Project (No. CS202030), Changshu Hospital Affiliated to Nanjing University of Chinese Medicine Youth Fund Supporting Project (No. Cszyy201912).
Conflict of interests:
The authors declare that there is no conflict of interest.
References
- Elizabeth L, Machado P, Zinöcker M, Baker P, Lawrence M. Ultra-processed foods and health outcomes: A narrative review. Nutrients 2020;12(7):1955.
[Crossref] [Google Scholar] [PubMed]
- Popa SL, Leucuta DC, Dumitrascu DL. Pressure management as an occupational stress risk factor in irritable bowel syndrome: A cross-sectional study. Medicine 2018;97(49):e13562.
[Crossref] [Google Scholar] [PubMed]
- Ford AC, Sperber AD, Corsetti M, Camilleri M. Functional gastrointestinal disorders 2 irritable bowel syndrome. Lancet 2020;396(10263):1675-88.
[Crossref] [Google Scholar] [PubMed]
- Dai L, Zhong LL, Ji G. Irritable bowel syndrome and functional constipation management with integrative medicine: A systematic review. World J Clin Cases 2019;7(21):3486.
[Crossref] [Google Scholar] [PubMed]
- El-Salhy M, Casen C, Valeur J, Hausken T, Hatlebakk JG. Responses to faecal microbiota transplantation in female and male patients with irritable bowel syndrome. World J Gastroenterol 2021;27(18):2219-37.
[Crossref] [Google Scholar] [PubMed]
- Hanel V, Schalla MA, Stengel A. Irritable bowel syndrome and functional dyspepsia in patients with eating disorders‐a systematic review. Eur Eat Disord Rev 2021;29(5):692-719.
[Crossref] [Google Scholar] [PubMed]
- Sabo CM, Grad S, Dumitrascu DL. Chronic abdominal pain in general practice. Dig Dis 2021;39(6):606-14.
[Crossref] [Google Scholar] [PubMed]
- Trinkley KE, Nahata MC. Treatment of irritable bowel syndrome. J Clin Pharm Ther 2011;36(3):275-82.
[Crossref] [Google Scholar] [PubMed]
- Ford AC, Moayyedi P, Chey WD, Harris LA, Lacy BE, Saito YA, et al. American college of gastroenterology monograph on management of irritable bowel syndrome. Am J Gastroenterol 2018;113(2):1-18.
[Crossref] [Google Scholar] [PubMed]
- Yao CJ, Li YL, Pu MJ, Luo LH, Feng PM. Traditional Chinese medicine for irritable bowel syndrome: A protocol for meta-analysis. Medicine 2020;99(48):e23394.
[Crossref] [Google Scholar] [PubMed]
- Rahimi R, Abdollahi M. Herbal medicines for the management of irritable bowel syndrome: A comprehensive review. World J Gastroenterol 2012;18(7):589-600.
[Crossref] [Google Scholar] [PubMed]
- Zhang SS. Thinking and strategy on the diagnosis and treatment of functional gastrointestinal disorders with integrative medicine. Chin J Integr Med 2009;15(2):83-5.
- Song XP, Chen XT, Jing XP. Curative effect of Jiawei Xiaoyao San combined with bifid-triple viable capsule and its effects on clinical symptoms, intestinal microecology function in treatment of patients with irritable bowel syndrome. Med Pharm J Chin People Liberation Army 2020;32(2):84-7.
- Zhang XR. Analysis of the clinical curative effect of Jiawei Xiaoyao San in the treatment of irritable bowel syndrome. China Naturop 2016;24(7):66-7.
- Yang SQ, Lin LX. Analysis of the key ideas of clinical experience in 52 cases of irritable bowel syndrome treated with modified prescriptions of Yaosan. Cardiovasc Dis J Integr Tradit Chin West Med 2019;7(27):167.
- Qin F, Huang X, Ren P. Chinese herbal medicine modified Xiaoyao San for functional dyspepsia: Meta-analysis of randomized controlled trials. J Gastroenterol Hepatol 2009;24(8):1320-5.
[Crossref] [Google Scholar] [PubMed]
- Liu Q, Mao XY, Zhang T, Du SJ, Guo S, Su XL, et al. Meta-analysis and trial sequential analysis of the treatment of irritable bowel syndrome with modified Xiaoyao powder. World J Integr Tradit West Med 2020;15(10):1772-17.
- Shi JJ, Dai YY, Xu FY. Effect of Xiaoyao powder on the corticosterone level in serum of irritable bowel syndrome rat. Shandong J Tradit Chin Med 2008;27(1):46-9.
- Ding F, Wu J, Liu C, Bian Q, Qiu W, Ma Q, et al. Effect of Xiaoyaosan on colon morphology and intestinal permeability in rats with chronic unpredictable mild stress. Front Pharmacol 2020;11:1069.
[Crossref] [Google Scholar] [PubMed]
- Hopkins AL. Network pharmacology: The next paradigm in drug discovery. Nat Chem Biol 2008;4(11):682-90.
[Crossref] [Google Scholar] [PubMed]
- Ru J, Li P, Wang J, Zhou W, Li B, Huang C, et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J Cheminform 2014;6(1):1-6.
[Crossref] [Google Scholar] [PubMed]
- Gong B, Kao Y, Zhang C, Sun F, Zhao H. Systematic investigation of Scutellaria barbata Herba for treating hepatocellular carcinoma based on network pharmacology. Evid Based Complement Alternat Med 2018;2018.
[Crossref] [Google Scholar] [PubMed]
- Xu X, Zhang W, Huang C, Li Y, Yu H, Wang Y, et al. A novel chemometric method for the prediction of human oral bioavailability. Int J Mol Sci 2012;13(6):6964-82.
[Crossref] [Google Scholar] [PubMed]
- Wang W, Liu T, Yang L, Ma Y, Dou F, Shi L, et al. Study on the multi-targets mechanism of triphala on cardio-cerebral vascular diseases based on network pharmacology. Biomed Pharmacother 2019;116:108994.
[Crossref] [Google Scholar] [PubMed]
- Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, et al. PubChem in 2021: New data content and improved web interfaces. Nucleic Acids Res 2021;49(D1):D1388-95.
[Crossref] [Google Scholar] [PubMed]
- Gfeller D, Michielin O, Zoete V. Shaping the interaction landscape of bioactive molecules. Bioinformatics 2013;29(23):3073-9.
[Crossref] [Google Scholar] [PubMed]
- Bateman A. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49:D480-9.
[Crossref] [Google Scholar] [PubMed]
- Tang Y, Li M, Wang J, Pan Y, Wu FX. CytoNCA: A cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems 2015;127:67-72.
- Zhang Y, Li Z, Yang M, Wang D, Yu L, Guo C, et al. Identification of GRB2 and GAB1 coexpression as an unfavorable prognostic factor for hepatocellular carcinoma by a combination of expression profile and network analysis. PLoS One 2013;8(12):e85170.
[Crossref] [Google Scholar] [PubMed]
- Hsin KY, Ghosh S, Kitano H. Combining machine learning systems and multiple docking simulation packages to improve docking prediction reliability for network pharmacology. PLoS One 2013;8(12):e83922.
[Crossref] [Google Scholar] [PubMed]
- Qin HY, Zang KH, Zuo X, Wu XA, Bian ZX. Quercetin attenuates visceral hypersensitivity and 5-hydroxytryptamine availability in postinflammatory irritable bowel syndrome rats: Role of enterochromaffin cells in the colon. J Med Food 2019;22(7):663-71.
- Liu LN, Sun ZG, Cai XT, Cao P, Lu Y, Shao M, et al. Quercetin improves TNF-α induced intestinal barrier dysfunction in Caco-2 cells. J China Pharm Univ 2012;43(6):541-5.
- Gharzouli K, Holzer P. Inhibition of guinea pig intestinal peristalsis by the flavonoids quercetin, naringenin, apigenin and genistein. Pharmacology 2004;70(1):5-14.
[Crossref] [Google Scholar] [PubMed]
- Cai TG, Cai Y. Triterpenes from the fungus Poria cocos and their inhibitory activity on nitric oxide production in mouse macrophages via blockade of activating protein‐1 pathway. Chem Biodivers 2011;8(11):2135-43.
[Crossref] [Google Scholar] [PubMed]
- Huang CH, Jan RL, Kuo CH, Chu YT, Wang WL, Lee MS, et al. Natural flavone kaempferol suppresses chemokines expression in human monocyte THP‐1 cells through MAPK pathways. J Food Sci 2010;75(8):H254-9.
- Fond G, Loundou A, Hamdani N, Boukouaci W, Dargel A, Oliveira J, et al. Anxiety and depression comorbidities in irritable bowel syndrome (IBS): A systematic review and meta-analysis. Eur Arch Psychiatry Clin Neurosci 2014;264(8):651-60.
[Crossref] [Google Scholar] [PubMed]
- Yu L, Cheng P, Liyang Z, Xiaofang X. Study on glycyrrhizic acid on visceral hypersensitivity irritable bowel syndrome. Pharmacol Clin Chin Mat Med 2012:1.
- Dang CV. MYC, metabolism, cell growth, and tumorigenesis. Cold Spring Harb Perspect Med 2013;3(8):a014217.
[Crossref] [Google Scholar] [PubMed]
- Dang CV, Kim JW, Gao P, Yustein J. The interplay between MYC and HIF in cancer. Nat Rev Cancer 2008;8(1):51-6.
[Crossref] [Google Scholar] [PubMed]
- Kim JW, Gao P, Liu YC, Semenza GL, Dang CV. Hypoxia-inducible factor 1 and dysregulated c-Myc cooperatively induce vascular endothelial growth factor and metabolic switches hexokinase 2 and pyruvate dehydrogenase kinase 1. Mol Cell Biol 2007;27(21):7381-93.
[Crossref] [Google Scholar] [PubMed]
- O'Keefe SJ. Nutrition and colonic health: The critical role of the microbiota. Curr Opin Gastroenterol 2008;24(1):51-8.
[Crossref] [Google Scholar] [PubMed]
- Posserud I, Stotzer PO, Björnsson ES, Abrahamsson H, Simren M. Small intestinal bacterial overgrowth in patients with irritable bowel syndrome. Gut 2007;56(6):802-8.
[Crossref] [Google Scholar] [PubMed]
- Tchivileva IE, Lim PF, Smith SB, Slade GD, Diatchenko L, McLean SA, et al. Effect of catechol-O-methyltransferase polymorphism on response to propranolol therapy in chronic musculoskeletal pain: A randomized, double–blind, placebo-controlled, crossover pilot study. Pharmacogenet Genomics 2010;20(4):239-48.
[Crossref] [Google Scholar] [PubMed]
- Nackley AG, Tan KS, Fecho K, Flood P, Diatchenko L, Maixner W. Catechol-O-methyltransferase inhibition increases pain sensitivity through activation of both β2-and β3-adrenergic receptors. Pain 2007;128(3):199-208.
[Crossref] [Google Scholar] [PubMed]
- Diatchenko L, Anderson AD, Slade GD, Fillingim RB, Shabalina SA, Higgins TJ, et al. Three major haplotypes of the β2 adrenergic receptor define psychological profile, blood pressure, and the risk for development of a common musculoskeletal pain disorder. Am J Med Genet B Neuropsychiatr Genet 2006;141(5):449-62.
[Crossref] [Google Scholar] [PubMed]
- Kushnir VM, Cassell B, Gyawali CP, Newberry RD, Kibe P, Nix BD, et al. Genetic variation in the beta‐2 adrenergic receptor (ADRB2) predicts functional gastrointestinal diagnoses and poorer health‐related quality of life. Aliment Pharmacol Ther 2013;38(3):313-23.
[Crossref] [Google Scholar] [PubMed]
- Zhang H. 177Lu-CHX-A''-DTPA-ABD-Affibody (ZHER2: 342) 2. In: Molecular Imaging and Contrast Agent Database (MICAD). National Center for Biotechnology Information (US); 2008.
- Faghfoori Z, Pourghassem GB, Saber A, Seyyedi M, Fazelian S, Khosroushahi AY. Prophylactic effects of secretion metabolites of dairy lactobacilli through downregulation of ErbB-2 and ErbB-3 genes on colon cancer cells. Eur J Cancer Prev 2020;29(3):201-9.
[Crossref] [Google Scholar] [PubMed]
- Jalanka-Tuovinen J, Salojärvi J, Salonen A, Immonen O, Garsed K, Kelly FM, et al. Faecal microbiota composition and host-microbe cross-talk following gastroenteritis and in postinfectious irritable bowel syndrome. Gut 2014;63(11):1737-45.
[Crossref] [Google Scholar] [PubMed]
- Hughes PA, Harrington AM, Castro J, Liebregts T, Adam B, Grasby DJ, et al. Sensory neuro-immune interactions differ between irritable bowel syndrome subtypes. Gut 2013;62(10):1456-65.
[Crossref] [Google Scholar] [PubMed]
- Yu Q, Liu X, Huang H, Zheng X, Pan X, Fang J, et al. Mass spectrometry-based metabolomics for irritable bowel syndrome biomarkers. Therap Adv Gastroenterol 2019;12:1756284819886425.
[Crossref] [Google Scholar] [PubMed]
- Lim I, Gibbons SJ, Lyford GL, Miller SM, Strege PR, Sarr MG, et al. Carbon monoxide activates human intestinal smooth muscle L-type Ca2+ channels through a nitric oxide-dependent mechanism. Am J Physiol Gastrointest Liver Physiol 2005;288(1):G7-14.
[Crossref] [Google Scholar] [PubMed]
- Shah ND, Chitkara DK, Locke GR, Meek PD, Talley NJ. Ambulatory care for constipation in the United States, 1993-2004. Am J Gastroenterol 2008;103(7):1746-53.
[Crossref] [Google Scholar] [PubMed]
- Huang CH, Jan RL, Kuo CH, Chu YT, Wang WL, Lee MS, et al. Natural flavone kaempferol suppresses chemokines expression in human monocyte THP‐1 cells through MAPK pathways. J Food Sci 2010;75(8):H254-9.
- Halvorson HA, Schlett CD, Riddle MS. Postinfectious irritable bowel syndrome-A meta-analysis. Am J Gastroenterol 2006;101(8):1894-9.
[Crossref] [Google Scholar] [PubMed]
- Thabane M, Kottachchi DT, Marshall JK. Systematic review and meta‐analysis: The incidence and prognosis of post‐infectious irritable bowel syndrome. Aliment Pharmacol Ther 2007;26(4):535-44.
[Crossref] [Google Scholar] [PubMed]