- *Corresponding Author:
- M. W. Zhang
Department of Neurosurgery,
The Affiliated Traditional Chinese Medicine Hospital of Southwest Medical University,
Luzhou Sichuan 646000,
China
E-mail: 239525268@swmu.edu.cn
| This article was originally published in a special issue,“New Advancements in Biomedical and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2022:84(2) Spl Issue “29-41” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
To investigate the molecular mechanism of Ziziphi spinosae semen and Schisandrae chinensis fructus in the treatment of insomnia is the objective of the study. First, the compounds and corresponding target genes of Ziziphi spinosae semen and Schisandrae chinensis fructus were obtained by traditional Chinese medicine systems pharmacology database and analysis platform database and genes associated with insomnia were obtained by GeneCards database. The results were imported into Cytoscape software to construct a network (Herbal-active compound- Target-disease related genes network). Then, gene ontology and Kyoto encyclopedia of genes and genomes enrichment analyses were performed for common genes in disease and compounds (Target-disease related genes). Finally, the key genes were identified by Cytoscape software and the molecular docking of key genes was verified by Autodock software. A total of 21 components and 43 target genes were screened and 41 target-disease related genes were identified. 191 gene ontology functional terms and 26 Kyoto encyclopedia of genes and genomes pathways show that neuroactive ligand-receptor interactions including cholinergic, norepinephrine and dopaminergic signals play an important role in the treatment of insomnia. Solute carrier family 6 member 3, cholinergic receptor nicotinic alpha 7 subunit, nuclear receptor subfamily 3 group C member 1, cholinergic receptor nicotinic alpha 2 subunit, estrogen receptor 1, cholinergic receptor muscarinic 1, acetylcholinesterase and progesterone receptor were identified as key genes, and their molecular docking results showed that all compounds could stably bind to the active pockets. Through these techniques, the multi-component synergistic mechanism of Ziziphi spinosae semen and Schisandrae chinensis fructus in the treatment of insomnia can be elucidated.
Keywords
Ziziphi spinosae semen, Schisandrae chinensis fructus, molecular docking, network pharmacology, insomnia
Insomnia has become a common phenomenon, accounting for up to 10 % of the total population, insomnias often have frequent difficulty in falling asleep and maintaining sleep even when they have sufficient sleep time [1,2]. Insomnia has a huge impact on patient’s quality of life and social economy. In a survey, they used Quality Adjusted Life Years (QALYs) as a measure of health status burden and the results showed that the loss of QALYs associated with insomnia in adults was significantly greater than that of other diseases (including arthritis, hypertension, etc.,). This suggests that insomnia is a major source of QALYs losses [3]. Insomnia is also a major risk factor for increased cardiovascular disease [4]. A meta-analysis of 11 prospective cohort studies involving 58 924 subjects with at least 1 y of interviews found that single or combined symptoms of insomnia were significantly associated with increased incidence of hypertension [5]. In addition, it is not difficult to find evidence that insomnia increases the risk of coronary heart disease and heart failure in several large prospective cohort studies [6,7]. Insomnia puts a lot of strain on social production and the economy, and research shows that insomnia is associated with the productivity loss index at work and insomnia is the powerful predictor of productivity [8]. The direct and indirect cost of treating insomnia alone is estimated to be as high as 100 billion United States (US) dollars per year [9], more surprisingly, the 10 billion US dollars economic loss due to insomnia was found in 2019-2020 which is equivalent to 0.73 % of Australia’s Gross Domestic Product (GDP) [10].
Insomnia often has a persistent negative psychosocial trigger, of which depression is an important factor [1,11,12]. Insomnia and depression often come together [11]. A meta-analysis of 172 077 participants in 34 prospective cohort studies shows that insomnia increases with the increase of depression risk [13]. In addition, insomnia is also a non-sexual symptom of brain degenerative diseases (including Alzheimer's disease, Parkinson's disease, and so on) in the elderly, which is likely to be associated with the damage of the brain area of the disease itself or sleep [14].
Although the current Cognitive Behavioral Therapy for Insomnia (CBT-I) is considered the most promising treatment for insomnia, there is also a problem of resource allocation, compliance, high recurrence rate and high cost [1]. A combination of CBT-I and medication is advocated, but the long-term use of prescription drugs often leads to serious adverse consequences such as dependency, mental nervous system and other accidents [1,15]. However, in China, the history of using traditional Chinese medicine to treat insomnia according to the actual situation has exceeded in 2000 y [16]. There is no doubt that this has a clear and unique advantage in treating insomnia and its research potential is huge. A recent meta-analysis on the treatment of primary insomnia showed that Shenqi Wuweizi tablets, Zao Ren An Shen capsules, Suanzaoren decoction, Bailemian capsules and other composite herb combinations had a significant advantage in different aspects of the effectiveness and safety in terms of sedative hypnosis [17]. Sufficient evidence can also be found in some in vivo studies, such as Suanzaoren decoction combined with Lorazepam can significantly improve the symptoms of chronic insomnia patients, its effect is better than conventional treatment [18]. Zao Ren An Shen capsules can significantly improve Pittsburgh Sleep Quality Index (PSQI) and hemorheology indexes of elderly insomnia patients [19]. Noteworthy, all of these classic composite herb combinations have one thing in common that they all use Ziziphi spinosae semen (ZSS) and Schisandrae chinensis fructus (SCF) as common compatibility, which is definitely no accident. Due to their mechanism of action in the body is hard to explain [20], we choose to use the network pharmacology method to explain their compounds and the relationship of the target gene in order to try to more complete and systematic understanding of ZSS and SCF in insomnia [21], and verify their relationship by simulated molecular docking.
Materials and Methods
Data collection and preparation:
Screening of ZSS-SCF drug compounds: Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) is a computer program for the study of Chinese herbal medicine and pharmacology of traditional Chinese medicine system database and analysis platform [22]. Only need to upload the name of ZSS and SCF, the ideal compound and its corresponding Oral Bioavailability (OB) and the Drug- Likeness (DL) will be easy to get. OB represents the compounds through the blood vessels into the cycle of drug ratio [23] and DL indicates the similarity of compounds to known drugs. OB and DL are important tools for pharmacokinetics [24]. Therefore, the retrieved herbal information is imported into Excel and qualified active compounds were obtained according to OB≥30 % and DL≥0.18 as screening conditions [25].
Acquisition of potential target genes: Similarly, all compounds and target genes can be found in TCMSP. The UniProt database contains extensive protein sequence resources and associated detailed annotations [26]. So only by entering the target gene names obtained in TCMSP into the UniProt database (http://www.uniprot.org/), we can find the target gene of the human species.
Acquisition of disease-related genes and Target- Disease Related Genes (T-DRGs): Genecards (https://www.genecards.org/) combines more than 150 web resources, that is a powerful human gene bank for genetic function analysis [27]. We use insomnia related search words which including “Disorders of initiating and maintaining Sleep”, “Early awakening”, “Nonorganic insomnia”, “Primary insomnia”, “Transient insomnia”, “Secondary insomnia”, “Sleep initiation dysfunction”, “Sleeplessness”, “Insomnia”, “Psychophysiological insomnia” and “DIMS” to search for insomnia related genes in Genecards. In addition, also add search for genetic databases such as PharmGKB (https://www.pharmgkb.org/), OMIM (https://omim.org/), Drugbank (https://db.idrblab.org/).
All target genes of identified compounds and insomnia are put into Bioinformatics and evolutionary genomics system(http://bioinformatics.psb.ugent.be/webtools/Venn/), and a Venn graph is created. The cross part of the Venn graph indicates the common genes in disease and compounds (T-DRGs). T-DRGs are thought to be an effective target for the treatment of insomnia.
Pharmacological network and enrichment analysis:
Establishment of pharmacological network: The integration of T-DRGs, active compounds and Chinese herbs were imported into Cytoscape Version 3.8.2 to construct a complex target network, which visualized the interaction law between active compounds and targets.
Enrichment analysis: Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis is mainly used to explain the connection between basic genes and core functions or pathways, which helps us to understand the key role of genes [28]. Database for Annotation, Visualization and Integrated Discovery (DAVID) (https://david.ncifcrf. gov/summary.jsp) database provides the function that widely collected data translated into the biological significance and contributes to the analysis of genomic data sets [29]. The use of DAVID for GO and KEGG analysis is of great significance to further explore the mechanism of ZSS-SCF in treating insomnia.
Protein-Protein Interaction (PPI) network construction and key gene screening:
PPI network construction: The T-DRGs are entered into the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database to generate a PPI network that helps to analyze protein interaction relationships.
Screening of Key Genes (KGs): The results were imported into Cytoscape (Version 3.8.2) and the interaction network parameters of each gene encoded protein were calculated by the algorithm of the CytoNCA plugin. As a centrality tool for biological network computation and analysis, CytoNCA plug-in can provide 8 centrality measures, including Betweenness Centrality (BC), Closeness Centrality (CC), Degree Centrality (DC), Eigenvector Centrality (EC), Local Average Connectivity-based method (LAC), Network Centrality (NC) scores [30]. The genes were screened according to the median value of the parameters of interest and the results were constructed into a sub-network. CytoHubba plug-in is another network node ordering system based on network characteristics [31]. Another sub-network is built by using cytoHubba plug- in. The gene shared by the two subnetworks is the KG, which is considered to be the key effective target of ZSS-SCF in the treatment of insomnia.
Molecular docking:
Molecular docking techniques were used to verify details of interactions between compounds and target genes [21]. We respectively collected data from Research Collaboratory for Structural Bioinformatics Protein Data Bank (RCSB PDB) database (https://www.rcsb. org/) to obtain protein two-dimensional structure and from PubChem database (https://pubchem.ncbi.nlm.nih. gov/) to obtain the corresponding compound structure. Since the more stable the docking and the less the binding energy [32], the small molecule of the compound was imported into ChemBio Three-Dimensional (3D) software to calculate the 3D structure of the minimum energy quantization. AutoDock (1.5.6) software was used to dehydrate and hydrogenate the receptor and the location of the center was calculated and an active binding pocket was formed. Ligand-receptor docking was performed by computer and finally PyMOL (2.5) software was used to visualize the docking relationship.
Results and Discussion
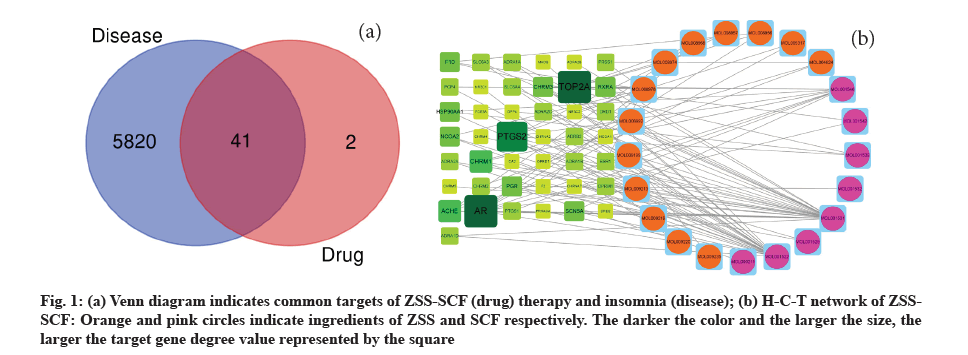
Results of compound screening and target gene prediction were shown below. After collation, it was found that a total of 21 active compounds (13 in ZSS and 8 in SCF) and 112 target genes were collected and relevant information was listed in the table (Table 1). The target genes and 5820 insomnia related genes from GeneCards database were made into Venn diagram (fig. 1a), and then 41 T-DRGs were found. Herbal- active Compound-T-DRGs network (H-C-T network, fig. 1b) shows the complex relationship between the active compound, T-DRG and herbal medicine, which is made up of 62 nodes (21 active compounds and 41 T-DRGs) and 98 edges. We found a compound that matched multiple target genes, and a target gene could also be associated with more than one compound, and the other, dl-nuciferine and (S)-coclaurine were the two most effective compounds.
Fig. 1: (a) Venn diagram indicates common targets of ZSS-SCF (drug) therapy and insomnia (disease); (b) H-C-T network of ZSSSCF: Orange and pink circles indicate ingredients of ZSS and SCF respectively. The darker the color and the larger the size, the larger the target gene degree value represented by the square
| Herb | Ingredient number | Ingredient name | OB % | DL | Target gene |
|---|---|---|---|---|---|
| ZSS | MOL001522 | (S)-Coclaurine | 42.35 | 0.24 | CHRM1, SCN5A, PTGS2, RXRA, PDE3A, ADRA1A, ADRA1B, SLC6A3, ADRB2, SLC6A4, HSP90AA1, MAOB, PCP4, DRD1, CHRM3, ADRA2A, CA2, sADRA2C, ADRA1D, OPRM1, PRKACA, NCOA2 |
| ZSS | MOL001525 | Daucosterol | 36.91 | 0.75 | PGR, NCOA2 |
| ZSS | MOL001531 | dl-Nuciferine | 29.26 | 0.4 | PTGS1, DRD1, CHRM3, CHRM1, AR, SCN5A, CHRM5, PTGS2, ADRA2A, ADRA2C, CHRM4, RXRA, OPRD1, AChE, ADRA1A, CHRM2, ADRA2B, ADRA1B, SLC6A3, ADRB2, ADRA1D, CHRNA2, SLC6A4, DRD2, OPRM1, CHRNA7 |
| ZSS | MOL001532 | Phytosterol | 36.91 | 0.75 | PGR |
| ZSS | MOL001539 | Sanjoinenine | 67.28 | 0.79 | F10, PTGS2, HSP90AA1 |
| ZSS | MOL001542 | Swertisin | 31.83 | 0.75 | AR, TOP2A |
| ZSS | MOL001546 | Zizyphusine | 41.53 | 0.55 | PTGS1, CHRM3, CHRM1, AR, SCN5A, PTGS2, RXRA, AChE, TOP2A, HSP90AA1, NCOA1, PCP4 |
| ZSS | MOL000211 | Mairin | 55.38 | 0.78 | PGR |
| SCF | MOL009199 | Interiotherin B | 31.76 | 0.77 | TOP2A |
| SCF | MOL009213 | Kadsulignan B | 30.63 | 0.84 | ESR1, AR |
| SCF | MOL009219 | Neokadsuranic acid C | 35.4 | 0.85 | NR3C1 |
| SCF | MOL009220 | Neokadsuranin | 33.35 | 0.88 | AR, PTGS2 |
| SCF | MOL009235 | Angusifolin B | 34.82 | 0.56 | TOP2A |
| SCF | MOL004624 | Longikaurin A | 47.72 | 0.53 | CHRM1, CHRM2, PRSS1 |
| SCF | MOL005317 | Deoxyharringtonine | 39.27 | 0.81 | AR, NR3C2 |
| SCF | MOL008956 | Angeloylgomisin O | 31.97 | 0.85 | AR, F10, TOP2A |
| SCF | MOL008957 | Schizandrer B | 30.7 | 0.83 | PTGS2, TOP2A |
| SCF | MOL008968 | Gomisin-A | 30.69 | 0.78 | F2, AChE |
| SCF | MOL008974 | Gomisin G | 32.68 | 0.83 | TOP2A |
| SCF | MOL008978 | Gomisin R | 34.84 | 0.86 | ESR1, AR, F10, PTGS2, TOP2A PRSS1, NCOA2 |
| SCF | MOL008992 | Wuweizisu C | 46.27 | 0.84 | AChE, DPP4 |
Table 1: The Active Compounds (OB≥30 % And DL≥0.18) and Corresponding Target Genes Were Screened
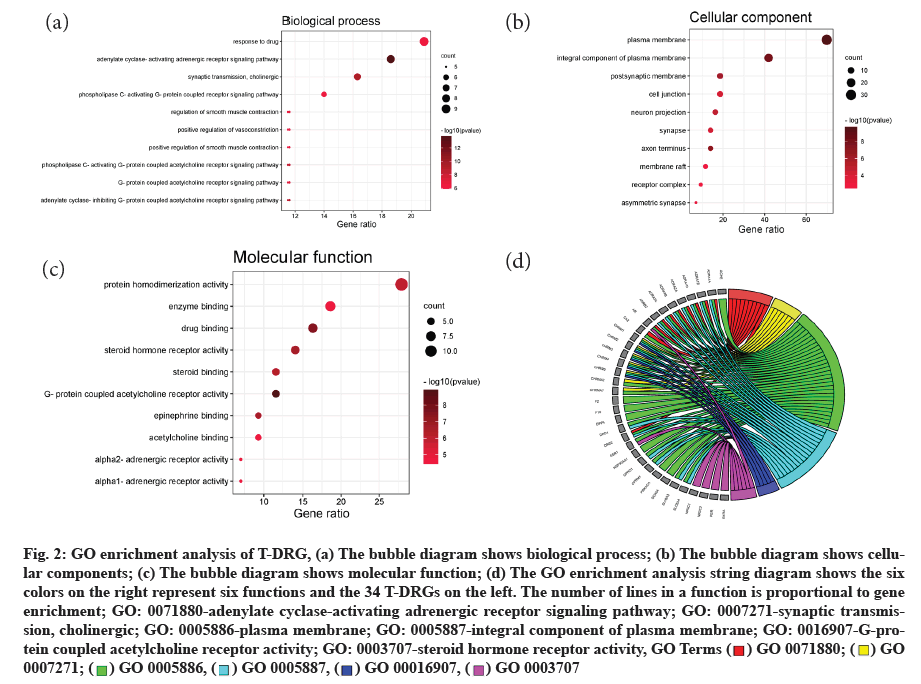
GO analysis is a system that is widely used to analyze the classification of genetic functions such as Biological Processes (BP), Cell Components (CC) and Molecular Functions (MF) [33]. 191 GO terms were identified from the DAVID database and bubble diagrams (fig. 2a-fig. 2c) for the first 10 terms of BP, CC and MF were made respectively by R language and Perl plugins. In these GO terms, adenylate cyclase activation of adrenergic receptors and cholinergic synaptic transmission are the most important processes in 125 BP. 24 CC shows that these active compounds are concentrated in the function of cell membrane, cytoplasmic membrane, nerve axon and postsynaptic membrane. In 42 MF, there are some neural receptors, including G-protein coupling acetylcholine receptor, alpha (α1/α2-adrenaline energy receptor, steroid hormone receptor, etc. The GO string diagram (fig. 2d) shows the two most significant terms in the BP, CC and MF respectively and the T-DRGs that participate in these functions. There are about 30 T-DRGs positioned in the cell membrane.
Fig. 2:GO enrichment analysis of T-DRG, (a) The bubble diagram shows biological process; (b) The bubble diagram shows cellular components; (c) The bubble diagram shows molecular function; (d) The GO enrichment analysis string diagram shows the six colors on the right represent six functions and the 34 T-DRGs on the left. The number of lines in a function is proportional to gene enrichment; GO: 0071880-adenylate cyclase-activating adrenergic receptor signaling pathway; GO: 0007271-synaptic transmission, cholinergic; GO: 0005886-plasma membrane; GO: 0005887-integral component of plasma membrane; GO: 0016907-G-protein coupled acetylcholine receptor activity; GO: 0003707-steroid hormone receptor activity, GO Terms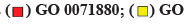
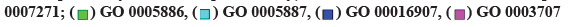
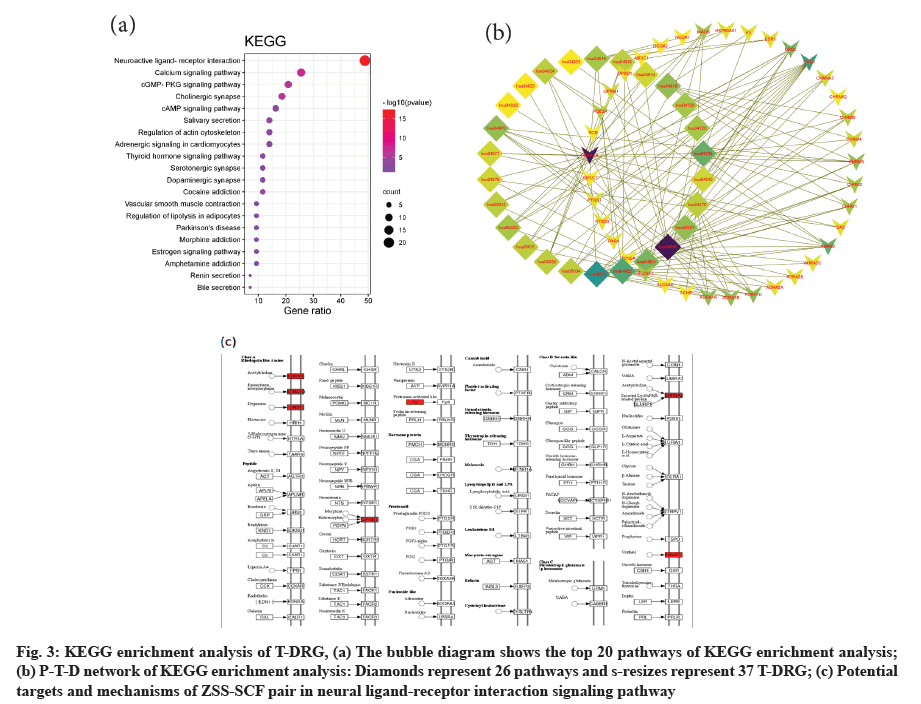
The signal pathway of 26 significant enrichment (p<0.01) was discovered through the KEGG analysis. It is noteworthy that the neural ligand- receptor interaction signaling pathway is the most enriched and the enrichment of T-DRGs is the largest. Calcium signal, cyclic Guanosine Monophosphate- Protein Kinase G (cGMP-PKG), cyclic Adenosine monophosphate (cAMP) and some synapses including cholinergic, serotonergic and dopaminergic synapses are also significantly enriched. The enrichment of the top 20 paths with small p values has been shown in the bubble graph (fig. 3a) and their names and corresponding term ID are listed in the table (Table 2). The corresponding relationship between significantly enriched signal pathways and T-DRGs are shown in the signal pathway-T-DRGs network (P-T-D network) (fig. 3b), which consists of 63 nodes (26 signaling pathways and 37 genes) and 137 edges. Without doubt, the neural ligand-receptor interaction signaling pathway (fig. 3c) is the object of our most attention and most of these T-DRGs are concentrated on receptors such as choline, adrenaline, Dopamine (DA), opioids and steroid hormones.
| Term | Path name |
|---|---|
| hsa04080 | Neuroactive ligand-receptor interaction |
| hsa04020 | Calcium signaling pathway |
| hsa04022 | cGMP-PKG signaling pathway |
| hsa04725 | Cholinergic synapse |
| hsa04970 | Salivary secretion |
| hsa05030 | Cocaine addiction |
| hsa04024 | cAMP signaling pathway |
| hsa04261 | Adrenergic signaling in cardiomyocytes |
| hsa04726 | Serotonergic synapse |
| hsa04923 | Regulation of lipolysis in adipocytes |
| hsa04919 | Thyroid hormone signaling pathway |
| hsa04728 | Dopaminergic synapse |
| hsa05031 | Amphetamine addiction |
| hsa04810 | Regulation of actin cytoskeleton |
| hsa05032 | Morphine addiction |
| hsa04915 | Estrogen signaling pathway |
| hsa04270 | Vascular smooth muscle contraction |
| hsa05012 | Parkinson’s disease |
| hsa04924 | Renin secretion |
| hsa04976 | Bile secretion |
| hsa04971 | Gastric acid secretion |
| hsa05034 | Alcoholism |
| hsa04914 | Progesterone-mediated oocyte maturation |
| hsa04540 | Gap junction |
Table 2: Names and Term ID of the First 20 KEGG Enriched Pathways
Fig. 3: KEGG enrichment analysis of T-DRG, (a) The bubble diagram shows the top 20 pathways of KEGG enrichment analysis; (b) P-T-D network of KEGG enrichment analysis: Diamonds represent 26 pathways and s-resizes represent 37 T-DRG; (c) Potential targets and mechanisms of ZSS-SCF pair in neural ligand-receptor interaction signaling pathway
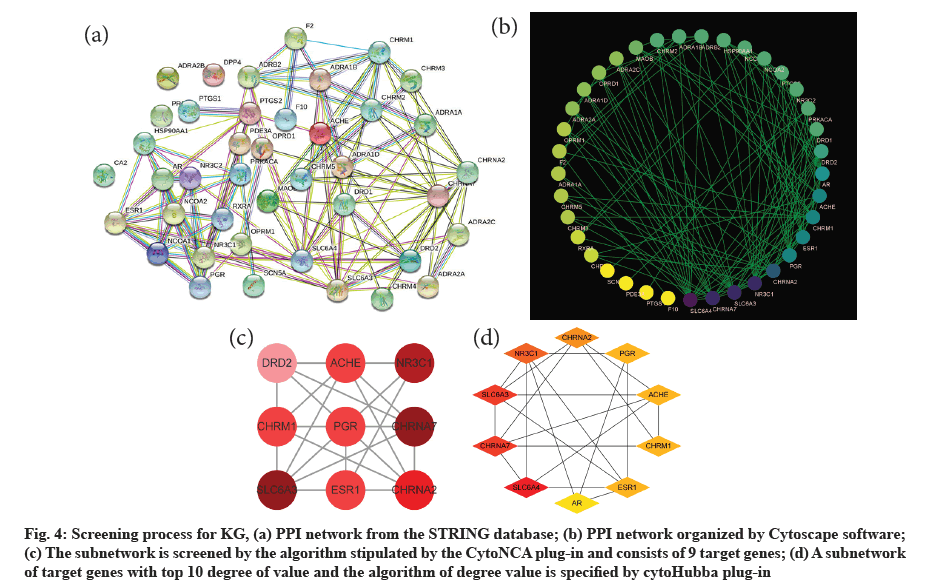
The PPI network exported from the STRING database is shown (fig. 4a). A new protein interaction network (fig. 4b) was constructed by Cytoscape software that consists of 38 nodes and 128 edges. Then, parameters (including BC, CC, DC, EC, LAC and NC) of each gene were calculated by CytoNca plug-in. The 9 T-DRGs larger than the median of the six parameters form a subnetwork (fig. 4c). All genes related parameters of the subnetwork are shown in the table (Table 3). In addition, another subnetwork consisting of 10 T-DRG was obtained by cytoHubba plug-in (fig. 4d). Finally, 8 KGs are generated, which included Solute Carrier Family 6 Member 3 (SLC6A3), Cholinergic Receptor Nicotinic Alpha 7 Subunit (CHRNA7), Nuclear Receptor Subfamily 3 Group C Member 1 (NR3C1), Cholinergic Receptor Nicotinic Alpha 2 Subunit (CHRNA2), Estrogen Receptor 1 (ESR1), Cholinergic Receptor Muscarinic 1 (CHRM1), Acetylcholinesterase (AChE) and Progesterone Receptor (PGR) according to sorting by CytoNca plug-in.
Fig. 4: Screening process for KG, (a) PPI network from the STRING database; (b) PPI network organized by Cytoscape software; (c) The subnetwork is screened by the algorithm stipulated by the CytoNCA plug-in and consists of 9 target genes; (d) A subnetwork of target genes with top 10 degree of value and the algorithm of degree value is specified by cytoHubba plug-in
| Name | BC | CC | DC | EC | LAC | NC |
|---|---|---|---|---|---|---|
| AChE | 94.58208568 | 0.507042254 | 10 | 0.24166 | 4 | 6.01429 |
| SLC6A3 | 192.1066686 | 0.590163934 | 15 | 0.30489 | 3.333333333 | 8.03343 |
| DRD2 | 35.54484118 | 0.514285714 | 8 | 0.22775 | 4.5 | 5.49048 |
| CHRNA7 | 114.5898475 | 0.514285714 | 15 | 0.30823 | 4.933333333 | 11.8016 |
| CHRM1 | 71.92516699 | 0.467532468 | 10 | 0.18999 | 3.4 | 6.16667 |
| CHRNA2 | 57.90267713 | 0.486486486 | 12 | 0.24888 | 4.5 | 9.11688 |
| NR3C1 | 304.4970998 | 0.580645161 | 14 | 0.24834 | 4.857142857 | 10.1361 |
| PGR | 36.65020886 | 0.444444444 | 10 | 0.15285 | 5.2 | 7.5 |
| ESR1 | 109.0580512 | 0.52173913 | 10 | 0.19274 | 4.6 | 6.375 |
Table 3: Parameters of 9 Target Genes Screened By the Algorithm of Cytonca Plug-In
The 8 KGs and the corresponding active compounds were simulated to the molecular docking respectively and the important parameters information of these receptors-ligand docking is listed in the table (Table 4), including the ligand and receptor names, receptor protein PDB ID, docking energy, central coordinate and grid size. The best docking mode is determined by the AutoDock software. All of the docking energy is less than -5 kcal/mol, which means that the docking is good [34].
| Targets | PDB-ID | Core active ingredient | Binding energy/(kcal/mol) | Grid center (xyz) | Grid size (xyz) |
|---|---|---|---|---|---|
| SLC6A3 | AF-Q01959-F1 | dl-Nuciferine | -6.9 | -14.239, -23.595, 3.181 | 106, 110, 72 |
| SLC6A3 | AF-Q01959-F1 | (S)-Coclaurine | -6.5 | -14.239, -23.595, 3.181 | 106, 110, 72 |
| CHRNA7 | 5AFN | dl-Nuciferine | -8.5 | 19.267, 5.881, 28.251 | 88, 80, 80 |
| NR3C1 | AF-P04150-F1 | Neokadsuranic acid C | -7.8 | -8.21, 0.852, -6.411 | 126, 122, 126 |
| CHRNA2 | AF-Q15822-F1 | dl-Nuciferine | -6.2 | -25.742, -9.133, 5.379 | 126, 86, 126 |
| ESR1 | 1xpc | Kadsulignan B | -5.9 | 23.553, 4.423, 20.424 | 46, 56, 54 |
| ESR1 | 1xpc | Gomisin R | -6.7 | 23.553?4.423?20.424 | 46, 56, 54 |
| CHRM1 | 6WJC | (S)-Coclaurine | -7.2 | -15.171, -18.987, 59.241 | 60, 74, 90 |
| CHRM1 | 6WJC | dl-Nuciferine | -8 | -15.171, -18.987, 59.241 | 60, 74, 90 |
| CHRM1 | 6WJC | Zizyphusin | -8.3 | -15.171, -18.987, 59.241 | 60, 74, 90 |
| CHRM1 | 6WJC | Longikaurin A | -9.1 | -15.171, -18.987, 59.241 | 60, 74, 90 |
| AChE | 6NTO | dl-Nuciferine | -8.7 | -33.918, -21.387, 56.538 | 64, 72, 86 |
| AChE | 6NTO | Zizyphusine | -9 | -33.918, -21.387, 56.538 | 64, 72, 86 |
| AChE | 6NTO | Gomisin-A | -6.7 | -33.918, -21.387, 56.538 | 64, 72, 86 |
| AChE | 6NTO | Wuweizisu C | -7.8 | -33.918, -21.387, 56.538 | 64, 72, 86 |
| PGR | AF-P06401-F1 | Daucosterol | -6.6 | -4.961, 4.815, 0.984 | 126, 126, 126 |
| PGR | AF-P06401-F1 | Phytosterol | -6.3 | -4.961, 4.815, 0.984 | 126, 126, 126 |
| PGR | AF-P06401-F1 | Mairin | -7.6 | -4.961, 4.815, 0.984 | 126, 126,126 |
Table 4: All KGs of Molecular Docking Model Related Information
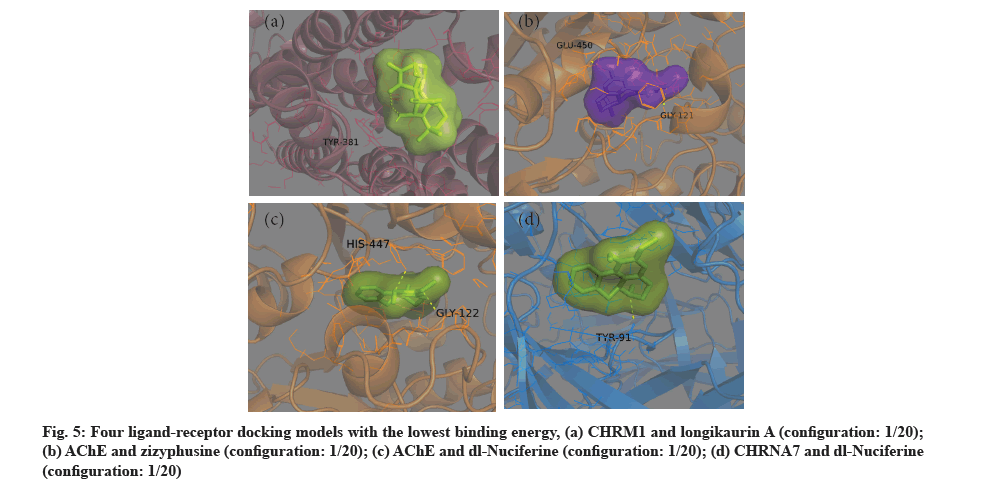
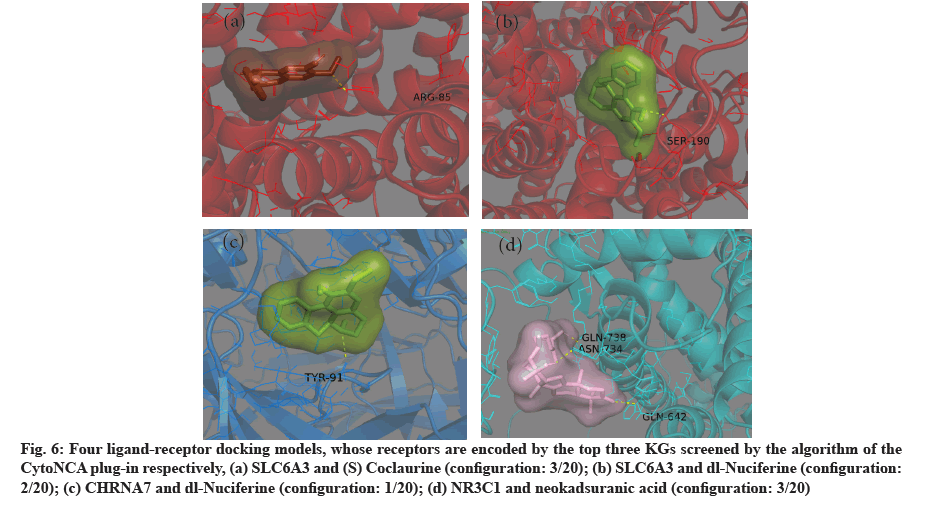
The four ligand-receptor docking models with the lowest binding energy are shown in fig. 5. The first three KGs were selected according to the sequence of parameters calculated by CytoNCA plug-in and their simulated molecular docking models were shown in fig. 6. The ligands of these models are fully wrapped in active pockets and at least one hydrogen bonding ligand configuration.
Fig. 6: Four ligand-receptor docking models, whose receptors are encoded by the top three KGs screened by the algorithm of the CytoNCA plug-in respectively, (a) SLC6A3 and (S) Coclaurine (configuration: 3/20); (b) SLC6A3 and dl-Nuciferine (configuration: 2/20); (c) CHRNA7 and dl-Nuciferine (configuration: 1/20); (d) NR3C1 and neokadsuranic acid (configuration: 3/20)
In this study, two herbs were identified with 21 compounds and 112 potential target genes. Then, 41 T-DRGs were predicted and they were rich in 191 GO terms and 26 KEGG pathways. Finally, eight KGs were predicted, these KGs are found to be closely and stably bond to the compound by simulating molecular docking.
The results show that ZSS can improve insomnia by regulation of Serotonin or 5-Hydroxytryptamine (5- HT), glutamate, gamma-Aminobutyric Acid (GABA), Norepinephrine (NE), DA and other monoamine and amino acid neurotransmitters [35]. SCF has the potential to alter the behaviour of the 5-serotonergic and aminobutyric systems and have sedative-hypnotic activity [36]. Dl-Nuciferine and (S)-Coclaurine were found to be the most active compounds by H-C-T network analysis. Studies have shown that smoking lotus leaves rich in dl-Nuciferine can bring feelings of relaxation [37]. The potential of dl-Nuciferine in the treatment of insomnia is enormous and it is a potent inhibitor of AChE and is also excitatory to 5-HT2C receptor. Dl-Nuciferine selectively binds tightly to several key amino acid residues of rat 5-HT2A receptor via the H bond [38,39]. The results of simulated molecular docking showed that dl-Nuciferine could bind closely to five KGs encoded proteins, including SLC6A3, CHRNA7, CHRNA2, CHRM1 and AChE(the binding energy <-6 and hydrogen bonding). These results indicate that dl-Nuciferine may play a role in regulating DA system and Acetylcholine (ACH)-associated receptors, which is consistent with previous studies. Coclaurine interacts with melatonin to produce sedative effects [40]. (S)-Coclaurine, as one of the configurations, has antagonistic activity against D1 and D2 DA receptors and is a potential target of psychiatric diseases [41]. In this study, Gomisin A and Wuweizisu C are active components of SCF, and act on ACH-associated receptors, which is what we focus on. Similar to dl-Nuciferine, Gomisin A significantly inhibits AChE activity and modulates ACH levels [42]. It also ameliorates nerve damage by reducing striatum toxicity induced by 3-Nitropropionic Acid (3-NPA) in Huntington’s Disease (HD) mouse models due to its anti- inflammatory and antioxidant activity [43]. Wuweizisu C has potential therapeutic effects on a variety of neuropsychiatric diseases through the regulation of NE, DA and 5-HT [44]. It also inhibits oxidative stress, modulates apoptotic signaling and cytotoxicity, and is effective in neurodegenerative diseases [45].
By combining KGs, KEGG and GO analyses, ACH- associated receptors are widely acted by multiple compounds of the two herbs and cholinergic synaptic transmission is the main process in which T-DRGs is involved, suggesting that the cholinergic system may be the most important pathway in the treatment of insomnia by ZSS and SCF. Using a recording method of channelrhodopsin-2-labeled neurons, they found that cholinergic neurons, along with glutamate and albumin-positive GABAergic neurons form circuits in the basal forebrain and participate in Sleep/Wake (S/W) regulation [46]. Cholinergic neurons are significantly active during Rapid Eye Movement (REM) sleep, but activated cholinergic neurons rapidly induce wakefulness [46]. Among the 8 KGs we predicted, ACH- associated receptors include CHRNA7, CHRNA2, CHRM1 and AChE. The simulated molecular docking showed that CHRM1 and AChE could stably and closely bind to a variety of compounds (binding energy <-6, with a large number of hydrogen-bonded configurations), suggesting that CHRM1 and AChE may be the most important targets of ZSS-SCF. Long- term use of AChE inhibitors significantly increases the risk of depression and alters sleep structure through the Hypothalamic-Pituitary-Adrenal-axis (HPA axis), such as increasing REM tension activity and decreasing REM duration [47]. CHRM1 is a key molecule in the regulation of REM sleep and the maintenance time of REM and Non-Rapid Eye Movement sleep (NREM) was moderately or more shortened in mice with this gene knocked out [48]. Nicotinic acetylcholine receptor (CHRNA) is a potential therapeutic target for major depression and CHRNA7 is the most reported. Adolescents who carry CHRNA7 tend to be more likely to have major depressive disorders [49]. Curiously, there is also evidence that CHRNA7 deficient mice are more likely to exhibit depressive symptoms [50] and the use of CHRNA7 agonist can promote the release of 5-HT and improve depressive behavior in depressed mice [51].
In addition to the cholinergic system, the noradrenergic and dopaminergic neuroregulatory systems play an important role in maintaining wakefulness [52]. We also found that Adrenergic Receptor A1 (α1AR), Adrenergic Receptor A2 (α2AR), Dopamine Receptor 1 (D12R) and Dopamine Receptor 2 (D22R) are the targets of active compounds in the neuroactive ligand receptor interaction pathway and GO analysis also suggested that adenylate cyclase activation of adrenergic receptors was a significantly enriched biological process, which revealed that noradrenergic and dopaminergic signaling pathways were key to the efficacy of the drug. Both of these signaling systems are important enhancers of cortical activation and behavioral arousal [53] and the signals transmitted by α1AR and α2AR are mainly from the S/W regulating LC network [53,54]. Using cortical electroencephalography combined with optrode recording, they found that α1AR primarily activating neurons that are active during waking, while sleep-active neurons receive inhibition of α2AR in ventrolateral preoptic nucleus (VLPO) [55]. D12R and D22R are related to the rhythm of sleep activity, recent results show that greater eveningness and physical inactivity are associated with high D12R in caudate nucleus and high D22R in nucleus accumbens, respectively [56]. SLC6A3 is the KG with the highest centrality predicted in this study. It mainly encodes DA transporters and reuptake of DA from synaptic cleft to presynaptic neurons, which is the main mechanism of DA system regulation in the striatum and the key to the pathogenesis of neurological and psychiatric diseases [57,58]. SLC6A3 and D22R genes jointly determine increased drowsiness, attention deficit, and θ/α wave energy ratio changes in healthy subjects after sleep deprivation [59]. In addition, delta (δ) and mu (µ) opioid receptors are also typical neuroactive receptors that are significantly enriched, which are associated with depression. The higher the availability of µ opioid receptors, the lower the depressive mood [60]. Activation of the δ opioid receptor, however, can be a rapid antidepressant [61].
In the combination of ZSS-SCF, although SCF does not seem to be as prominent as ZSS, it is exciting to note that ESR1 in KGs is the main target of SCF. A questionnaire showed a significant positive correlation between ESR1 messenger Ribonucleic acid (mRNA) expression level and the personality characteristics of female depressed patients [62] and a recent study also showed that inhibition of ESR1 mRNA can play an antidepressant role, especially in female population [63]. NR3C1 is one of the KGs of SCF acts alone and plays an important role in depression regulation through the HPA axis [64]. They found that increased promoter methylation of the NR3C1 exon accelerated the encoding of Glucocorticoid Receptor (GR), further influencing depression in children with early adversity [65].
As with other network pharmacology, the selection of the intersection of ZSS-FCS and insomnia targets in this study not only follows the operational process of network pharmacology, but also makes virtual screening results more reliable [66]. However, there are some unavoidable problems. Some of their potential effects may have been overlooked in this classic combination, such as OB and DL not having the desired active ingredient, target genes that may have been missed, whether two herbs cooked together will react to form new active compounds that enhance their efficacy and mitigate their toxic side effects, etc.
As described above, the complex relationship between the active components of ZSS and SCF and their targets has been elucidated by means of network pharmacology and simulated molecular docking. The molecular mechanism explains the use of ZSS and SCF in the treatment of insomnia and similar results have been obtained from previous in vivo experiments. The system analysis in this paper can provide a comprehensive idea for further research. The datasets presented in this study can be found in online repositories. (https://data.4tu.nl/ account/home).
Conflict of interests:
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Zhou ES, Gardiner P, Bertisch SM. Integrative medicine for insomnia. Med Clin North Am 2017;101(5):865-79.
[Crossref] [Google Scholar] [PubMed]
- Bianchi MT. Chronic insomnia. Semin Neurol 2017;37(4):433-438.
[Crossref] [Google Scholar] [PubMed]
- Olfson M, Wall M, Liu SM, Morin CM, Blanco C. Insomnia and impaired quality of life in the United States. J Clin Psychiatry 2018;79(5):9151.
[Crossref] [Google Scholar] [PubMed]
- Javaheri S, Redline S. Insomnia and risk of cardiovascular disease. Chest 2017;152(2):435-44.
[Crossref] [Google Scholar] [PubMed]
- Meng L, Zheng Y, Hui R. The relationship of sleep duration and insomnia to risk of hypertension incidence: A meta-analysis of prospective cohort studies. Hypertens Res 2013;36(11):985-95.
[Crossref] [Google Scholar] [PubMed]
- Strand LB, Tsai MK, Gunnell D, Janszky I, Wen CP, Chang SS. Self-reported sleep duration and coronary heart disease mortality: A large cohort study of 400 000 Taiwanese adults. Int J Cardiol 2016;207:246-51.
[Crossref] [Google Scholar] [PubMed]
- Laugsand LE, Strand LB, Platou C, Vatten LJ, Janszky I. Insomnia and the risk of incident heart failure: A population study. Eur Heart J 2014;35(21):1382-93.
[Crossref] [Google Scholar] [PubMed]
- Espie CA, Pawlecki B, Waterfield D, Fitton K, Radocchia M, Luik AI. Insomnia symptoms and their association with workplace productivity: Cross-sectional and pre-post intervention analyses from a large multinational manufacturing company. Sleep Health 2018;4(3):307-12.
[Crossref] [Google Scholar] [PubMed]
- Taddei-Allen P. Economic burden and managed care considerations for the treatment of insomnia. Am J Manag Care 2020;26(4):S91-6.
[Crossref] [Google Scholar] [PubMed]
- Streatfeild J, Smith J, Mansfield D, Pezzullo L, Hillman D. The social and economic cost of sleep disorders. Sleep 2021;44(11):132.
[Crossref] [Google Scholar] [PubMed]
- Medrano-Martínez P, Ramos-Platón MJ. Cognitive and emotional alterations in chronic insomnia. Rev Neurol 2016;62(4):170-78.
[Crossref] [Google Scholar] [PubMed]
- Riemann D, Krone LB, Wulff K, Nissen C. Sleep, insomnia and depression. Neuropsychopharmacology 2020;45(1):74-89.
[Crossref] [Google Scholar] [PubMed]
- Li L, Wu C, Gan Y, Qu X, Lu Z. Insomnia and the risk of depression: A meta-analysis of prospective cohort studies. BMC Psychiatry 2016;16(1):1-6.
[Crossref] [Google Scholar] [PubMed]
- Malhotra RK. Neurodegenerative disorders and sleep. Sleep Med Clin 2018;13(1):63-70.
[Crossref] [Google Scholar] [PubMed]
- Dujardin S, Pijpers A, Pevernagie D. Prescription drugs used in insomnia. Sleep Med Clin 2018;13(2):169-82.
[Crossref] [Google Scholar] [PubMed]
- Singh A, Zhao K. Treatment of insomnia with traditional Chinese herbal medicine. Int Rev Neurobiol 2017;135:97-115.
[Crossref] [Google Scholar] [PubMed]
- Yao WQ, Zhao YH, Zheng YW, Zhang Q, Han X. Network meta-analysis of Chinese patent medicines of Ziziphi spinosae semen in treatment of primary insomnia. Zhongguo Zhong Yao Za Zhi 2021;46(17):4541-54.
[Crossref] [Google Scholar] [PubMed]
- Song MF, Chen LQ, Shao QY, Hu LL, Liu WJ, Zhang YH. Efficacy and safety of Jiawei Suanzaoren decoction combined with lorazepam for chronic insomnia: A parallel-group randomized controlled trial. Evid Based Complement Alternat Med 2020;2020.
[Crossref] [Google Scholar] [PubMed]
- Gan JG, Tian GQ, Qin GX. Study on efficacy of zaoren anshen capsules in treating senile insomnia and changes in its hemorheology. Zhongguo Zhong Yao Za Zhi 2013;38(2):273-5.
[Google Scholar] [PubMed]
- Tang H, He S, Zhang X, Luo S, Zhang B, Duan X, et al. A network pharmacology approach to uncover the pharmacological mechanism of XuanHuSuo powder on osteoarthritis. Evid Based Complement Alternat Med 2016;2016.
[Crossref] [Google Scholar] [PubMed]
- Lee AY, Lee JY, Chun JM. Exploring the mechanism of Gyejibokryeong-hwan against atherosclerosis using network pharmacology and molecular docking. Plants 2020;9(12):1750.
[Crossref] [Google Scholar] [PubMed]
- Ru J, Li P, Wang J, Zhou W, Li B, Huang C, et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J Cheminform 2014;6(1):1-6.
[Crossref] [Google Scholar] [PubMed]
- Di L, Kerns EH, Carter GT. Drug-like property concepts in pharmaceutical design. Curr Pharm Des 2009;15(19):2184-94.
[Crossref] [Google Scholar] [PubMed]
- Prentis RA, Lis YV, Walker SR. Pharmaceutical innovation by the seven UK?owned pharmaceutical companies (1964?1985). Br J Clin Pharmacol 1988;25(3):387-96.
[Crossref] [Google Scholar] [PubMed]
- Tsaioun K, Blaauboer BJ, Hartung T. Evidence-based absorption, distribution, metabolism, excretion (ADME) and its interplay with alternative toxicity methods. ALTEX 2016;33(4):343-58.
[Crossref] [Google Scholar] [PubMed]
- The UniProt consortium. UniProt: The universal protein knowledgebase. Nucleic Acids Res 2017;45:158-169.
[Crossref] [Google Scholar] [PubMed]
- Safran M, Solomon I, Shmueli O, Lapidot M, Shen-Orr S, Adato A, et al. GeneCards™ 2002: Towards a complete, object-oriented, human gene compendium. Bioinformatics 2002;18(11):1542-3.
[Crossref] [Google Scholar] [PubMed]
- Chen L, Zhang YH, Wang S, Zhang Y, Huang T, Cai YD. Prediction and analysis of essential genes using the enrichments of gene ontology and KEGG pathways. PLoS One. 2017;12(9):e0184129.
[Crossref] [Google Scholar] [PubMed]
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, et al. DAVID: Database for annotation, visualization and integrated discovery. Genome Biol 2003;4(9):1-11.
[Crossref] [Google Scholar] [PubMed]
- Tang Y, Li M, Wang J, Pan Y, Wu FX. CytoNCA: A cytoscape plugin for centrality analysis and evaluation of protein interaction networks. Biosystems 2015;127:67-72.
[Crossref] [Google Scholar] [PubMed]
- Chin CH, Chen SH, Wu HH, Ho CW, Ko MT, Lin CY. cytoHubba: Identifying hub objects and sub-networks from complex interactome. BMC Syst Biol 2014;8(4):1-7.
[Crossref] [Google Scholar] [PubMed]
- Morris GM, Huey R, Olson AJ. Using autodock for ligand?receptor docking. Curr Protoc Bioinformatics 2008;24(1):8-14.
[Crossref] [Google Scholar] [PubMed]
- Wu Q, Hu Y. Integrated network pharmacology and molecular docking strategy to explore the mechanism of medicinal and edible Astragali Radix?Atractylodis Macrocephalae Rhizoma acting on pneumonia via immunomodulation. J Food Biochem 2020;44(12):e13510.
[Crossref] [Google Scholar] [PubMed]
- Wu L, Chen Y, Chen M, Yang Y, Che Z, Li Q, et al. Application of network pharmacology and molecular docking to elucidate the potential mechanism of Astragalus-Scorpion against prostate cancer. Andrologia 2021;53(9):e14165.
[Crossref] [Google Scholar] [PubMed]
- Yan YA, Qiang LI, Hui-Zhi DU, Chen-Xi SH, Ai-Ping LI, Xiang-Ping PE, et al. Determination of five neurotransmitters in the rat brain for the study of the hypnotic effects of Ziziphi spinosae semen aqueous extract on insomnia rat model by UPLC-MS/MS. Chin J Nat Med 2019;17(7):551-60.
[Crossref] [Google Scholar] [PubMed]
- Zhang C, Mao X, Zhao X, Liu Z, Liu B, Li H, et al. Gomisin N isolated from Schisandra chinensis augments pentobarbital-induced sleep behaviors through the modification of the serotonergic and GABAergic system. Fitoterapia 2014;96:123-30.
[Crossref] [Google Scholar] [PubMed]
- Kumarihamy M, León F, Pettaway S, Wilson L, Lambert JA, Wang M, et al. In vitro opioid receptor affinity and in vivo behavioral studies of Nelumbo nucifera flower. J Ethnopharmacol 2015;174:57-65.
[Crossref] [Google Scholar] [PubMed]
- Mathew M, Subramanian S. In vitro screening for anti-cholinesterase and antioxidant activity of methanolic extracts of ayurvedic medicinal plants used for cognitive disorders. PLoS One 2014;9(1):e86804.
[Crossref] [Google Scholar] [PubMed]
- Munusamy V, Yap BK, Buckle MJ, Doughty SW, Chung LY. Structure?based identification of aporphines with selective 5?HT2A receptor-binding activity. Chem Biol Drug Des 2013;81(2):250-6.
[Crossref] [Google Scholar] [PubMed]
- de la Peña JB, Lee HL, Yoon SY, Kim GH, Lee YS, Cheong JH. The involvement of magnoflorine in the sedative and anxiolytic effects of Sinomeni Caulis et Rhizoma in mice. J Nat Med 2013;67(4):814-21.
[Crossref] [Google Scholar] [PubMed]
- Zhou H, Hou T, Gao Z, Guo X, Wang C, Wang J, et al. Discovery of eight alkaloids with D1 and D2 antagonist activity in leaves of Nelumbo nucifera Gaertn. using FLIPR assays. J Ethnopharmacol 2021;278:114335.
[Crossref] [Google Scholar] [PubMed]
- Hung TM, Na M, Min BS, Ngoc TM, Lee I, Zhang X, et al. Acetylcholinesterase inhibitory effect of lignans isolated from Schizandra chinensis. Arch Pharm Res 2007;30(6):685-90.
[Crossref] [Google Scholar] [PubMed]
- Kim EJ, Jang M, Lee MJ, Choi JH, Lee SJ, Kim SK, et al. Schisandra chinensis stem ameliorates 3-nitropropionic acid-induced striatal toxicity via activation of the nrf2 pathway and inhibition of the MAPKs and NF-κB pathways. Front Pharmacol 2017;8:673.
[Crossref] [Google Scholar] [PubMed]
- Zhang Y, Lv X, Liu R, Zhang M, Liu H, Gao H, et al. An integrated strategy for ascertaining quality marker of Schisandra chinensis (Turcz.) Baill based on correlation analysis between depression-related monoaminergic metabolites and chemical components profiling. J Chromatogr A 2019;1598:122-31.
[Crossref] [Google Scholar] [PubMed]
- Song JX, Lin X, Wong RN, Sze SC, Tong Y, Shaw PC, et al. Protective effects of dibenzocyclooctadiene lignans from Schisandra chinensis against beta?amyloid and homocysteine neurotoxicity in PC12 cells. Phytother Res 2011;25(3):435-43.
[Crossref] [Google Scholar] [PubMed]
- Xu M, Chung S, Zhang S, Zhong P, Ma C, Chang WC, et al. Basal forebrain circuit for sleep-wake control. Nat Neurosci 2015;18(11):1641-7.
[Crossref] [Google Scholar] [PubMed]
- Dulawa SC, Janowsky DS. Cholinergic regulation of mood: From basic and clinical studies to emerging therapeutics. Mol Psychiatry 2019;24(5):694-709.
[Crossref] [Google Scholar] [PubMed]
- Niwa Y, Kanda GN, Yamada RG, Shi S, Sunagawa GA, Ukai-Tadenuma M, et al. Muscarinic acetylcholine receptors Chrm1 and Chrm3 are essential for REM sleep. Cell Rep 2018;24(9):2231-47.
[Crossref] [Google Scholar] [PubMed]
- Gillentine MA, Lozoya R, Yin J, Grochowski CM, White JJ, Schaaf CP, et al. CHRNA7 copy number gains are enriched in adolescents with major depressive and anxiety disorders. J Affect Disord 2018;239:247-52.
[Crossref] [Google Scholar] [PubMed]
- Yin J, Chen W, Yang H, Xue M, Schaaf CP. Chrna7 deficient mice manifest no consistent neuropsychiatric and behavioral phenotypes. Sci Rep 2017;7(1):1-10.
[Crossref] [Google Scholar] [PubMed]
- Andreasen JT, Redrobe JP, Nielsen EØ. Combined α7 nicotinic acetylcholine receptor agonism and partial serotonin transporter inhibition produce antidepressant-like effects in the mouse forced swim and tail suspension tests: A comparison of SSR180711 and PNU-282987. Pharmacol Biochem Behav 2012;100(3):624-9.
[Crossref] [Google Scholar] [PubMed]
- Yu X, Franks NP, Wisden W. Sleep and sedative states induced by targeting the histamine and noradrenergic systems. Front Neural Circuits 2018;12:4.
[Crossref] [Google Scholar] [PubMed]
- Jones BE. Arousal and sleep circuits. Neuropsychopharmacology 2020;45(1):6-20.
[Crossref] [Google Scholar] [PubMed]
- Szabadi E. Functional neuroanatomy of the central noradrenergic system. J Psychopharmacol 2013;27(8):659-93.
[Crossref] [Google Scholar] [PubMed]
- Liang Y, Shi W, Xiang A, Hu D, Wang L, Zhang L. The NAergic locus coeruleus-ventrolateral preoptic area neural circuit mediates rapid arousal from sleep. Curr Biol 2021;31(17):3729-42.
[Crossref] [Google Scholar] [PubMed]
- Zhang R, Manza P, Tomasi D, Kim SW, Shokri-Kojori E, Demiral SB, et al. Dopamine D1 and D2 receptors are distinctly associated with rest-activity rhythms and drug reward. J Clin Invest 2021;131(18) e149722.
[Crossref] [Google Scholar] [PubMed]
- Klein M, Van Donkelaar M, Verhoef E, Franke B. Imaging genetics in neurodevelopmental psychopathology. Am J Med Genet B Neuropsychiatr Genet 2017;174(5):485-537.
[Crossref] [Google Scholar] [PubMed]
- Salatino?Oliveira A, Rohde LA, Hutz MH. The dopamine transporter role in psychiatric phenotypes. Am J Med Genet B Neuropsychiatr Genet 2018;177(2):211-31.
[Crossref] [Google Scholar] [PubMed]
- Holst SC, Müller T, Valomon A, Seebauer B, Berger W, Landolt HP. Functional polymorphisms in dopaminergic genes modulate neurobehavioral and neurophysiological consequences of sleep deprivation. Sci Rep 2017;7(1):1-11.
[Crossref] [Google Scholar] [PubMed]
- Nummenmaa L, Karjalainen T, Isojärvi J, Kantonen T, Tuisku J, Kaasinen V, et al. Lowered endogenous mu-opioid receptor availability in subclinical depression and anxiety. Neuropsychopharmacology 2020;45(11):1953-9.
[Crossref] [Google Scholar] [PubMed]
- Nagase H, Saitoh A. Research and development of κ opioid receptor agonists and δ opioid receptor agonists. Pharmacol Ther 2020;205:107427.
[Crossref] [Google Scholar] [PubMed]
- Talarowska ME, Szemraj J, Kuan-Pin S. Expression of ESR1 and ESR2 oestrogen receptor encoding gene and personality traits–preliminary study. Prz Menopauzalny 2019;18(3):133-40.
[Crossref] [Google Scholar] [PubMed]
- Li L, Lu JJ, Wang WT, Wang N, Wang LX, Ma YM, et al. Potential targets of Euodiae Fructus in treatment of insomnia based on network pharmacology. Zhongguo Zhong Yao Za Zhi 2021;46(12):3016-23.
[Crossref] [Google Scholar] [PubMed]
- Chen D, Meng L, Pei F, Zheng Y, Leng J. A review of DNA methylation in depression. J Clin Neurosci 2017;43:39-46.
[Crossref] [Google Scholar] [PubMed]
- Farrell C, Doolin K, O’Leary N, Jairaj C, Roddy D, Tozzi L, et al. DNA methylation differences at the glucocorticoid receptor gene in depression are related to functional alterations in hypothalamic–pituitary–adrenal axis activity and to early life emotional abuse. Psychiatry Res 2018;265:341-8.
[Crossref] [Google Scholar] [PubMed]
- Liang Y, Liang B, Wu XR, Chen W, Zhao LZ. Network pharmacology-based systematic analysis of molecular mechanisms of dingji fumai decoction for ventricular arrhythmia. Evid Based Complement Alternat Med 2021;2021.
[Crossref] [Google Scholar] [PubMed]