- *Corresponding Author:
- H. Huang
Department of Rehabilitation Medicine
E-mail: 60120006@sdutcm.edu.cn
| This article was originally published in a special issue, “Current Trends in Pharmaceutical and Biomedical Sciences” |
| Indian J Pharm Sci 2022:84(5) Spl Issue “199-216” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Colchicine is an alkaloid with antitumor effect isolated from Colchicum autumnale plants, it has been reported in multiple studies as a potential treatment for coronavirus disease-19 and this study applied network pharmacology and bioinformatics analysis to explore the potential mechanism of colchicine against non-small cell lung cancer and coronavirus disease-19. The R software was used to determine the differentially expressed genes of non-small cell lung cancer/coronavirus disease-19, and carry out prognostic analysis and clinical analysis of the differentially expressed genes, the targets of colchicine were obtained from various databases, the protein-protein interaction network of intersection targets of colchicine and non-small cell lung cancer/coronavirus disease-19 was constructed, enrichment analysis of gene ontology and kyoto encyclopedia of genes and genomes pathways was performed by Metascape database and the molecular docking and molecular dynamics simulation were completed. We obtained a total of 716 differentially expressed genes and identified 13 potential prognostic genes associated with the clinical characterization of non-small cell lung cancer/coronavirus disease-19 patients. C-C motif chemokine ligand 2, toll like receptor 4, intercellular adhesion molecule 1, peroxisome proliferator activated receptor gamma, interleukin 17A, interferon gamma, angiotensin I converting enzyme, fos proto-oncogene, nuclear factor kappa B inhibitor alpha, TIMP metallopeptidase inhibitor 1 and secreted phosphoprotein 1 were core targets. The intersection targets of colchicine and non-small cell lung cancer/coronavirus disease-19 were mainly enriched in biological processes such as inflammatory response, response to cytokine and response to lipopolysaccharide, as well as signal pathways such as interleukin 17 signaling pathway, hypoxia inducible factor 1 signaling pathway and nucleotide binding oligomerization domain-like receptor signaling pathway. The results of molecular docking showed that the colchicine is better combined with the core targets. Analysis of molecular dynamics simulation showed that the protein and ligand form a stabilizing effect. Based on bioinformatics analysis and network pharmacology, we obtained biomarkers that may be used for the prognosis of non-small cell lung cancer/coronavirus disease-19 patients and revealed that colchicine may play a potential role in the treatment of non-small cell lung cancer/coronavirus disease-19 by regulating multiple targets and multiple signaling pathways and participating in multiple biological processes.
Keywords
Colchicine, coronavirus disease-19, non-small cell lung cancer, network pharmacology, bioinformatics analysis, molecular docking, molecular dynamics simulation
Currently, Coronavirus Disease 2019 (COVID-19), caused by Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2), is spreading globally with high morbidity and mortality[1], it is the biggest health problem perplexing the world nowadays. It has been reported that cancer patients are more susceptible to SARS-CoV-2 infection than the general population[2], lung cancer is the third most common cancer in the world and the leading cause of cancer death worldwide[3], Non-Small Cell Lung Cancer (NSCLC) is the most common pathological type of lung cancer, accounting for 85 % of all lung cancers[4], with high morbidity and mortality[5]. Unfortunately, due to immunosuppression, increased comorbidities and underlying lung damage, lung cancer patients have a higher mortality rate than normal individuals infected with SARS-CoV-2[6,7] and they have more severe complications[8]. COVID-19 is still spreading rapidly around the world and the number of NSCLC patients is also increasing, the treatment of NSCLC/COVID-19 presents enormous challenges for researchers, so there is an urgent need to develop potential therapeutic drugs for NSCLC/COVID-19. Colchicine is an alkaloid isolated from Colchicum autumnale plants[9], it is a Food and Drug Administration (FDA)-approved treatment for inflammatory diseases such as gout and familial Mediterranean fever[10,11] and has anti-inflammatory, anti-fibrotic, immunomodulatory, and anti-tumor effects, etc. A clinical study by Deftereos showed that compared with patients receiving a placebo, patients who received colchicine gave up oxygen supplementation earlier and after 1 w of treatment, significantly reduced C-Reactive Protein (CRP) levels[12]. A retrospective cohort study showed that colchicine administration can reduce the mortality of COVID-19 patients and contribute to the improvement of patients’ clinical symptoms[13]. Another study reported that patients treated with colchicine were approximately 5 times more likely to be discharged from the hospital and had a lower mortality rate compared with the standard group[14]. The timing and dose of colchicine are still in the exploratory stage, Pourdowlat’s study showed[15] that colchicine could improve the clinical outcome of COVID-19 patients and reduce pulmonary infiltration under the condition of reasonable selection of the time and dose of therapeutic intervention. In addition, the latest meta-analysis showed[16] that colchicine treatment reduced CRP levels and COVID-19 severity in patients. These studies appear to reveal colchicine as a potential treatment for COVID-19. In addition, colchicine is a well-known anticancer compound, this may be primarily due to its ability to inhibit mitosis by inhibiting microtubule formation, leading to cell cycle arrest, apoptosis and cell death[17]; however, abnormal cell proliferation and apoptosis are the most basic characteristics of tumor cells and inhibition of cancer cell proliferation and induction of cancer cell apoptosis are important anti-tumor mechanisms. Colchicine has shown anticancer effects in Hepatocellular Carcinoma (HCC), cholangiocarcinoma, cervical cancer, gastric cancer, breast cancer[18-21], etc. The anti-proliferative effect of colchicine on HCC cells is related to the expression of anti-proliferative genes A-Kinase Anchoring Protein 12 (AKAP12) and Transforming Growth Factor Beta 2 (TGFB2)[22], clinically acceptable concentrations of colchicine have anticancer effects on HCC. Zhang’s experimental study showed that colchicine induced apoptosis of gastric cancer cells by activating the caspase-3-mediated mitochondrial apoptosis pathway[23]. In a cohort study[24], a reduction in all-cause cancer risk in gout patients was associated with colchicine use (p=0.001). In addition, Bhattacharya, et al.[25] found that colchicine can induce autophagy and senescence in lung cancer cells and combined with autophagy inhibitors can become an effective treatment strategy for lung cancer. However, there is currently no research on the mechanism of colchicine against COVID-19 and NSCLC and further exploration is needed.
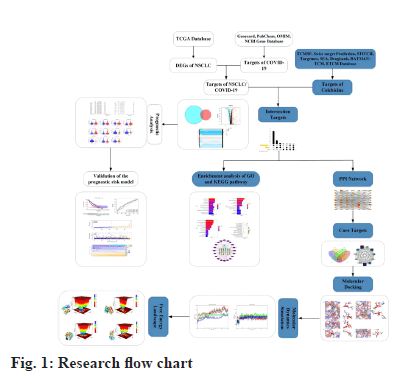
Network pharmacology is an emerging discipline based on the theory of systems biology, which is widely used to predict and analyze the molecular mechanism of drug action and identify potential drug targets[26]. Bioinformatics analysis is a valuable strategy for comprehensive analysis of large databases including complex genetic information[27] and is widely used to identify interactions between gene signatures and tumors[28]. Molecular docking is commonly used to study protein and ligand interaction modes and to calculate affinities, and molecular dynamics simulates various ligand and receptor motions through Newtonian mechanics to assess stability and flexibility[29,30]. In this study, we used network pharmacology, bioinformatics analysis, molecular docking and Molecular Dynamics Simulation (MDS) to help more systematically predict the potential mechanism and targets of colchicine in anti-COVID-19 and NSCLC at the molecular level. The flow chart of this study was shown in fig. 1.
Materials and Methods
Acquisition of disease-related genes:
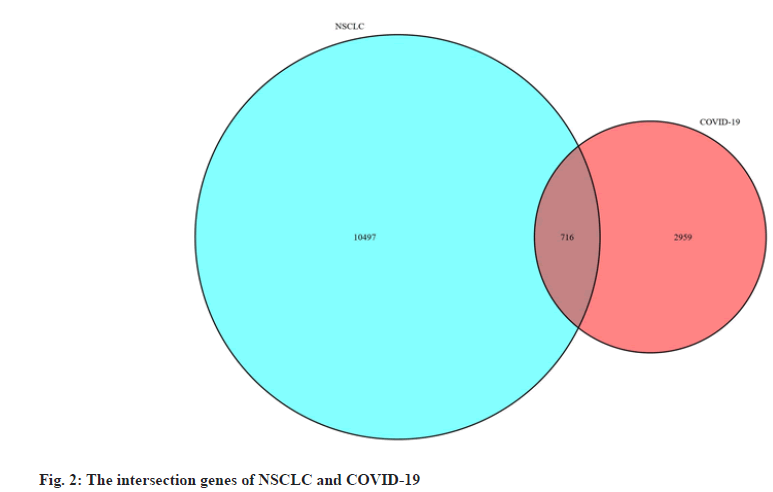
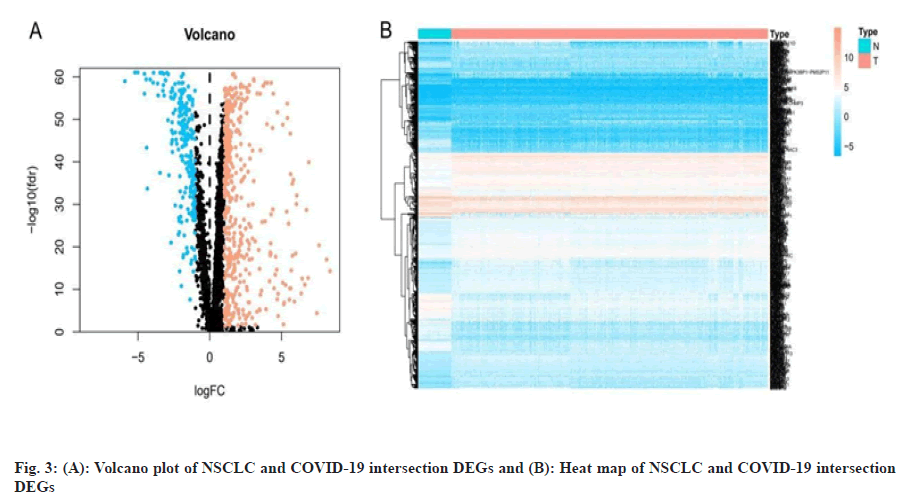
We obtained data of NSCLC from The Cancer Genome Atlas (TCGA) data portal (https://tcga-data.nci.nih.gov/tcga/), downloaded the Ribonucleic Acid (RNA) sequencing data and clinical data of Lung Adenocarcinoma (LUAD) and Lung Squamous Cell Carcinoma (LUSC) patient. Differentially Expressed Genes (DEGs) of NSCLC were obtained by screening with the “limma” package of the R 3.6.3 software (https://www.r-project.org/), the threshold was False Discovery Rate (FDR) <0.05 and |logFold Change (FC)|>1. We obtained targets of COVID-19 from Genecards database[31] (https://www.genecards.org/), PubChem database[32] (https://pubchem.ncbi.nlm.nih.gov/), Online Mendelian Inheritance in Man (OMIM) database[33] (https://omim.org/) and National Center for Biotechnology Information (NCBI) Gene database (https://www.ncbi.nlm.nih.gov/) and then we listed the targets in the UniProt database[34] (http://www. uniprot.org/) incorporates the “Human” search term and normalizes the naming. Finally, we used the “VennDiagram” package to construct a Venn diagram to obtain the intersection genes of NSCLC and COVID-19 and used the “pheatmap” package to construct the volcano and heat map of the intersection DEGs.
Construction of prognostic models and clinical analysis:
We used the “survival” package of R software to perform NSCLC and COVID-19 intersection genes univariate and multivariate cox regression analysis and draw forest plots and then used multivariate cox regression analysis to combine risk scores with different clinical characteristics of patients to perform related prognostic analysis, with the “beeswarm” package for this process.
Validation of the prognostic risk model:
We performed survival analysis and risk analysis based on the results of multivariate cox regression analysis, divided into high-risk and low-risk groups according to the average risk score and further determined the relationship between survival probability, survival time, survival status and gene expression between the two groups, with R packages such as “survival”, “survminer”, “time Receiver Operating Characteristic (ROC)”, “pheatmap”, etc.
Acquisition of colchicine-related targets:
We obtained the detail information of colchicine from the Pubchem database and we collected candidate targets of colchicine from the Traditional Chinese Medicine Systems Pharmacology (TCMSP) database[35] (http:// lsp.nwu.edu.cn/tcmsp.php/), the Swiss target Prediction database[36] (http://www. swisstargetprediction.ch/), STITCH database[37] (http://stitch.embl.de/), Genecards database (https://www.genecards.org/), SEA database[38] (https://sea .bkslab.org/), Drugbank database[39] (https:// www.drugbank.ca/), BATMAN-TCM database[40] (http://bionet.ncpsb.org.cn/batman-tcm/) and ETCM database[41] (http://www.tcmip.cn/ETCM/). We used the Uniprot database to merge the “human” search to correct the candidate target protein to the official name and then deduplicate to obtain the candidate target of colchicine after merging. We used the TBtools[42], tool to map the candidate targets of colchicine and the intersection genes of NSCLC and COVID-19 onto the UpSet map and identified the intersection targets of drugs and diseases.
Construction of Protein-Protein Interaction (PPI) network and prediction of core targets:
In order to better understand the PPI between colchicine and NSCLC/COVID-19 intersection targets, we used the STRING database[43] (https://www.string-db. org/) to construct the PPI network of the intersection targets. We visualized it using Cytoscape 3.8.2[44] software and screened core targets with the cytohubba plugin’s “Maximal Clique Centrality (MCC)”, “Density of Maximum Neighborhood Component (DMNC)”, “Maximum Neighborhood Component (MNC)”, “Degree” algorithms and Venn diagrams[45].
Enrichment analysis of Gene Ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway:
We used the Metascape database[46] (https://metascape.org/gp/index.html) to perform enrichment analysis of GO function and KEGG pathway for the intersection targets of colchicine and NSCLC/COVID-19, with a threshold of enrichment of the number of set genes ≥3 and p<0.01. We used the bioinformatics online platform (http://www.bioinformatics.com.cn/) to complete the visualization of the significantly enriched results and used the Cytoscape 3.8.2 software to construct the chemical component-target-pathways interaction network.
Molecular docking:
We obtained the protein structure of core targets and reported potential anti-COVID-19 targets SARSCoV- 2 3CL hydrolase (Protein Data Bank (PDB) ID: 6LU7) and Angiotensin Converting Enzyme 2 (ACE2) (PDB ID: 1R42) from the PDB database (https://www.rcsb.org/)[47]. We used the Chem Three Dimensional (3D) module of Chem Office software to minimize the energy of ligand compound and save it in 2 mol format, imported the receptor protein into Pymol software to remove water and other ligands, used the Autodock tools v.1.5.6 software to add hydrogen to the receptor protein and ligand, select the twisted key of the ligand, saved it in PDBQT format and used Autodock Vina[48] to select the most suitable binding site to complete the molecular docking, and then used Pymol software to visualize the results of molecular docking.
MDS:
We used Gromacs 5.1.2 software, GROMOS96 43a1 force field and the Simple Point-Charge (SPC) water model to perform MDS. We obtained topology files of proteins and ligands, defined unit boxes, filled solvent water molecules, added ions to ensure charge neutral of the simulated system, heated the entire system for 50 000 steps and simulated temperatures from 0 K to 300 K, used the steepest descent method to minimize energy. We used the position constraint to perform the NVT and NPT ensemble equilibrium with a step size of 2 fs and a total time of 100 ps for the energy-minimized system, lifted the constraints after simulating the dynamic equilibrium and performed 10 ns MDS.
The results were visualized by QtGrace software. In order to understand the conformational changes and stability of protein-ligand complexes during MDS, we introduced the gmx trjconv and gmx sham programs of Gromacs and plotted Two Dimensional (2D) and Three Dimensional (3D) Free Energy Landscape (FEL) to analyze MDS trajectories.
Results and Discussion
We obtained the NSCLC, RNA sequencing data and clinical data from the TCGA data portal, including 594 LUAD-related gene expression files (54 normal samples and 535 tumor samples) and 515 cases, 551 LUSC-related files (49 normal samples and 502 tumor samples) and 501 cases. We used R software to obtain a total of 11 213 differentially expressed genes related to NSCLC, including 8383 up-regulated genes and 2830 down-regulated genes. By searching with Genecards database, PubChem database, OMIM database and NCBI Gene database, merging and deduplicating, we obtained a total of 3675 genes related to COVID-19. The two parts of genes were mapped to the Venn diagram, as shown in fig. 2 and a total of 716 intersection DEGs were obtained, including 469 up-regulated genes and 247 down-regulated genes and we draw the volcano plot and heat map of intersection DEGs, as shown in fig. 3.
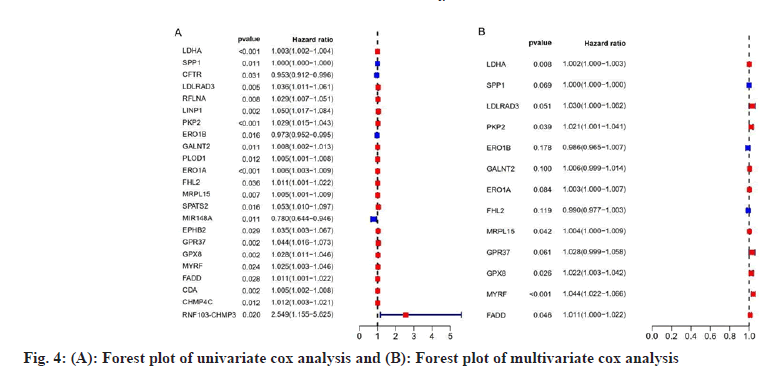
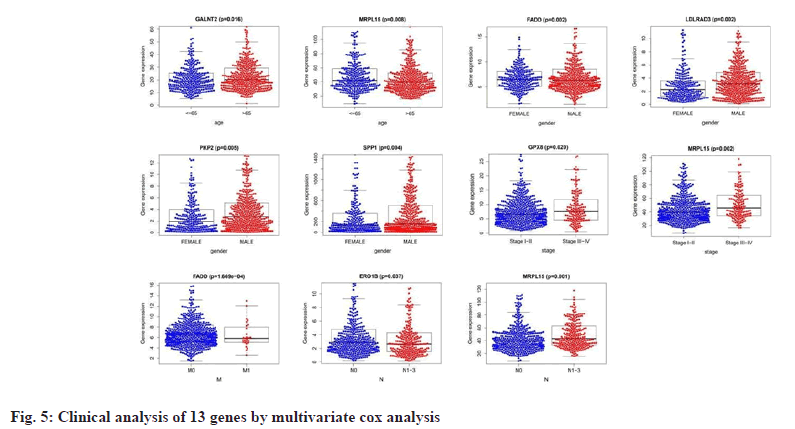
We performed a univariate Cox proportional Hazard Ratio (HR) regression analysis by the “survival” package based on information such as gene expression, survival time and survival status of NSCLC and COVID-19 intersecting genes, and identified 23 genes, as shown in fig. 4A, based on the KM value and p<0.05, 13 key genes were identified by multivariate Cox proportional HR regression analysis, such as Lactate Dehydrogenase A (LDHA), Secreted Phosphoprotein 1 (SPP1), Low Density Lipoprotein Receptor Class A Domain 3 (LDLRAD3), Plakophilin 2 (PKP2), Endoplasmic Reticulum Oxidoreductase 1 Beta (ERO1B), Polypeptide N-Acetylgalactosaminyltransferase 2 (GALNT2), ERO1A, Four and a Half LIM Domains 2 (FHL2), Mitochondrial Ribosomal Protein L15 (MRPL15), G Protein-Coupled Receptor 37 (GPR37), Glutathione Peroxidase 8 (GPX8), Myelin Regulatory Factor (MYRF) and Fas Associated via Death Domain (FADD), as shown in fig. 4B and listed in Table 1. Prognostic analysis of relevant clinical characteristics was performed based on the results of multivariate Cox proportional HR regression analysis and clinical information, as shown in fig. 5 and listed in Table 2. The expression of GALNT2 and MRPL15 was significantly correlated with age, the expression of FADD, LDLRAD3, PKP2, SPP1 was significantly correlated with gender, the expression of GPX8 and MRPL15 was significantly correlated with the pathological stage of NUSCLC and the expression of FADD was significantly correlated with M stage, ERO1B and MRPL15 were significantly associated with N stage.
| Symbol | Confidence interval | HR | HR.95 l | HR.95 H | p value |
|---|---|---|---|---|---|
| LDHA | 0.00175 | 1.00175 | 1.00046 | 1.00305 | 0.00807 |
| SPP1 | 0.00018 | 1.00018 | 0.99999 | 1.00037 | 0.0693 |
| LDLRAD3 | 0.02998 | 1.03043 | 0.9999 | 1.0619 | 0.05076 |
| PKP2 | 0.02061 | 1.02082 | 1.00103 | 1.04101 | 0.03915 |
| ERO1B | -0.0143 | 0.98578 | 0.96544 | 1.00655 | 0.17811 |
| GALNT2 | 0.00626 | 1.00628 | 0.99881 | 1.01382 | 0.0997 |
| ERO1A | 0.00341 | 1.00341 | 0.99955 | 1.0073 | 0.08357 |
| FHL2 | -0.0104 | 0.98963 | 0.97675 | 1.00269 | 0.11918 |
| MRPL15 | 0.00445 | 1.00446 | 1.00016 | 1.00878 | 0.04217 |
| GPR37 | 0.02768 | 1.02807 | 0.9987 | 1.0583 | 0.06118 |
| GPX8 | 0.02198 | 1.02223 | 1.00257 | 1.04227 | 0.02649 |
| MYRF | 0.04319 | 1.04414 | 1.02239 | 1.06634 | 5.8E-05 |
| FADD | 0.01078 | 1.01084 | 1.00018 | 1.02161 | 0.04628 |
Table 1: P-Value, Risk Factor HR and Confidence Interval of Multivariate COX Analysis
| Symbol | Age | Gender | Stage | T | M | N |
|---|---|---|---|---|---|---|
| SPP1 | -0.727 (0.468) | -2.917 (0.004) | -0.48 (0.632) | -0.746 (0.457) | 0.627 (0.536) | 0.368 (0.713) |
| LDLRAD3 | -0.266 (0.790) | -3.044 (0.002) | -0.211 (0.833) | -1.634 (0.104) | 0.191 (0.850) | -0.609 (0.543) |
| PKP2 | -0.827 (0.409) | -2.829 (0.005) | -1.933 (0.055) | -0.578 (0.564) | -0.658 (0.516) | -1.554 (0.121) |
| ERO1B | -0.224 (0.822) | 0.063 (0.950) | 0.714 (0.476) | -0.177 (0.860) | 0.483 (0.632) | 2.087 (0.037) |
| GALNT2 | -2.417 (0.016) | -1.668 (0.096) | -1.249 (0.213) | -1.617 (0.108) | -0.501 (0.621) | -0.723 (0.470) |
| ERO1A | -1.047 (0.296) | -1.905 (0.057) | -1.799 (0.073) | -1.34 (0.182) | -0.192 (0.849) | -1.266 (0.206) |
| FHL2 | 0.083 (0.934) | 1.293 (0.197) | -1.671 (0.096) | -1.148 (0.253) | -0.855 (0.400) | 0.229 (0.819) |
| MRPL15 | 2.655 (0.008) | -1.455 (0.146) | -3.119 (0.002) | -0.84 (0.402) | -1.165 (0.254) | -3.242 (0.001) |
| GPR37 | 0.34 (0.734) | 0.764 (0.445) | -1.236 (0.218) | 0.437 (0.663) | -0.777 (0.444) | -1.035 (0.301) |
| GPX8 | -0.616 (0.538) | 1.505 (0.133) | -2.348 (0.020) | -1.594 (0.113) | -1.136 (0.266) | -1.733 (0.084) |
| MYRF | -1.473 (0.141) | 1.631 (0.104) | -0.244 (0.808) | -0.158 (0.875) | -1.474 (0.151) | 1.067 (0.287) |
| FADD | -0.819 (0.413) | -3.176 (0.002) | 0.297 (0.767) | -0.973 (0.332) | 3.972 (1.669e-04) | -1.368 (0.172) |
| RiskScore | 0.664 (0.507) | -1.805 (0.072) | -1.765 (0.080) | -1.441 (0.152) | -0.754 (0.457) | -1.904 (0.058) |
Table 2: Clinical Analysis of Different Characteristics of 13 Genes
Based on risk scores, we divided patients into high-risk and low-risk groups. Survival analysis was performed on the 13 genes of the multivariate Cox proportional HR regression analysis and the results showed that there was a significant difference between the survival probability of the low-risk group and the high-risk group, as shown in fig. 6A, time-dependent ROC curve of the survival probability risk score for 1 y, 3 y, 5 y Area Under the Curve (AUC) were 0.677, 0.645 and 0.607, respectively, the model has good accuracy, as shown in fig. 6B. We constructed a heat map of the risk score distribution, survival status and gene expression for 13 genes, showing the correlation between the two groups, as shown in fig. 7.
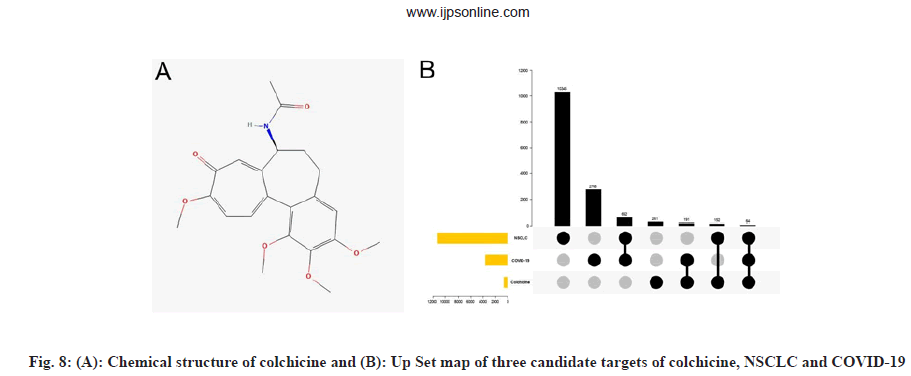
We obtained the chemical structure of colchicine, as shown in fig. 8A and comprehensively searched and screened the candidate targets of colchicine from TCMSP, Swiss target prediction, STITCH, Targetnet, SEA, Drugbank, Batman and ETCM database. After data correction using the Uniprot database, a total of 668 candidate targets of colchicine were obtained after merging and deduplication. The NSCLC and COVID-19 intersection genes and colchicine targets obtained from 2.1 were mapped onto the UpSet graph, as shown in fig. 8B and 64 intersection targets of colchicine and NSCLC/COVID-19 were obtained.
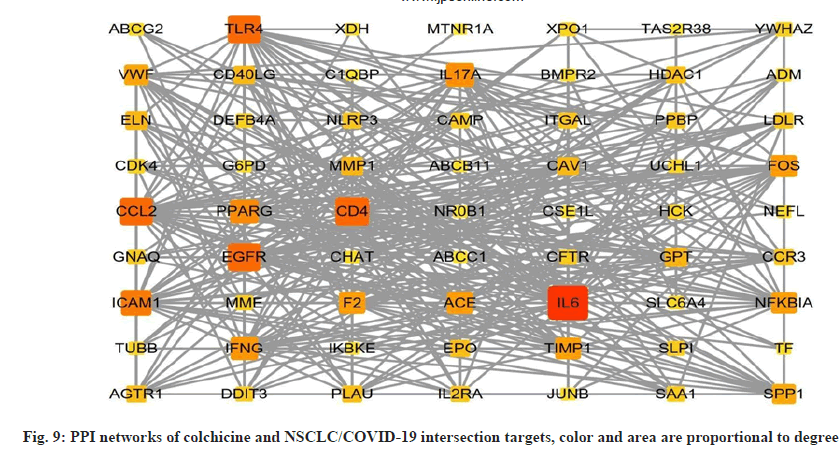
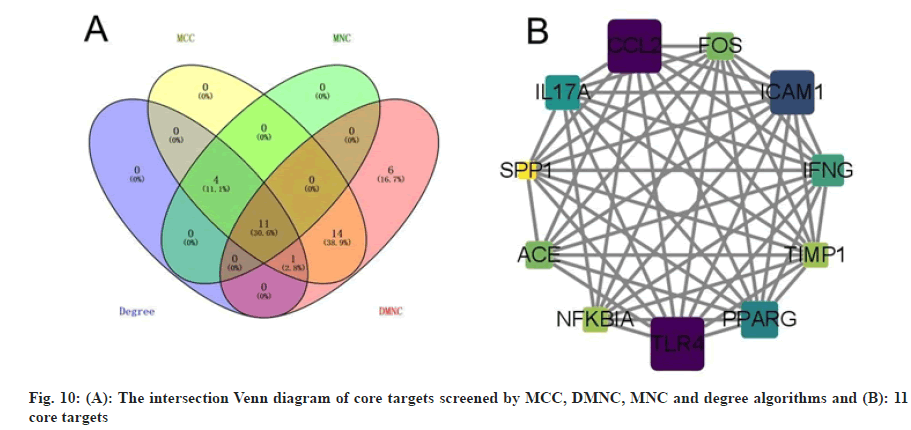
In order to better understand the protein-protein interactions of the cross-targets, we used the STRING database and Cytoscape 3.8.2 software to construct the PPI network of the cross-targets of colchicine and NSCLC/COVID-19, as shown in fig. 9, with a total of 63 nodes and 381 edges. The four algorithms of the cytohubba plugin were used to screen the core target genes. The screening conditions were that the MCC value was greater than or equal to twice the median of 10 368, the DMNC value was greater than or equal to the median of 0.548813253832958, the MNC value was greater than or equal to twice the median of 18 and the degree value was greater than or equal to twice the median of 18 the candidate targets screened by the 4 algorithms were mapped to the Venn diagram and 11 core targets were identified, as shown in fig. 10, such as C-C Motif Chemokine Ligand 2 (CCL2), Toll Like Receptor 4 (TLR4), Intercellular Adhesion Molecule 1 (ICAM1), Peroxisome Proliferator Activated Receptor Gamma (PPARG), Interleukin 17A (IL17A), Interferon Gamma (IFNG), ACE, Fos Proto-Oncogene (FOS), Nuclear Factor-Kappa-B-Inhibitor Alpha (NFKBIA), TIMP Metallopeptidase Inhibitor 1 (TIMP1), SPP1 were listed in Table 3.
| Rank | Uniprot | Symbol | Degree | MCC | DMNC | MNC | logFC | FDR | Regulation |
|---|---|---|---|---|---|---|---|---|---|
| 1 | P13500 | CCL2 | 33 | 1.80E+08 | 0.626504512 | 33 | -1.851890715 | 2.89E-18 | Down |
| 2 | O00206 | TLR4 | 33 | 1.80E+08 | 0.574077356 | 33 | -1.642107552 | 7.98E-52 | Down |
| 3 | P05362 | ICAM1 | 29 | 1.80E+08 | 0.675917591 | 29 | -1.434852987 | 1.30E-28 | Down |
| 4 | P37231 | PPARG | 26 | 1.67E+08 | 0.573981856 | 26 | -1.624728173 | 8.80E-49 | Down |
| 5 | Q16552 | IL17A | 25 | 1.28E+08 | 0.659783784 | 25 | 1.581943936 | 5.95E-05 | Up |
| 6 | P01579 | IFNG | 24 | 1.76E+08 | 0.774763828 | 24 | 1.078131793 | 0.039079099 | Up |
| 7 | P12821 | ACE | 22 | 5.20E+07 | 0.699819824 | 22 | -1.958230062 | 1.06E-56 | Down |
| 8 | P01100 | FOS | 22 | 1.31E+08 | 0.655669811 | 21 | -1.876376017 | 2.37E-32 | Down |
| 9 | P25963 | NFKBIA | 21 | 8.39E+07 | 0.604798878 | 21 | -1.0864282 | 2.67E-29 | Down |
| 10 | P01033 | TIMP1 | 21 | 1.79E+08 | 0.802630286 | 21 | 1.026642657 | 7.77E-21 | Up |
| 11 | P10451 | SPP1 | 18 | 9.19E+07 | 0.866799693 | 18 | 4.715613019 | 2.13E-52 | Up |
Table 3: The Detailed Information of 11 Core Targets
We used the Metascape database to perform the enrichment analysis of GO function and KEGG pathway for the intersection targets of colchicine and NSCLC/COVID-19, as shown in fig. 11, the number of enriched genes ≥3 and p<0.01, a total of 289 enriched GO entries were obtained. In terms of biological processes, the intersection targets are mainly enriched in the inflammatory response, response to bacterium, response to cytokine, response to lipopolysaccharide and response to molecule of bacterial origin, etc. In terms of cellular components, the intersection targets are mainly enriched in side of membrane, external side of plasma membrane, membrane raft, membrane microdomain, receptor complex, etc. In terms of molecular functions, the intersection targets are mainly enriched in signaling receptor activator activity, signaling receptor regulator activity, receptor ligand activity, G protein-coupled receptor binding, cytokine activity, etc.
Fig. 11:GO functional enrichment and KEGG pathway enrichment analysis of the intersection targets of colchicine and NSCLC/ COVID-19, (A): GO functional enrichment of intersection targets in TOP10 biological processes; (B): GO functional enrichment of intersection targets in TOP10 cellular components; (C) GO functional enrichment of intersection targets in TOP10 molecular functions and (D): KEGG pathway enrichment of intersection targets
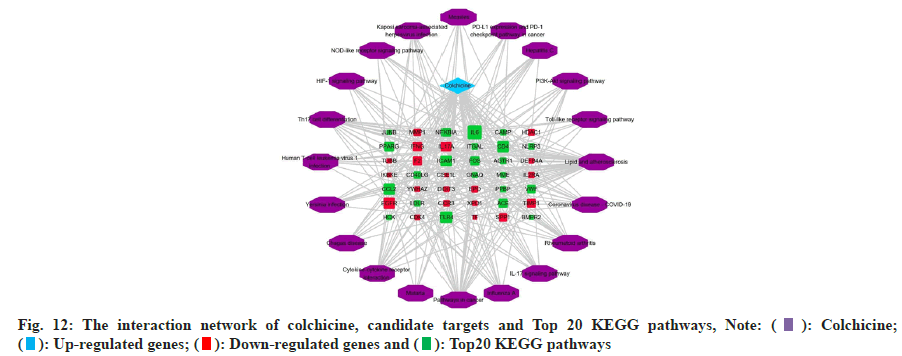
The KEGG pathway enrichment analysis of intersection targets are enriched in 98 pathways, sorted by p-value, intersection targets are mainly enriched in lipid and atherosclerosis, COVID-19, rheumatoid arthritis, IL- 17 signaling pathway, influenza A, pathways in cancer, malaria, Cytokine-cytokine receptor interaction, Chagas disease, Yersinia infection, human T-cell leukemia virus 1 infection, Th17 cell differentiation, Hypoxia Inducible Factor 1 (HIF-1) signaling pathway, Nucleotide Binding Oligomerization Domain (NOD)- like receptor signaling pathway, etc. In order to understand the interaction between colchicine, KEGG pathways and candidate targets, we used Cytoscape 3.8.2 software to construct the interaction network about colchicine, candidate targets and KEGG pathways, as shown in fig. 12.
We used Autodock Tools v.1.5.6 and AutoDock Vina software to conduct molecular docking of colchicine with the core targets and screened the conformation with the highest binding energy for analysis. The binding energy is less than 0, which means that the protein and the ligand can bind in the natural state. The lower the docking score, the higher the binding energy, and more stable the binding between the protein and ligand. The binding energies of colchicine to core targets CCL2, TLR4, ICAM1, PPARG, IL17A, IFNG, ACE, FOS, NFKBIA, TIMP1 and SPP1 were all less than -6 kcal/mol and the binding activity is good, as listed in Table 4.
| No. | Target name | Energy (kcal/mol) | Compound |
|---|---|---|---|
| 1 | CCL2 | -6.6 | |
| 2 | TLR4 | -7.4 | |
| 3 | ICAM1 | -6.8 | |
| 4 | PPARG | -8.2 | |
| 5 | IL17A | -6 | |
| 6 | IFNG | -7.1 | |
| 7 | ACE | -7.4 | Colchicine |
| 8 | FOS | -6.5 | |
| 9 | NFKBIA | -6.9 | |
| 10 | TIMP1 | -7.6 | |
| 11 | SPP1 | -6.4 | |
| SARS-CoV-2 | 3CL | -6.4 | |
| ACE2 | -7.5 |
Table 4: Molecular Docking of Colchicine with Core Target Proteins and Potential Anti-Covid-19 Targets
We plotted the docking pattern of colchicine with binding energy less than -7 kcal/mol to PPARG, TIMP1, TLR4, ACE, IFNG as shown in fig. 13A. At the same time, we obtained the reported potential anti-COVID-19 targets SARS-CoV-2 3CL hydrolase and ACE2 and then molecularly docked them with colchicine. The highest binding energy of them was also less than -6 kcal/mol and the binding activity were good, as listed in Table 4. Colchicine binds to SARS-CoV-2 3CL hydrolase, forms hydrogen-bonding interactions with residues THR-199 and MET-276, binds to ACE2, forms hydrogen-bonding interactions with residues GLN-442, GLU-406, THR-371, as shown in fig. 13B.
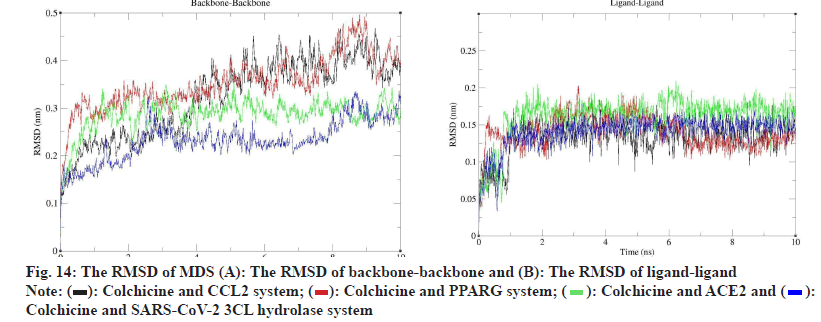
To evaluate the molecular docking results, we performed MDS of 4 complexes of colchicine with CCL2, PPARG, SARS-CoV-2 3CL hydrolase and ACE2 and obtained the square Root of Mean Square Deviation (RMSD) of the protein backbone and ligands of the MDS complexes, as shown in fig. 14, in terms of the RMSD of protein backbone-protein backbone, the mean±Standard Deviation (SD) of colchicine and CCL2 was 0.322±0.084 (nm), the mean±SD of colchicine and PPARG was 0.353±0.056 (nm), the mean±SD of ACE2 was 0.286±0.034 (nm), and the mean±SD of SARS-CoV-2 3CL hydrolase was 0.232±0.042 (nm).
In terms of ligand-ligand RMSD, the mean±SD of colchicine and CCL2 was 0.133±0.023 (nm), the mean±SD of colchicine and PPARG was 0.140±0.022 (nm), the mean±SD of ACE2 was 0.141±0.021 (nm) and the mean±SD of SARS-CoV-2 3CL hydrolase was 0.141±0.021 (nm), according to the RMSD results, both the protein and the ligand could form stable interactions during the 10 ns MDS.
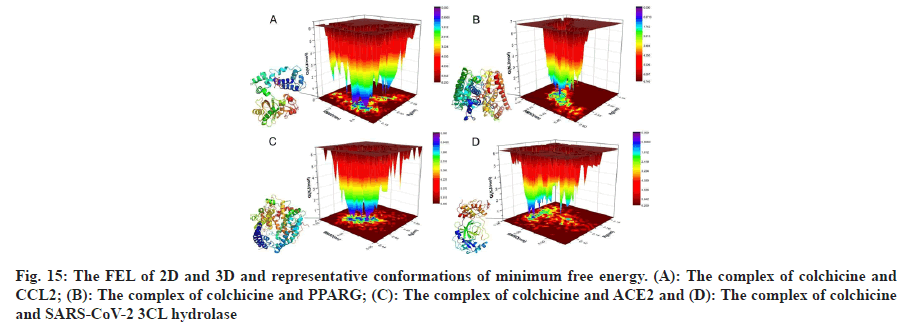
According to the analysis of MDS results, we learned that the MDS of 4 complexes of colchicine with CCL2, PPARG, SARS-CoV-2 3CL hydrolase and ACE2 gradually became stable after 3 ns, therefore, we extracted the MDS trajectory of the 3-10 ns proteinligand complexes of the 4 systems, extracted the RMSD and radius of gyration of the protein, obtained the free energy of the conformation according to these two variables and plotted the FEL of 2D and 3D, as shown in fig. 15, to learn about free energy changes and stability of complex and protein conformations MDS, according to the FEL calculation, deep purple energy wells appeared in the FEL of the 4 complexes, indicating that the 4 complexes are stable, we extracted representative conformations of the 4 complexes with the smallest free energy and the most stable conformations for visualization, as shown in fig. 15.
At present, the devastating impact of the COVID-19 pandemic caused by SARS-CoV-2 is unprecedented[49,50] and the development of the disease is receiving great attention from the global community. Inflammation is linked to the progression of COVID-19 disease, systemic inflammation is a hallmark of moderate-tosevere COVID-19 disease[51], which leads to an excessive inflammatory response of the innate and adaptive immune systems, termed Cytokine Release Syndrome (CRS)[52]. Inflammation is also an important part of the tumor environment and plays an important role in the development and progression of malignant tumors[53]. Cancer patients, as one of the most susceptible populations, are facing severe disadvantages during the COVID-19 epidemic, lung cancer patients, in particular[54], have been reported to be more susceptible to SARS-Cov-2 infection and they are at higher risk of developing pneumonia and severe complications of COVID-19 on admission[55]. ACE2 is a functional receptor for SARS-CoV-2 to enter and replicate in host cells[56], and its expression level is thought to indicate susceptibility to COVID-19[57], the expression level of ACE2 in lung tissue is increased[58,59] and according to the analysis of a study, it is found that the messenger RNA (mRNA) expression of ACE2 is higher in lung and colorectal cancer than other types of cancer[60], which may be another explanation for lung cancer patients being more susceptible to SARS-CoV-2 infection and at higher risk of severe COVID-19. Vaccination is effective in controlling COVID-19 outbreaks, but studies have reported that vaccination in lung cancer patients is less effective than in healthy people[61,62]. In conclusion, the link between lung cancer and COVID-19 has been shown, at present, the number of patients with COVID-19 is still increasing rapidly nowadays, how to conduct research on the mechanism of association between the two diseases and the treatment of NSCLC patients infected with SARSCoV- 2 will be a major challenge now and in the future. The clinical studies have shown[63,64] that colchicine has achieved promising results in the treatment of COVID-19, possibly related to its antiviral and antiinflammatory properties[65,66] and according to our analysis of the references cited above, colchicine has significant anti-cancer effects on a variety of cancer cells and can be used to develop potential anticancer chemotherapeutic drugs[67], so we hypothesized that colchicine might be a potential treatment for NSCLC/ COVID-19 patients. This study applied bioinformatics analysis, network pharmacology, molecular docking and MDS combined analysis to predict the potential mechanism of action of colchicine against NSCLC/ COVID-19. The identification of potential biomarkers plays an important role in the prognosis of NSCLC/ COVID-19 patients and drug intervention. Using bioinformatics analysis in this study, we identified 716 DEGs related to NSCLC/COVID-19, including 469 upregulated genes and 247 down-regulated genes, the DEGs are associated with the clinical characterization of NSCLC/COVID-19 patients and may be potential therapeutic targets for NSCLC/COVID-19, we performed multivariate Cox proportional HR regression analysis on this part of genes, LDHA, SPP1, LDLRAD3, PKP2, ERO1B, GALNT2, ERO1A, FHL2, MRPL15, GPR37, GPX8, MYRF and FADD were identified as key genes, evaluated the correlation between risk characteristics and clinical characteristics such as age, gender, pathological stage and tumor stage of NSCLC/ COVID-19 patients and combined with the validation results, we speculate that these 13 key genes may be associated with the development of NSCLC/COVID-19 patients and may serve as diagnostic and prognostic biomarkers. Using network pharmacology, we obtained 64 intersection targets of colchicine and NSCLC/ COVID-19. By analyzing the PPI network of intersection targets, we identified 11 core targets such as CCL2, TLR4, ICAM1, PPARG, IL17A, IFNG, ACE, FOS, NFKBIA, TIMP1 and SPP1. Among them, TLR4 plays an important role in pathogen recognition and innate immune activation, is closely related to the inflammatory response and is a key mediator during SARS-CoV-2 infection[68,69], according to relevant clinical reports[70], TLR4 enhances the activation of the NF-κB cascade signaling pathway and downstream genes and it is involved in the hyperinflammatory response of COVID-19 patients. Among the proinflammatory cytokines closely related to COVID-19, both IL-6 and TNF-α are located downstream of the TLR4 signaling pathway[71,72] and they are closely related to the production of the cytokine storm. Other proinflammatory cytokines like IL-1β, IL- 18, Interferon gamma (IFN-γ) and chemokines like CXCL-9, CXCL-10, CCL-2[73] were detected at higher levels in COVID-19 patients[74], these inflammatory factors migrate to other organs and tissues, damage the lungs and other organs and cause systemic inflammation. In addition, high levels of SPP1 increase in severe COVID-19 patients, the study speculates that SPP1 may be an upstream activator of abnormal innate responses to severe COVID-19 and may predict the pathological trajectory of COVID-19[75], ICAM1 and NFKBIA are key genes associated with COVID-19[76]. The ACE2 receptor is a homolog of the ACE receptor[77], which together balances the Renin-Angiotensin (RAS) system. Since the SARS-CoV-2 virus reduces the availability of ACE2 through ACE2 binding and entry, researchers believe that the RAS system and the imbalance of ACE1 and ACE2 activity (enhanced Ang II) play an important role in the stage of COVID-19 infection[78,79]. In studies related to NSCLC, we learned that TLR4 is a functional receptor for resistin migration and invasion in LUAD cells, promoting LUAD metastasis through TLR4/NF-κB related pathway[80,81], TLR4 activation also protects the lung from inflammation during any underlying tumorigenesis[82]. Chemokine CCL2 can affect the development of lung cancer cell metastasis[83], its knockdown or autophagy induction can successfully reverse drug resistance in tumor cells[84], and may serve as potential predictors and therapeutic targets for survival in NSCLC patients[85]. IFN-γ often appears together with Programmed Death-Ligand 1 (PD-L1) in studies on the role of IFN-γ in lung cancer[86,87], IFN-γ plays an important role in cancer-related immunity[88], therapeutic strategies for Programmed Cell Death 1 (PD-1) or its ligand PD-L1 have been developed as immunotherapy against tumor progression in various cancer types including NSCLC[89], IFN-γ can induce the expression of PD-L1, interestingly, patients with tumors expressing both IFN-γ and PD-L1 had the best prognosis compared to either. TIMPs are tissue inhibitors of Matrix Metalloproteinases (MMPs) and TIMP1 has been the most studied, it mainly plays the role of promoting cell growth and anti-apoptosis in tumor tissue, TIMP1 is overexpressed in NSCLC tumor tissues and was later shown to have prognostic value in NSCLC[90,91]. SPP1 is an oncogene that is upregulated in NSCLC patients[92], involved in lung cancer proliferation, migration, invasion and cisplatin resistance, targeting SPP1 may be a potential lung cancer treatment strategy[93]. The activation of PPARG expression may be involved in inhibiting the development and progression of LUSC by regulating the upstream regulators and downstream marker genes of LUSC[94]. From the above, we speculate that these core targets may play a role in the potential treatment of NSCLC/COVID-19 patients by colchicine. In the GO functional enrichment results, the main enriched biological processes of colchicine and NSCLC/ COVID-19 intersection targets were related to inflammation, such as inflammatory response, response to cytokines, response to lipopolysaccharide, etc. The enrichment analysis of the KEGG pathway showed that 64 intersection targets were mainly enriched in the IL- 17 signaling pathway (DEFB4A, FOS, IFNG, IL6, IL17A, MMP1, NFKBIA, CCL2, IKBKE), HIF-1 signaling pathway (EGFR, EPO, IFNG, IL6, TF, TIMP1, TLR4), NOD-like receptor signaling pathway (CAMP, DEFB4A, IL6, NFKBIA, CCL2, TLR4, IKBKE, NLRP3) and so on. IL-17 is a pro-inflammatory cytokine group ligand, mainly produced by activated CD4+ T helper cells (Th17 cells)[95] and is involved in the pathological process of various diseases. It plays an important role in chronic inflammatory diseases, autoimmune diseases and various malignant tumors[96,97], it can up-regulate the production of Vascular Endothelial Growth Factor (VEGF) in cancer cells through the activation of the STAT3/GIV signaling pathway and promote tumor angiogenesis in NSCLC[98]. In addition, IL-17 is associated with cytokine storm[99], which occurs in severe COVID-19 patients, leading to Acute Respiratory Distress Syndrome (ARDS) and multiple organ failure[100,101], Mendoza believes[102] that IL-17 plays a key role in the immune deregulation observed in COVID-19 and other severe coronavirusmediated respiratory syndromes and modulation of IL 17 may be a potential treatment strategy for NSCLC/ COVID-19. Hypoxia is a common feature of both COVID-19 and lung cancer, and as a major regulator of oxygen homeostasis, HIF-1-related pathways may play an important role in promoting the development of lung cancer after infection with COVID-19[103], the hypoxia and inflammatory microenvironment of COVID-19 upregulates the expression of Hypoxia- Inducible Factor 1-alpha (HIF-1α) and converts cell metabolism to aerobic glycolysis, promoting the occurrence and metastasis of lung cancer cells[104,105], strengthening research on HIF-1 may have important implications for the treatment of COVID-19 and NSCLC. Another significantly enriched KEGG pathway is the NOD-like receptor signaling pathway, which is involved in the regulation of multiple biological roles such as host injury, immunity and inflammatory responses[106,107], NLRP3 is an important member of this pathway and is one of the most characteristic inflammasomes, studies have shown[108] that the NLRP3 inflammasome can activate the production of IL-1β and 18, the main components of the inflammatory response in the LUAD cell line A549, but its role in the pathogenesis of lung cancer needs further study. Based on current evidence, down regulation of the NLRP3 inflammasome may attenuate the inflammatory cascade induced by SARS-CoV-2 infection[109] and correlate with severity in COVID-19 patients[110,111], interestingly, multiple studies have shown[112-114] that colchicine has an inhibitory effect on the NLRP3 inflammasome, blocking NLRP3, reducing the release of downstream cytokines like IL-1β and IL- 18 to reduce the inflammatory burden, this may be another explanation for the potential role of colchicine in COVID-19, but the specific mechanism is not completely clear and it still has great research value. Through molecular docking verification, we found that the binding energies of colchicine to 11 core targets were all less than -6 kcal/mol, which indicates that colchicine can effectively bind to these targets, among them, the binding energy of PPARG to colchicine is the highest among the core targets and the degree value of CCL2 is the highest among the core targets, so we predict that these two targets may play important roles in the anti-NSCLC/COVID-19 of colchicine, we chose the complexes of these two targets with colchicine for MDS, and calculated the RMSD and FEL, the results show that the two targets and colchicine can form stable effects and the two complexes are stable. Therefore, colchicine may exert its anti-NSCLC/COVID-19 effect by regulating core targets such as CCL2 and PPARG.
3CL hydrolase, the main protease of SARS-CoV-2, plays a key role in mediating viral replication and transcription[115], the experimental study has shown[116] that potent 3CL hydrolase inhibitors can significantly reduce SARS-CoV-2 viral titers, reduce body weight and improve survival in mice, inhibition of this ACE2 and 3CL hydrolase is a common strategy used to screen treatments for COVID-19[117,118], so we added the molecular docking of ACE2 and 3CL hydrolase with colchicine and the binding energies were -7.5 kcal/mol and -6.4 kcal/mol, respectively, combined with the results of MDS, colchicine can stably bind to these two targets against SARS-CoV-2, this result provides some theoretical evidence for colchicine against COVID-19. Combining computer network technology to predict the underlying mechanism of drug action disease can help improve the development efficiency of potential treatment methods. Although our study may provide a potential rationale for colchicine against NSCLC/ COVID-19, but further in vitro and in vivo experiments are still needed to verify the above findings in our study.
In this study, by using network pharmacology and bioinformatics analysis, we identified potential biomarkers associated with prognosis in NSCLC/ COVID-19 patients and we found that colchicine may against NSCLC/COVID-19 by regulating multiple targets and IL-17 signaling pathway, HIF-1 signaling pathway, NOD-like receptor signaling pathway and other signaling pathways. We hope to provide a reference for the development of colchicine and the treatment of NSCLC/COVID-19 patients.
Data availability statement:
Publicly available datasets were analyzed in this study. The RNA sequencing data and clinical data of TCGALUAD and TCGA-LUSC patients were obtained from the TCGA data portal (https://tcga-data.nci.nih.gov/tcga/). The targets of COVID-19 were obtained from Genecards database (https://www.genecards.org/), PubChem database (https://pubchem.ncbi.nlm.nih.gov/), OMIM database (https://omim.org/) and NCBI Gene database (https://www.ncbi.nlm.nih.gov/). The targets of Colchicine were obtained from the TCMSP database (http://lsp.nwu.edu.cn/tcmsp.php/), the Swiss target Prediction database (http://www.swisstargetprediction.ch/), STITCH database (http://stitch.embl.de/), Genecards database (https://www. genecards.org/), SEA database (https://sea.bkslab.org/), Drugbank database (https://www.drugbank.ca/), BATMAN-TCM database (http://bionet.ncpsb.org.cn/batman-tcm/) and ETCM database (http://www.tcmip.cn/ETCM/).
Author contributions:
Zhiyao Liu designed and wrote the manuscript. Ying Yu and Yuqi Jia collected the data. Lingling Li, Xin Shi and Fangqi Wang analyzed the data. Hailiang Huang reviewed the manuscript and made comments. All authors contributed to the article and approved the submitted version.
Conflict of interests:
The authors declared no conflict of interests.
References
- Zhou F, Yu T, Du R, Fan G, Liu Y, Liu Z, et al. Clinical course and risk factors for mortality of adult inpatients with COVID-19 in Wuhan, China: A retrospective cohort study. Lancet 2020;395(10229):1054-62.
- Vivarelli S, Falzone L, Grillo CM, Scandurra G, Torino F, Libra M. Cancer management during COVID-19 pandemic: Is immune checkpoint inhibitors-based immunotherapy harmful or beneficial? Cancers 2020;12(8):2237.
[Crossref] [Google Scholar] [PubMed]
- Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018;68(6):394-424.
[Crossref] [Google Scholar] [PubMed]
- Reck M, Heigener DF, Mok T, Soria JC, Rabe KF. Management of non-small-cell lung cancer: Recent developments. Lancet 2013;382(9893):709-19.
[Crossref] [Google Scholar] [PubMed]
- Teng Y, Wang B, Shang D, Yang N. Identification and validation of an immune and ferroptosis-combined index for non–small cell lung cancer. Front Genet 2021;12:764869.
[Crossref] [Google Scholar] [PubMed]
- Mehta V, Goel S, Kabarriti R, Cole D, Goldfinger M, Acuna-Villaorduna A, et al. Case fatality rate of cancer patients with COVID-19 in a New York hospital system. Cancer Discov 2020;10(7):935-41.
[Crossref] [Google Scholar] [PubMed]
- Rugge M, Zorzi M, Guzzinati S. SARS-CoV-2 infection in the Italian Veneto region: Adverse outcomes in patients with cancer. Nat Cancer 2020;1(8):784-8.
[Crossref] [Google Scholar] [PubMed]
- Luo J, Rizvi H, Preeshagul IR, Egger JV, Hoyos D, Bandlamudi C, et al. COVID-19 in patients with lung cancer. Ann Oncol 2020;31(10):1386-96.
[Crossref] [Google Scholar] [PubMed]
- Dasgeb B, Kornreich D, McGuinn K, Okon L, Brownell I, Sackett DL. Colchicine: An ancient drug with novel applications. Br J Dermatol 2018;178(2):350-6.
[Crossref] [Google Scholar] [PubMed]
- Nett RS, Lau W, Sattely ES. Discovery and engineering of colchicine alkaloid biosynthesis. Nature 2020;584(7819):148-53.
[Crossref] [Google Scholar] [PubMed]
- Slobodnick A, Shah B, Pillinger MH, Krasnokutsky S. Colchicine: Old and new. Am J Med 2015;128(5):461-70.
[Crossref] [Google Scholar] [PubMed]
- Deftereos SG, Giannopoulos G, Vrachatis DA, Siasos GD, Giotaki SG, Gargalianos P, et al. Effect of colchicine vs. standard care on cardiac and inflammatory biomarkers and clinical outcomes in patients hospitalized with coronavirus disease 2019: The GRECCO-19 randomized clinical trial. JAMA Netw Open 2020;3(6):e2013136.
[Crossref] [Google Scholar] [PubMed]
- Manenti L, Maggiore U, Fiaccadori E, Meschi T, Antoni AD, Nouvenne A, et al. Reduced mortality in COVID-19 patients treated with colchicine: Results from a retrospective, observational study. PLoS One 2021;16(3):e0248276.
[Crossref] [Google Scholar] [PubMed]
- Brunetti L, Diawara O, Tsai A, Firestein BL, Nahass RG, Poiani G, et al. Colchicine to weather the cytokine storm in hospitalized patients with COVID-19. J Clin Med 2020;9(9):2961.
[Crossref] [Google Scholar] [PubMed]
- Pourdowlat G, Saghafi F, Mozafari A, Sahebnasagh A, Abedini A, Nabi Meybodi M, et al. Efficacy and safety of colchicine treatment in patients with COVID-19: A prospective, multicenter, randomized clinical trial. Phytother Res 2022;36(2):891-8.
[Crossref] [Google Scholar] [PubMed]
- Yasmin F, Najeeb H, Moeed A, Hassan W, Khatri M, Asghar MS, et al. Safety and efficacy of colchicine in COVID-19 patients: A systematic review and meta-analysis of randomized control trials. PloS one 2022;17(4):e0266245.
[Crossref] [Google Scholar] [PubMed]
- Bhattacharyya B, Panda D, Gupta S, Banerjee M. Anti-mitotic activity of colchicine and the structural basis for its interaction with tubulin. Med Res Rev 2008;28(1):155-83.
[Crossref] [Google Scholar] [PubMed]
- Wu CC, Lin ZY, Kuoc CH, Chuang WL. Clinically acceptable colchicine concentrations have potential for the palliative treatment of human cholangiocarcinoma. Kaohsiung J Med Sci 2015;31(5):229-34.
[Crossref] [Google Scholar] [PubMed]
- Yan L, Huang H, Zhang Y, Yuan X, Yan Z, Cao C, et al. Involvement of p53-dependent apoptosis signal in antitumor effect of colchicine on human papilloma virus (HPV)-positive human cervical cancer cells. Biosci Rep 2020;40(3).
[Crossref] [Google Scholar] [PubMed]
- Lin ZY, Kuo CH, Wu DC, Chuang WL. Anticancer effects of clinically acceptable colchicine concentrations on human gastric cancer cell lines. Kaohsiung J Med Sci 2016;32(2):68-73.
[Crossref] [Google Scholar] [PubMed]
- Sun Y, Lin X, Chang H. Proliferation inhibition and apoptosis of breast cancer MCF-7 cells under the influence of colchicine. J. BUON 2016;21(3):570-5.
[Google Scholar] [PubMed]
- Lin ZY, Wu CC, Chuang YH, Chuang WL. Anti-cancer mechanisms of clinically acceptable colchicine concentrations on hepatocellular carcinoma. Life Sci 2013;93(8):323-8.
[Crossref] [Google Scholar] [PubMed]
- Zhang T, Chen W, Jiang X, Liu L, Wei K, Du H, et al. Anticancer effects and underlying mechanism of colchicine on human gastric cancer cell lines in vitro and in vivo. Biosci Rep 2019;39(1).
[Crossref] [Google Scholar] [PubMed]
- Kuo MC, Chang SJ, Hsieh MC. Colchicine significantly reduces incident cancer in gout male patients: A 12-year cohort study. Medicine 2015;94(50).
[Crossref] [Google Scholar] [PubMed]
- Bhattacharya S, Das A, Datta S, Ganguli A, Chakrabarti G. Colchicine induces autophagy and senescence in lung cancer cells at clinically admissible concentration: Potential use of colchicine in combination with autophagy inhibitor in cancer therapy. Tumour Biol 2016;37(8):10653-64.
[Crossref] [Google Scholar] [PubMed]
- Hopkins AL. Network pharmacology: The next paradigm in drug discovery. Nat Chem Biol 2008;4(11):682-90.
[Crossref] [Google Scholar] [PubMed]
- Zeng X, Shi G, He Q, Zhu P. Screening and predicted value of potential biomarkers for breast cancer using bioinformatics analysis. Sci Rep 2021;11(1):1-11.
[Crossref] [Google Scholar] [PubMed]
- Qiu T, Wang H, Wang Y, Zhang Y, Hui Q, Tao K. Identification of genes associated with melanoma metastasis. Kaohsiung J Med Sci 2015;31(11):553-61.
[Crossref] [Google Scholar] [PubMed]
- Zhang X, Wang M, Qiao Y, Shan Z, Yang M, Li G, et al. Exploring the mechanisms of action of Cordyceps sinensis for the treatment of depression using network pharmacology and molecular docking. Ann Transl Med 2022;10(6).
[Crossref] [Google Scholar] [PubMed]
- Ye J, Li L, Hu Z. Exploring the molecular mechanism of action of Yinchen Wuling powder for the treatment of hyperlipidemia, using network pharmacology, molecular docking and molecular dynamics simulation. Biomed Res Int 2021;2021.
[Crossref] [Google Scholar] [PubMed]
- Safran M, Chalifa-Caspi V, Shmueli O, Olender T, Lapidot M, Rosen N, et al. Human gene-centric databases at the Weizmann Institute of Science: GeneCards, UDB, CroW 21 and HORDE. Nucleic Acids Res 2003;31(1):142-6.
[Crossref] [Google Scholar] [PubMed]
- Kim S, Chen J, Cheng T, Gindulyte A, He J, He S, et al. PubChem 2019 update: Improved access to chemical data. Nucleic Acids Res 2019;47(D1):D1102-9.
[Crossref] [Google Scholar] [PubMed]
- Amberger JS, Bocchini CA, Schiettecatte F, Scott AF, Hamosh A. OMIM. Org: Online Mendelian Inheritance in Man (OMIM®), an online catalog of human genes and genetic disorders. Nucleic Acids Res 2015;43(D1):D789-98.
[Crossref] [Google Scholar] [PubMed]
- UniProt Consortium. UniProt: A worldwide hub of protein knowledge. Nucleic Acids Res 2019;47(D1):D506-15.
[Crossref] [Google Scholar] [PubMed]
- Ru J, Li P, Wang J, Zhou W, Li B, Huang C, et al. TCMSP: A database of systems pharmacology for drug discovery from herbal medicines. J Cheminform 2014;6(1):1-6.
[Crossref] [Google Scholar] [PubMed]
- Daina A, Michielin O, Zoete V. SwissTargetPrediction: Updated data and new features for efficient prediction of protein targets of small molecules. Nucleic Acids Res 2019;47(W1):W357-64.
[Crossref] [Google Scholar] [PubMed]
- Szklarczyk D, Santos A, von Mering C, Jensen LJ, Bork P, Kuhn M. STITCH 5: Augmenting protein–chemical interaction networks with tissue and affinity data. Nucleic Acids Res 2016;44(D1):D380-4.
[Crossref] [Google Scholar] [PubMed]
- Keiser MJ, Roth BL, Armbruster BN, Ernsberger P, Irwin JJ, Shoichet BK. Relating protein pharmacology by ligand chemistry. Nat Biotechnol 2007;25(2):197-206.
[Crossref] [Google Scholar] [PubMed]
- Wishart DS, Feunang YD, Guo AC, Lo EJ, Marcu A, Grant JR, et al. DrugBank 5.0: A major update to the DrugBank database for 2018. Nucleic Acids Res 2018;46(D1):D1074-82.
[Crossref] [Google Scholar] [PubMed]
- Liu Z, Guo F, Wang Y, Li C, Zhang X, Li H, et al. BATMAN-TCM: A bioinformatics analysis tool for molecular mechanism of traditional Chinese medicine. Sci Rep 2016;6(1):1-11.
[Crossref] [Google Scholar] [PubMed]
- Xu HY, Zhang YQ, Liu ZM, Chen T, Lv CY, Tang SH, et al. ETCM: An encyclopaedia of traditional Chinese medicine. Nucleic Acids Res 2019;47(D1):D976-82.
[Crossref] [Google Scholar] [PubMed]
- Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, et al. TBtools: An integrative toolkit developed for interactive analyses of big biological data. Mol Plant 2020;13(8):1194-202.
[Crossref] [Google Scholar] [PubMed]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: Protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47(D1):D607-13.
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13(11):2498-504.
[Crossref] [Google Scholar] [PubMed]
- Venny OJ. An interactive tool for comparing lists with Venn Diagrams. BioinfoGP, CNB-CSIC 2007;6994833.
- Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun 2019;10(1):1.
[Crossref] [Google Scholar] [PubMed]
- Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, et al. The protein data bank. Nucleic Acids Res 2000;28(1):235-42.
[Crossref] [Google Scholar] [PubMed]
- Trott O, Olson AJ. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization and multithreading. J Comput Chem 2010;31(2):455-61.
[Crossref] [Google Scholar] [PubMed]
- Sohrabi C, Alsafi Z, O'neill N, Khan M, Kerwan A, Al-Jabir A, et al. World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19). Int J Surg 2020;76:71-6.
[Crossref] [Google Scholar] [PubMed]
- Whitworth J. COVID-19: A fast evolving pandemic. Trans R Soc Trop Med Hyg 2020;114(4):241-8.
[Crossref] [Google Scholar] [PubMed]
- Ruan Q, Yang K, Wang W, Jiang L, Song J. Clinical predictors of mortality due to COVID-19 based on an analysis of data of 150 patients from Wuhan, China. Intensive Care Med 2020;46(5):846-8.
[Crossref] [Google Scholar] [PubMed]
- Jose RJ, Manuel A. COVID-19 cytokine storm: The interplay between inflammation and coagulation. Lancet Respir Med 2020;8(6):e46-7.
[Crossref] [Google Scholar] [PubMed]
- Keit E, Coutu B, Zhen W, Zhang C, Lin C, Bennion N, et al. Systemic inflammation is associated with inferior disease control and survival in stage III non-small cell lung cancer. Ann Transl Med 2021;9(3):227.
[Crossref] [Google Scholar] [PubMed]
- Tagliamento M, Agostinetto E, Bruzzone M, Ceppi M, Saini KS, de Azambuja E, et al. Mortality in adult patients with solid or hematological malignancies and SARS-CoV-2 infection with a specific focus on lung and breast cancers: A systematic review and meta-analysis. Crit Rev Oncol Hematol 2021;163:103365.
[Crossref] [Google Scholar] [PubMed]
- Kalinka E, Chmielewska I, Wojas-Krawczyk K. Viral infection and lung cancer immunotherapy. Front Oncol 2021;11:577514.
[Crossref] [Google Scholar] [PubMed]
- Zhou P, Yang XL, Wang XG, Hu B, Zhang L, Zhang W, et al. A pneumonia outbreak associated with a new coronavirus of probable bat origin. Nature 2020;579(7798):270-3.
[Crossref] [Google Scholar] [PubMed]
- Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science 2020;367(6485):1444-8.
[Crossref] [Google Scholar] [PubMed]
- Huang X, Liang H, Zhang H, Tian L, Cong P, Wu T, et al. The potential mechanism of cancer patients appearing more vulnerable to SARS-CoV-2 and poor outcomes: A pan-cancer bioinformatics analysis. Front Immunol 2021;12:804387.
[Crossref] [Google Scholar] [PubMed]
- Chen L, Liu Y, Wu J, Deng C, Tan J, Liu H, et al. Lung adenocarcinoma patients have higher risk of SARS-CoV-2 infection. Aging (Albany NY) 2021;13(2):1620-32.
[Crossref] [Google Scholar] [PubMed]
- Wang B, Huang Y. Which type of cancer patients are more susceptible to the SARS-COX-2: Evidence from a meta-analysis and bioinformatics analysis. Crit Rev Oncol Hematol 2020;153:103032.
[Crossref] [Google Scholar] [PubMed]
- Rolfo C, Meshulami N, Russo A, Krammer F, García-Sastre A, Mack PC, et al. Lung cancer and severe acute respiratory syndrome Coronavirus 2 infection: Identifying important knowledge gaps for investigation. J Thorac Oncol 2021;17(2):214-27.
[Crossref] [Google Scholar] [PubMed]
- Massarweh A, Eliakim-Raz N, Stemmer A, Levy-Barda A, Yust-Katz S, Zer A, et al. Evaluation of seropositivity following BNT162b2 messenger RNA vaccination for SARS-CoV-2 in patients undergoing treatment for cancer. JAMA Oncol 2021;7(8):1133-40.
[Crossref] [Google Scholar] [PubMed]
- Lopes MI, Bonjorno LP, Giannini MC, Amaral NB, Menezes PI, Dib SM, et al. Beneficial effects of colchicine for moderate to severe COVID-19: A randomised, double-blinded, placebo-controlled clinical trial. RMD Open 2021;7(1):e001455.
[Crossref] [Google Scholar] [PubMed]
- Nawangsih EN, Kusmala YY, Rakhmat II, Handayani DR, Juliastuti H, Wibowo A, et al. Colchicine and mortality in patients with coronavirus disease 2019 (COVID-19) pneumonia: A systematic review, meta-analysis and meta-regression. Int Immunopharmacol 2021;96:107723.
- Della-Torre E, Della-Torre F, Kusanovic M, Scotti R, Ramirez GA, Dagna L, et al. Treating COVID-19 with colchicine in community healthcare setting. Clin Immunol 2020;217:108490.
[Crossref] [Google Scholar] [PubMed]
- Rojas-Villarraga A, Parra-Medina R, Gómez-López A. Colchicine treatment in COVID-19: The remaining unsolved question. Lancet Respir Med 2021;9(12):1351-3.
[Crossref] [Google Scholar] [PubMed]
- Stein A, Hilken née Thomopoulou P, Frias C, Hopff SM, Varela P, Wilke N, et al. B-nor-methylene colchicinoid PT-100 selectively induces apoptosis in multidrug-resistant human cancer cells via an intrinsic pathway in a caspase-independent manner. ACS Omega 2022;7(3):2591-603.
- Zhao Y, Kuang M, Li J, Zhu L, Jia Z, Guo X, et al. SARS-CoV-2 spike protein interacts with and activates TLR41. Cell Res 2021;31(7):818-20.
[Crossref] [Google Scholar] [PubMed]
- Choudhury A, Mukherjee S. In silico studies on the comparative characterization of the interactions of SARS-CoV-2 spike glycoprotein with ACE-2 receptor homologs and human TLRs. J Med Virol 2020;92(10):2105-13.
[Crossref] [Google Scholar] [PubMed]
- Sohn KM, Lee SG, Kim HJ, Cheon S, Jeong H, Lee J, et al. COVID-19 patients upregulate toll-like receptor 4-mediated inflammatory signaling that mimics bacterial sepsis. J Korean Med Sci 2020;35(38):e343.
[Crossref] [Google Scholar] [PubMed]
- Mukherjee S, Karmakar S, Babu SP. TLR2 and TLR4 mediated host immune responses in major infectious diseases: A review. Braz J Infect Dis 2016;20:193-204.
[Crossref] [Google Scholar] [PubMed]
- Pearce L, Davidson SM, Yellon DM. The cytokine storm of COVID-19: A spotlight on prevention and protection. Expert Opin Ther Targets 2020;24(8):723-30.
[Crossref] [Google Scholar] [PubMed]
- Manik M, Singh RK. Role of toll-like receptors in modulation of cytokine storm signaling in SARS-CoV-2-induced COVID-19. J Med Virol 2022;94(3):869-77.
[Crossref] [Google Scholar] [PubMed]
- Teixeira PC, Dorneles GP, Santana Filho PC, da Silva IM, Schipper LL, Postiga IA, et al. Increased LPS levels coexist with systemic inflammation and result in monocyte activation in severe COVID-19 patients. Int Immunopharmacol 2021;100:108125.
[Crossref] [Google Scholar] [PubMed]
- MacDonald L, Alivernini S, Tolusso B, Elmesmari A, Somma D, Perniola S, et al. COVID-19 and RA share an SPP1 myeloid pathway that drives PD-L1+ neutrophils and CD14+ monocytes. JCI Insight 2021;6(13):e147413.
[Crossref] [Google Scholar] [PubMed]
- Sharma P, Pandey AK, Bhattacharyya DK. Determining crucial genes associated with COVID-19 based on COPD findings✶,✶,✶. Comput Biol Med 2021;128:104126.
[Crossref] [Google Scholar] [PubMed]
- LaClair HJ, Khosrodad N, Sule AA, Koehler T, Krishnamoorthy G. Effect of ACE inhibitors and angiotensin receptor blockers on in-hospital mortality and length of stay in hospitalized COVID-19 patients. Vascul Pharmacol 2021;141:106902.
[Crossref] [Google Scholar] [PubMed]
- Sarangarajan R, Winn R, Kiebish MA, Bountra C, Granger E, Narain NR. Ethnic prevalence of angiotensin-converting enzyme deletion (D) polymorphism and COVID-19 risk: Rationale for use of angiotensin-converting enzyme inhibitors/angiotensin receptor blockers. J Racial Ethn Health Disparities 2021;8(4):973-80.
[Crossref] [Google Scholar] [PubMed]
- Ferrara F, Vitiello A. Scientific hypothesis for treatment of COVID-19’s lung lesions by adjusting ACE/ACE2 imbalance. Cardiovasc Toxicol 2021;21(6):498-503.
[Crossref] [Google Scholar] [PubMed]
- Feng L, Cheng P, Feng Z, Zhang X. Transmembrane p24 trafficking protein 2 regulates inflammation through the TLR4/NF-κB signaling pathway in lung adenocarcinoma. World J Surg Oncol 2022;20(1):1-32.
[Crossref] [Google Scholar] [PubMed]
- Gong WJ, Liu JY, Yin JY, Cui JJ, Xiao D, Zhuo W, et al. Resistin facilitates metastasis of lung adenocarcinoma through the TLR 4/Src/EGFR/PI 3K/NF-κB pathway. Cancer Sci 2018;109(8):2391-400.
[Crossref] [Google Scholar] [PubMed]
- Bauer AK, Fostel J, Degraff LM, Rondini EA, Walker C, Grissom SF, et al. Transcriptomic analysis of pathways regulated by toll-like receptor 4 in a murine model of chronic pulmonary inflammation and carcinogenesis. Mol Cancer 2009;8(1):107.
[Crossref] [Google Scholar] [PubMed]
- Cai Z, Chen Q, Chen J, Lu Y, Xiao G, Wu Z, et al. Monocyte chemotactic protein 1 promotes lung cancer–induced bone resorptive lesions in vivo. Neoplasia 2009;11(3):228-36.
[Crossref] [Google Scholar] [PubMed]
- Xu W, Wei Q, Han M, Zhou B, Wang H, Zhang J, et al. CCL2-SQSTM1 positive feedback loop suppresses autophagy to promote chemoresistance in gastric cancer. Int J Biol Sci 2018;14(9):1054.
[Crossref] [Google Scholar] [PubMed]
- Li L, Liu YD, Zhan YT, Zhu YH, Li Y, Xie D, et al. High levels of CCL2 or CCL4 in the tumor microenvironment predict unfavorable survival in lung adenocarcinoma. Thoracic Cancer 2018;9(7):775-84.
[Crossref] [Google Scholar] [PubMed]
- Karachaliou N, Gonzalez-Cao M, Crespo G, Drozdowskyj A, Aldeguer E, Gimenez-Capitan A, et al. Interferon gamma, an important marker of response to immune checkpoint blockade in non-small cell lung cancer and melanoma patients. Ther Adv Med Oncol 2018;10:1758834017749748.
[Crossref] [Google Scholar] [PubMed]
- Gao Y, Yang J, Cai Y, Fu S, Zhang N, Fu X, et al. IFN-γ-mediated inhibition of lung cancer correlates with PD-L1 expression and is regulated by PI3K-AKT signaling. Int J Cancer 2018;143(4):931-43.
[Crossref] [Google Scholar] [PubMed]
- Critchley-Thorne RJ, Simons DL, Yan N, Miyahira AK, Dirbas FM, Johnson DL, et al. Impaired interferon signaling is a common immune defect in human cancer. Proc Natl Acad Sci 2009;106(22):9010-5.
[Crossref] [Google Scholar] [PubMed]
- Kumar R, Collins D, Dolly S, McDonald F, O’Brien ME, Yap TA. Targeting the PD-1/PD-L1 axis in non–small cell lung cancer. Curr Problems Cancer 2017;41(2):111-24.
[Crossref] [Google Scholar] [PubMed]
- Fong KM, Kida Y, Zimmerman PV, Smith PJ. TIMP1 and adverse prognosis in non-small cell lung cancer. Clin Cancer Res 1996;2(8):1369-72.
[Google Scholar] [PubMed]
- Pesta M, Kulda V, Kucera R, Pesek M, Vrzalova J, Liska V, et al. Prognostic significance of TIMP-1 in non-small cell lung cancer. Anticancer Res 2011;31(11):4031-8.
[Google Scholar] [PubMed]
- Miao TW, Xiao W, Du LY, Mao B, Huang W, Chen XM, et al. High expression of SPP1 in patients with chronic obstructive pulmonary disease (COPD) is correlated with increased risk of lung cancer. FEBS Open Bio 2021;11(4):1237-49.
- Tang H, Chen J, Han X, Feng Y, Wang F. Up regulation of SPP1 is a marker for poor lung cancer prognosis and contributes to cancer progression and cisplatin resistance. Front Cell Dev Biol 2021;9:1109.
[Crossref] [Google Scholar] [PubMed]
- Shi S, Yu G, Huang B, Mi Y, Kang Y, Simon JP. PPARG could work as a valid therapeutic strategy for the treatment of lung squamous cell carcinoma. PPAR Res 2020;2020:25120951.
[Crossref] [Google Scholar] [PubMed]
- Moseley TA, Haudenschild DR, Rose L, Reddi AH. Interleukin-17 family and IL-17 receptors. Cytokine Growth Factor Rev 2003;14(2):155-74.
[Crossref] [Google Scholar] [PubMed]
- Beringer A, Noack M, Miossec P. IL-17 in chronic inflammation: From discovery to targeting. Trends Mol Med 2016;22(3):230-41.
[Crossref] [Google Scholar] [PubMed]
- Zhao J, Chen X, Herjan T, Li X. The role of interleukin-17 in tumor development and progression. J Exp Med 2020;217(1):20190297.
[Crossref] [Google Scholar] [PubMed]
- Pan B, Shen J, Cao J, Zhou Y, Shang L, Jin S, et al. Interleukin-17 promotes angiogenesis by stimulating VEGF production of cancer cells via the STAT3/GIV signaling pathway in non-small-cell lung cancer. Sci Rep 2015;5(1):16053.
[Crossref] [Google Scholar] [PubMed]
- Crowe CR, Chen K, Pociask DA, Alcorn JF, Krivich C, Enelow RI, et al. Critical role of IL-17RA in immunopathology of influenza infection. J Immunol 2009;183(8):5301-10.
[Crossref] [Google Scholar] [PubMed]
- Mangalmurti N, Hunter CA. Cytokine storms: Understanding COVID-19. Immunity 2020;53(1):19-25.
[Crossref] [Google Scholar] [PubMed]
- Vaninov N. In the eye of the COVID-19 cytokine storm. Nat Rev Immunol 2020;20(5):277.
[Crossref] [Google Scholar] [PubMed]
- Mendoza VM. Interleukin-17: A potential therapeutic target in COVID-19. J Infect 2020;81(2):e136-8.
[Crossref] [Google Scholar] [PubMed]
- Zhu Z, Zheng Z, Liu J. Comparison of COVID-19 and lung cancer via reactive oxygen species Signaling. Front Oncol 2021;11:708263.
[Crossref] [Google Scholar] [PubMed]
- Moreno Roig E, Groot AJ, Yaromina A, Hendrickx TC, Barbeau LM, Giuranno L, et al. HIF-1α and HIF-2α differently regulate the radiation sensitivity of NSCLC cells. Cells 2019;8(1):45.
[Crossref] [Google Scholar] [PubMed]
- Jan YH, Lai TC, Yang CJ, Lin YF, Huang MS, Hsiao M. Adenylate kinase 4 modulates oxidative stress and stabilizes HIF-1α to drive lung adenocarcinoma metastasis. J Hematol Oncol 2019;12(1):1-4.
[Crossref] [Google Scholar] [PubMed]
- Tannahill GM, O’Neill LA. The emerging role of metabolic regulation in the functioning of Toll-like receptors and the NOD-like receptor Nlrp3. FEBS Lett 2011;585(11):1568-72.
[Crossref] [Google Scholar] [PubMed]
- Patel S. Inflammasomes, the cardinal pathology mediators are activated by pathogens, allergens and mutagens: A critical review with focus on NLRP3. Biomed Pharmacother 2017;92:819-25.
[Crossref] [Google Scholar] [PubMed]
- Schmidt RL, Lenz LL. Distinct licensing of IL-18 and IL-1β secretion in response to NLRP3 inflammasome activation. PLoS One 2012;7(9):e45186.
[Crossref] [Google Scholar] [PubMed]
- Bonaventura A, Vecchié A, Dagna L, Tangianu F, Abbate A, Dentali F. Colchicine for COVID-19: Targeting NLRP3 inflammasome to blunt hyperinflammation. Inflamm Res 2022:71(3):297-307.
[Crossref] [Google Scholar] [PubMed]
- Rodrigues TS, de Sá KS, Ishimoto AY, Becerra A, Oliveira S, Almeida L, et al. Inflammasomes are activated in response to SARS-CoV-2 infection and are associated with COVID-19 severity in patients. J Exp Med 2021;218(3).
[Crossref] [Google Scholar] [PubMed]
- Toldo S, Bussani R, Nuzzi V, Bonaventura A, Mauro AG, Cannatà A, et al. Inflammasome formation in the lungs of patients with fatal COVID-19. Inflamm Res 2021;70(1):7-10.
[Crossref] [Google Scholar] [PubMed]
- Martinon F, Pétrilli V, Mayor A, Tardivel A, Tschopp J. Gout-associated uric acid crystals activate the NALP3 inflammasome. Nature 2006;440(7081):237-41.
[Crossref] [Google Scholar] [PubMed]
- Silvis MJ, Fiolet AT, Opstal TS, Dekker M, Suquilanda D, Zivkovic M, et al. Colchicine reduces extracellular vesicle NLRP3 inflammasome protein levels in chronic coronary disease: A LoDoCo2 biomarker substudy. Atherosclerosis 2021;334:93-100.
[Crossref] [Google Scholar] [PubMed]
- Robertson S, Martínez GJ, Payet CA, Barraclough JY, Celermajer DS, Bursill C, et al. Colchicine therapy in acute coronary syndrome patients acts on caspase-1 to suppress NLRP3 inflammasome monocyte activation. Clin Sci 2016;130(14):1237-46.
[Crossref] [Google Scholar] [PubMed]
- Dai W, Zhang B, Jiang XM, Su H, Li J, Zhao Y, et al. Structure-based design of antiviral drug candidates targeting the SARS-CoV-2 main protease. Science 2020;368(6497):1331-5.
[Crossref] [Google Scholar] [PubMed]
- Rathnayake AD, Zheng J, Kim Y, Perera KD, Mackin S, Meyerholz DK, et al. 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV–infected mice. Sci Transl Med 2020;12(557):eabc5332.
[Crossref] [Google Scholar] [PubMed]
- Giofrè SV, Napoli E, Iraci N, Speciale A, Cimino F, Muscarà C, et al. Interaction of selected terpenoids with two SARS-CoV-2 key therapeutic targets: An in silico study through molecular docking and dynamics simulations. Comput Biol Med 2021;134:104538.
[Crossref] [Google Scholar] [PubMed]
- de Vries M, Mohamed AS, Prescott RA, Valero-Jimenez AM, Desvignes L, O’Connor R, et al. A comparative analysis of SARS-CoV-2 antivirals characterizes 3CLpro inhibitor PF-00835231 as a potential new treatment for COVID-19. J Virol 2021;95(10):e01819-20.
[Crossref] [Google Scholar] [PubMed]
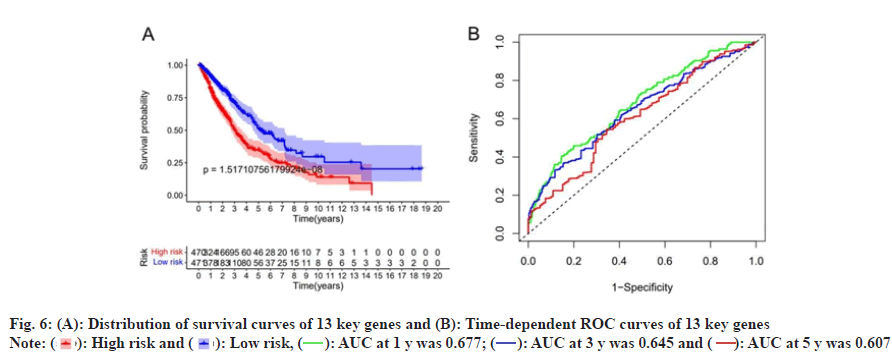
 ): High risk and (
): High risk and ( ): Low risk, (
): Low risk, ( ): AUC at 1 y was 0.677; (
): AUC at 1 y was 0.677; ( ): AUC at 3 y was 0.645 and (
): AUC at 3 y was 0.645 and ( ): AUC at 5 y was 0.607
): AUC at 5 y was 0.607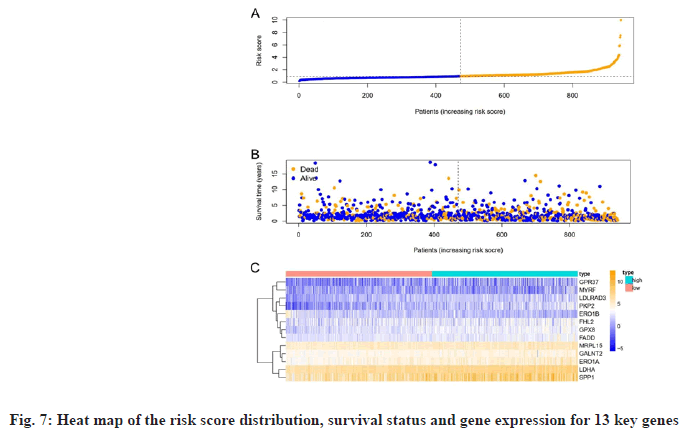

 ): Colchicine;
(
): Colchicine;
( ): Up-regulated genes; (
): Up-regulated genes; ( ): Down-regulated genes and (
): Down-regulated genes and ( ): Top20 KEGG pathways
): Top20 KEGG pathways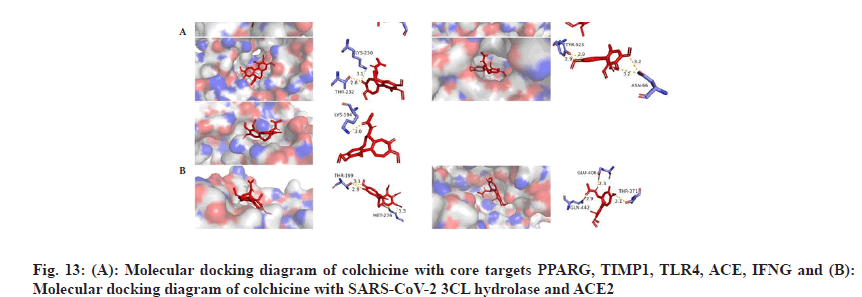
 ): Colchicine and CCL2 system; (
): Colchicine and CCL2 system; ( ): Colchicine and PPARG system; (
): Colchicine and PPARG system; ( ): Colchicine and ACE2 and (
): Colchicine and ACE2 and ( ):
Colchicine and SARS-CoV-2 3CL hydrolase system
):
Colchicine and SARS-CoV-2 3CL hydrolase system