- *Corresponding Author:
- Boyang Liu
Department of Clinical Medicine, College of Medicine, Zhejiang University, Hangzhou, Zhejiang 310058, China
E-mail: 3180105305@zju.edu.cn
| This article was originally published in a special issue, “Modern Applications in Biomedical Research and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2022:84(3) Spl Issue “190-201” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Phellodendrine is a Phellodendri Cortex-derived isoquinoline alkaloids, has been shown to have various activities, especially hypoglycemic effect in mice, predicting its medicinal value on diabetes mellitus. To further understand the pharmacological effect of phellodendrine on diabetes mellitus, network pharmacological techniques have been used to elaborate the involved mechanisms. 84 common target molecules were screened, based on the chemical structure of phellodendrine molecule and disease database. These proteins were enriched in insulin resistance, insulin secretion and inflammatory response, mainly focus on the phosphatidylinositol 3-kinase/protein kinase B signaling pathway, mitogen-activated protein kinase signaling pathway and interleukin-17 signaling pathway. Moreover, enrichment analysis suggested that the targets of phellodendrine such as phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha and mitogen-activated protein kinases 8 were associated with coronavirus disease 2019. To verify the results, molecular docking technique was used to evaluate the interaction between phellodendrine and key targets in the signaling pathway. The calculated binding energy indicates that phellodendrine can form stable complex with insulin receptor, mitogen-activated protein kinases 8, phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit alpha and glycogen synthase kinase 3 beta. These data suggest that phellodendrine should be beneficial for treatment of diabetes mellitus.
Keywords
Phellodendrine, diabetes mellitus, network pharmacology, molecular docking
Phellodendrine (PHE) is one of the most important ingredients in the Phellodendri Cortex that is the dried bark of Phellodendron, traditionally used as herbal medicine widely distributed in East Asia. Phellodendri Cortex was proven to have anti-inflammatory, antimicrobial, anticancer, hypotensive, antiarrhythmic, antioxidant and antipyretic properties[1]. PHE is a kind of isoquinoline alkaloids, which can be absorbed into blood and tissues rapidly and the major distribution tissues is kidney[2]. Previous studies reported that PHE has anti-inflammatory effect and it may be due to its ability to suppress the cellular immune response[3-5]. This pharmacological effect also manifests as inhibiting-phospholipase A2 activity[6]. Noteworthily, PHE produces a marked effect in many types of inflammation, including bacterial vaginosis in mice[7], acute kidney injury in sepsis rats[8], inflammatory response of BV2 microglia induced by Lipopolysaccharide (LPS)[9] and the Reactive Oxygen Species (ROS)-mediated inflammatory response in zebrafish embryo[10]. All these studies came to the same conclusion that the anti-inflammatory effect may be related to the inhibition of Toll Like Receptor-4 (TLR-4)/Nuclear Factor Kappa B (NF-κB) pathway activation and the release of inflammatory factors. PHE can also possess hypoglycemic effect in vitro[11], but little in vivo as the hepatic disposition prevents the transfer of 96.1 % of it[12]. PHE has an inhibitory effect on in vitro acetylcholinesterase activity, proven to be dose-dependent[13]. Also, it has been observed in Ulcerative Colitis (UC) model rats that PHE could reduce the intestinal damage through activating the phosphorylated-AMP Activated Protein Kinase (p-AMPK)/mammalian Target of Rapamycin (mTOR) signaling pathway, as well as autophagy[14]. In addition, PHE suppresses proliferation of KRAS proto-oncogene mutated pancreatic cancer cells through inhibition of nutrients uptake via micropinocytosis[15], which shows the potential of anticancer effects.
Diabetes Mellitus (DM) is a group of chronic diseases characterized by increased blood sugar associated with abnormal insulin production and action. DM and its complications pose a major global health threat. The International Diabetes Federation (IDF) calculates that 9.3 % of adult’s aged 20-79 y, a staggering 463 million people, is living with diabetes in 2019. Prediction from IDF projects to rise to 700 million by 2045. IDF also estimates the annual global health expenditure on diabetes at USD 760 billion[16]. DM is characterized by chronic increase of blood glucose level, which is an important factor leading to blindness, kidney failure, heart failure, cardiovascular and cerebrovascular diseases. At present, there are many effective drugs for the treatment of diabetes, especially type 2 diabetes, including long-acting insulin, metformin, Dipeptidyl Peptidase-4 (DPP-4) pathway inhibitors, sulfonylureas, Glucagon-Like Peptide-1 (GLP-1) analogues, alpha (α)-glucosidase inhibitors, thiazolidinediones, etc[17]. These drugs mainly promote anabolism and lower blood glucose by reducing glycogen production, increasing peripheral glucose utilization or acting on intermediate metabolism. In the meantime, these therapeutic drugs are not effective enough, associating with a variety of adverse reactions, including hypoglycemia, weight gain or lactic acidosis. DM is a complex disease under the interaction of multiple factors, requiring intervention of multiple targets and multiple pathways to achieve ideal therapeutic effects[18]. In the prevention and treatment of DM, Traditional Chinese Medicine (TCM) and its active ingredients have the characteristics of stable efficacy, few adverse reactions, long-term use, multi-target and multi-channel regulation of metabolic disorders and protection of the structure and function of pancreas, liver and kidney, and other related organs[19]. In view of the hypoglycemic effect of PHE to some extent, in order to further understand the pharmacological effect of PHE on DM, this study predicted the possible mechanism based on network pharmacology strategy.
Materials and Methods
Identification of PHE anti-DM targets:
The Structural Data File (SDF) file with PHE Three- Dimensional (3D) structure was downloaded from the PubChem database (https://pubchem.ncbi.nlm.nih.gov/). PHE targets in Homo sapiens based on 3D similarity of chemical structure were screened on the Swiss target prediction platform (http://www.swisstargetprediction.ch/index.php/). Also, the Pharm Mapper platform (http://www.lilab-ecust.cn/pharmmapper/) was used to search PHE targets via pharmacophore mapping approach. The duplication was eliminated and all the targets captured from the two prediction methods were merged.
Medical Subject Headings (MeSH) of DM was entered, namely DM (MeSH Unique ID: D003920) and retrieved the related disease genes on DisGeNET databases (https://www.disgenet.org/). The same molecular targets were picked up between DM associated target genes and PHE target molecules.
Bioinformatics annotation:
Classify target proteins of PHE in Protein Analysis through Evolutionary Relationships (PANTHER) classification system (http://pantherdb.org/). Perform enrichment analysis of PHE anti-DM targets in g:Profiler platform (http://biit.cs.ut.ee/gprofiler/gost). Map the key regulatory pathways in Kyoto Encyclopedia of Genes and Genomes (KEGG) database (https://www.kegg.jp/kegg/). Analyze the interaction between PHE affinity targets and DM-related proteins on STRING platform (https://string-db.org/) and construct a visual network by Cytoscape v3.8.2.
Molecular docking:
Download crystal structures of the interest PHE molecular targets, including Insulin Receptor (INSR) (Protein Database (PDB) ID: 4IBM), Mitogen- Activated Protein Kinase (MAPK) (PDB ID: 3PZE), Phosphatidylinositol-4,5-Bisphosphate 3-Kinase Catalytic Subunit Alpha (PIK3CA) (PDB ID: 4JPS), Glycogen Synthase Kinase 3 Beta (β) (GSK3B) (PDB ID: 1Q41) from the PDB (http://www.rcsb.org/). PyMOL software (The PyMOL Molecular Graphics System, Version 2.0 Schrödinger, LLC.) was used to peel the original ligand from the target protein. Ligand and macromolecule files for molecular docking were prepared by auto dock tool. The docking results (PDB files) were uploaded to Protein Ligand Interaction Profiler (PLIP) platform (https://plip-tool.biotec.tu-dresden.de/plip-web/plip/index) to analyze the interaction between small molecules and proteins.
Results and Discussion
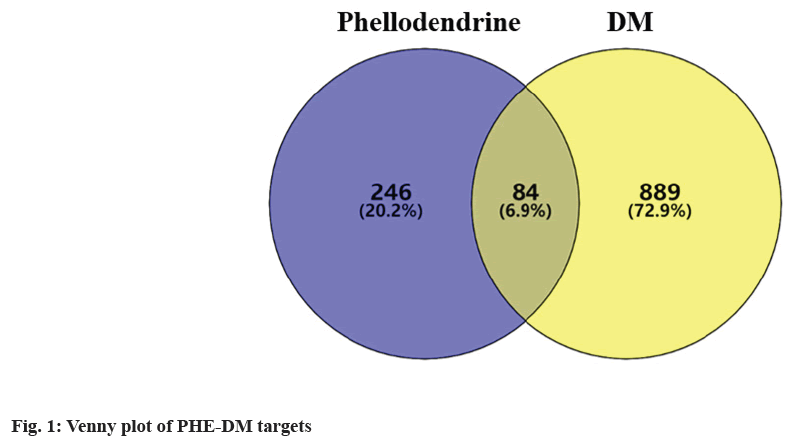
Based on cheminformatics strategies, 330 PHE affinity targets were captured from PharmMapper and Swiss Target Prediction platform in the aggregate. 2619 DM-related genes were selected from g: Profiler databases, 973 genes were finally picked up through filtering the median. As shown in the Venn diagram (fig. 1), an intersection of 84 targets was revealed. By functional classification of proteins, 22 objects were incorporated into metabolite interconversion enzyme, including kinase (Phosphoenolpyruvate Carboxykinase 1 (PCK1), PIK3CA), Phosphodiesterase (PDE4D, PDE5A), dehydrogenase (Hydroxysteroid 11-Beta Dehydrogenase 1 (HSD11B1), Alcohol Dehydrogenase 1B (AHD1B)), glycosyltransferase (ABO), transferase (Hematopoietic Prostaglandin D Synthase (HPGDS), Glutathione S-Transferase Pi 1 (GSTP1), Glutathione S-Transferase Mu 1 (GSTM1)), oxidoreductase (Nitric oxide Synthase 3 (NOS3), Superoxide Dismutase 2 (SOD2)), lyase (Glyoxalase I (GLO1)), reductase (Aldo-Keto Reductase Family 1 Member B (AKR1B1), 3-Hydroxy-3-Methylglutaryl-CoA Reductase (HMGCR), Glutathione-Disulfide Reductase (GSR)), hydrolase (Transthyretin (TTR)), esterase (Acetylcholinesterase (ACHE), Butyrylcholinesterase (BCHE)), oxygenase (Cytochrome P450 Family 2 Subfamily C Member 9 (CYP2C9), Cytochrome P450 Family 19 Subfamily A Member 1 (CYP19A1)) and phospholipase (Phospholipase A2 Group IIA (PLA2G2A)). 20 targets were classified as protein modifying enzyme, including metalloprotease (Matrix Metallopeptidase (MMP) 1, MMP2, MMP3, MMP8, MMP9, Membrane Metalloendopeptidase (MME)), protein phosphatase (Protein Tyrosine Phosphatase Non-Receptor Type 1 (PTPN1)), protease (Caspase 1 (CASP1), CASP3), non-receptor serine/threonine protein kinase (GSK3B, MAPK1, MAPK8, MAPK14, RAF1), serine protease (Elastase Neutrophil Expressed (ELANE), DPP4, F2, F7) and cysteine protease (Cathepsin (CTS) S, CTSB). 11 targets were transmembrane signal receptor, including G-protein coupled receptor (Melatonin Receptor 1B (MTNR1B), Adrenoceptor Beta 1 (ADRB1), ADRB2, ADRB3) and transmembrane signal receptor (Epidermal Growth Factor Receptor (EGFR), Tyrosine Kinase (TEK), MET proto-oncogene, INSR, Kinase Insert Domain Receptor (KDR), F3, Insulin Like Growth Factor 1 Receptor (IGF1R)). Also, 4 gene-specific transcriptional regulators were classified, turned out to be C4 zinc finger nuclear receptor (Peroxisome Proliferator Activated Receptor Alpha (PPARA), Nuclear Receptor Subfamily 1 Group I Member 2 (NR1I2), Vitamin D Receptor (VDR), Peroxisome Proliferator Activated Receptor Gamma (PPARG)). 4 targets were classified as transfer/carrier protein (Tocopherol Transfer Protein Alpha (TTPA), Albumin (ALB), Lipocalin 2 (LCN2), Retinol Binding Protein 4 (RBP4)). Another 4 belonged to the transporter family, including Adenosine Triphosphate (ATP) binding cassette transporter (ATP Binding Cassette Subfamily G Member 2 (ABCG2)), primary active transporter (Solute Carrier Family 6 Member 3 (SLC6A3), SLC6A4) and transporter (SLC47A1). 2 targets were sorted out to be chaperone (Heat Shock Protein 90 Alpha Family Class A Member 1 (HSP90AA1), Peptidylprolyl Isomerase A (PPIA)). 2 objects were incorporated into nucleic acid metabolism protein, including endoribonuclease (Endoplasmic Reticulum to Nucleus Signaling 1 (ERN1)) and Deoxyribonucleic Acid (DNA) metabolism protein (Poly ADP-Ribose Polymerase 1 (PARP1)). Proteinbinding activity modulator family contained 2 targets, including protease inhibitor (Alpha-1-antitrypsin (SERPINA1)) and kinase modulator (Phosphoinositide- 3-Kinase Regulatory 1 (PIK3R1)). 1 was classified as calcium-binding protein, which related to calmodulin (S100A9). Finally, 12 targets could not be classified as any protein family, which were reduced to unclassified protein (Nuclear Receptor 3 Group C Member 2 (NR3C2), Fatty Acid Binding Protein 4 (FABP4), Renin (REN), Carbonic Anhydrase 2 (CA2), Androgen Receptor (AR), Glucocorticoid receptor gene (NR3C1), Sex Hormone Binding Globulin (SHBG), Dual Specificity Tyrosine Phosphorylation Regulated Kinase 1A (DYRK1A), Janus Kinase 2 (JAK2), Lysine Acetyltransferase 2B (KAT2B), Estrogen Receptor 1 (ESR1), Interleukin (IL)-2 as shown in Table 1.
| Target class | Protein class | Gene symbol (description) | Relevance score of DM |
|---|---|---|---|
| Metabolite interconversion enzyme | Kinase | PCK1 (Phosphoenolpyruvate carboxykinase, cytosolic) | 0.05 |
| PIK3CA (Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform) | 0.1 | ||
| Phosphodiesterase | PDE4D (cAMP-specific 3',5'-cyclic phosphodiesterase 4D) | 0.11 | |
| PDE5A (cGMP-specific 3',5'-cyclic phosphodiesterase) | 0.05 | ||
| Dehydrogenase | HSD11B1 (Corticosteroid 11-beta-dehydrogenase isozyme 1) | 0.08 | |
| AHD1B (All-trans-retinol dehydrogenase) | 0.05 | ||
| Glycosyltransferase | ABO (Histo-blood group ABO system transferase) | 0.1 | |
| Transferase | HPGDS (Hematopoietic prostaglandin D synthase) | 0.08 | |
| GSTP1 (Glutathione S-transferase P) | 0.04 | ||
| GSTM1 (Glutathione S-transferase Mu 1) | 0.1 | ||
| Oxidoreductase | NOS3 (Nitric oxide synthase, endothelial) | 0.1 | |
| SOD2 (Superoxide dismutase [Mn], mitochondrial) | 0.1 | ||
| Lyase | GLO1 (Lactoylglutathione lyase) | 0.1 | |
| Reductase | AKR1B1 (Aldo-keto reductase family 1 member B1) | 0.1 | |
| HMGCR (3-hydroxy-3-methylglutaryl-coenzyme A reductase) | 0.09 | ||
| GSR (Glutathione reductase, mitochondrial) | 0.08 | ||
| Hydrolase | TTR (Transthyretin) | 0.04 | |
| Esterase | BCHE (Cholinesterase) | 0.1 | |
| ACHE (Acetylcholinesterase) | 0.08 | ||
| Oxygenase | CYP2C9 (Cytochrome P450 2C9) | 0.04 | |
| CYP19A1 (Aromatase) | 0.05 | ||
| Phospholipase | PLA2G2A (Phospholipase A2, membrane associated) | 0.03 | |
| Protein modifying enzyme | Metalloprotease | MMP3 (Stromelysin-1) | 0.04 |
| MMP1 (Interstitial collagenase) | 0.04 | ||
| MMP2 (72 kDa type IV collagenase) | 0.1 | ||
| MMP9 (Matrix metalloproteinase-9) | 0.1 | ||
| MME (Neprilysin) | 0.07 | ||
| MMP8 (Neutrophil collagenase) | 0.03 | ||
| Protein phosphatase | PTPN1 (Tyrosine-protein phosphatase non-receptor type 1) | 0.1 | |
| Protease | CASP1 (Caspase-1) | 0.03 | |
| CASP3 (Caspase-3) | 0.1 | ||
| Non-receptor serine/threonine protein kinase | GSK3B (Glycogen synthase kinase-3 beta) | 0.1 | |
| MAPK1 (Mitogen-activated protein kinase 1) | 0.1 | ||
| MAPK8 (Mitogen-activated protein kinase 8) | 0.1 | ||
| MAPK14 (Mitogen-activated protein kinase 14) | 0.1 | ||
| RAF1 (RAF proto-oncogene serine/threonine-protein kinase) | 0.03 | ||
| Serine protease | ELANE (Neutrophil elastase) | 0.03 | |
| DPP4 | 0.1 | ||
| F7 (Coagulation factor VII) | 0.2 | ||
| F2 (Prothrombin) | 0.06 | ||
| Cysteine protease | CTSS (Cathepsin S) | 0.03 | |
| CTSB (Cathepsin B) | 0.03 | ||
| Transmembrane signal receptor | G-protein coupled receptor | MTNR1B (Melatonin receptor type 1B) | 0.1 |
| ADRB1 (Beta-1 adrenergic receptor) | 0.3 | ||
| ADRB3 (Beta-3 adrenergic receptor) | 0.1 | ||
| ADRB2 (Beta-2 adrenergic receptor) | 0.05 | ||
| Transmembrane signal receptor | EGFR (Epidermal growth factor receptor) | 0.09 | |
| TEK (Angiopoietin-1 receptor) | 0.03 | ||
| MET (Hepatocyte growth factor receptor) | 0.05 | ||
| INSR | 0.2 | ||
| KDR (Vascular endothelial growth factor receptor 2) | 0.06 | ||
| F3 (Tissue factor) | 0.07 | ||
| IGF1R (Insulin-like growth factor 1 receptor) | 0.2 | ||
| Gene-specific transcriptional regulator | C4 zinc finger nuclear receptor | PPARA (Peroxisome proliferator-activated receptor alpha) | 0.01 |
| NR1I2 (Nuclear receptor subfamily 1 group I member 2) | 0.4 | ||
| VDR (Vitamin D3 receptor) | 0.1 | ||
| PPARG (Peroxisome proliferator-activated receptor gamma) | 0.5 | ||
| Transfer/carrier protein | Transfer/carrier protein | TTPA (Alpha-tocopherol transfer protein) | 0.1 |
| ALB(Albumin) | 0.1 | ||
| LCN2 (Neutrophil gelatinase-associated lipocalin) | 0.1 | ||
| RBP4 (Retinol-binding protein 4) | 0.1 | ||
| Transporter | Transporter | SLC47A1 (Multidrug and toxin extrusion protein 1) | 0.03 |
| ATP-binding cassette transporter | ABCG2 (Broad substrate specificity ATP-binding cassette transporter ABCG2) | 0.25 | |
| Primary active transporter | SLC6A4 (Sodium-dependent serotonin transporter) | 0.03 | |
| SLC6A3 (Sodium-dependent dopamine transporter) | 0.03 | ||
| Chaperone | Hsp90 family chaperone | HSP90AA1 (Heat shock protein HSP 90-alpha) | 0.05 |
| chaperone | PPIA (Peptidyl-prolyl cis-trans isomerase A) | 0.03 | |
| Nucleic acid metabolism protein | Endoribonuclease | ERN1 (Serine/threonine-protein kinase/endoribonuclease IRE1) | 0.04 |
| DNA metabolism protein | PARP1 (Poly [ADP-ribose] polymerase 1) | 0.1 | |
| Protein-binding activity modulator | Protease inhibitor | SERPINA1 (Alpha-1-antitrypsin) | 0.03 |
| Kinase modulator | PIK3R1 (Phosphatidylinositol 3-kinase regulatory subunit alpha) | 0.11 | |
| calcium-binding protein | Calmodulin-related | S100A9 (Protein S100-A9) | 0.05 |
| Unclassified protein | Unclassified protein | NR3C2 (Mineralocorticoid receptor) | 0.1 |
| FABP4 (Fatty acid-binding protein, adipocyte) | 0.1 | ||
| REN (Renin) | 0.1 | ||
| CA2( Carbonic anhydrase 2) | 0.03 | ||
| AR (Androgen receptor) | 0.07 | ||
| NR3C1 (Glucocorticoid receptor) | 0.05 | ||
| SHBG (Sex hormone-binding globulin) | 0.1 | ||
| DYRK1A (Dual specificity tyrosine-phosphorylation-regulated kinase) | 0.03 | ||
| JAK2 (Tyrosine-protein kinase JAK2) | 0.05 | ||
| KAT2B (Histone acetyltransferase KAT2B) | 0.03 | ||
| ESR1(Estrogen receptor) | 0.1 | ||
| IL-2 (Interleukin-2) | 0.03 |
Table 1: Phe Associated Target Molecules
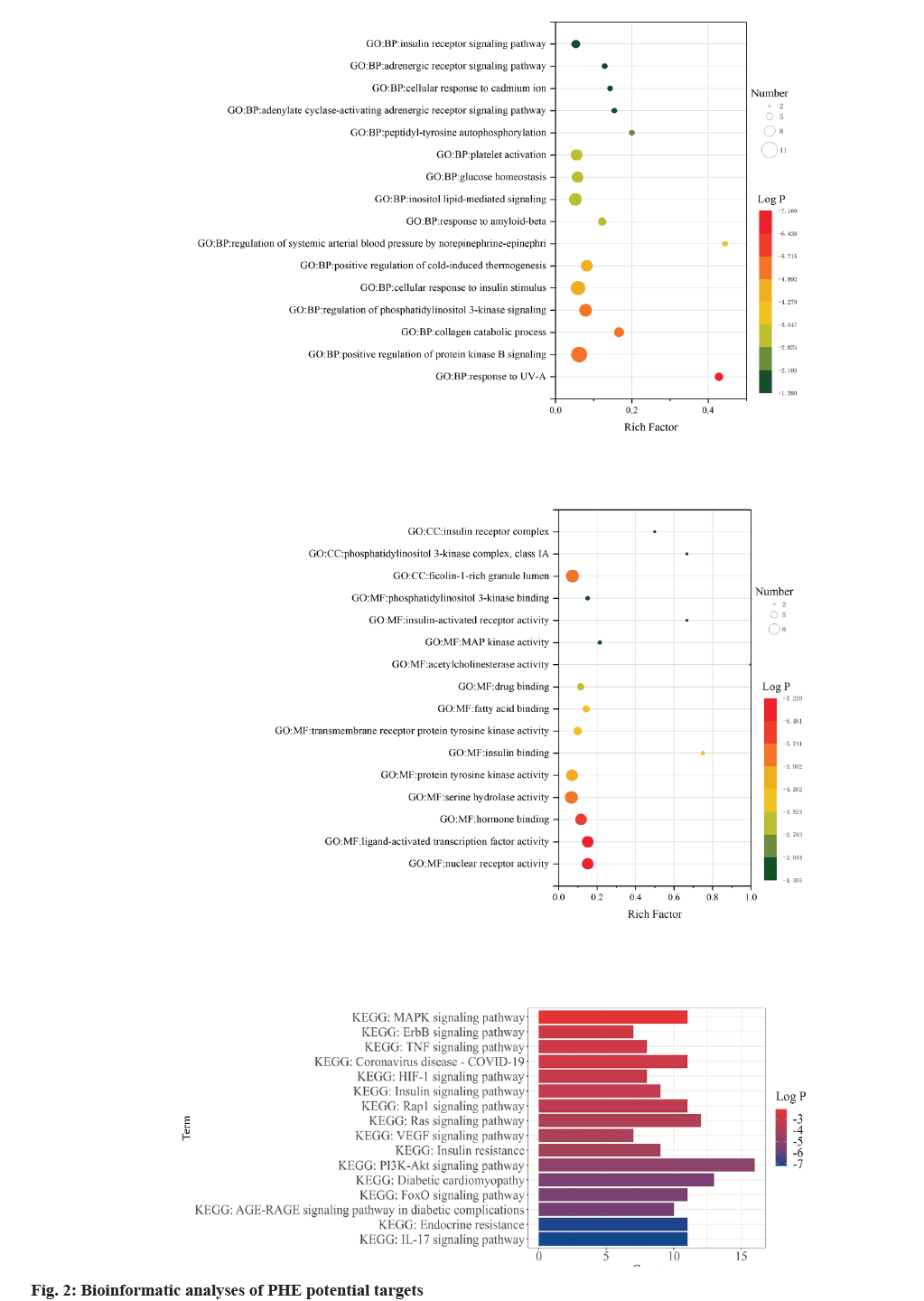
Gene Ontology (GO) enrichment analysis was performed on key target genes to study the potential mechanism of PHE in the treatment of DM. Through the classification and annotation of functional genes of Biological Process (BP), Molecular Biological Function (MF) and Cellular Component (CC), the project name of each type of expression of PHE acting on DM genes was screened out, and the reliability of the conclusion was demonstrated with log p value as the standard (fig. 2). In the enrichment analysis of BP, PHE potential targets had the highest significant level with response to Ultraviolet (UV)-A, with log p value of -7.4 and a total of 6 PHE targets were involved. Phosphatidylinositol 3-Kinase (PI3K) signal path and AKT/Protein Kinase B (PKB) signal path have been shown to be closely related to the development of DM[16,17] that insulin and other growth factors are involved in the activation of PKB through PI3K[18]. In the study, the term regulation of PI3K signaling (log p=-5.02) and positive regulation of PKB signaling (log p=-5.66) showed high significant level. The term cellular response to insulin stimulus also showed high significant level with log p value of -4.70 and the potential PHE targets associated with this process were INSR, PI3K, PTP1B, c-Jun N-terminal Kinase (JNK), GSK-3β and RAF1. In the enrichment analysis of MF, the term nuclear receptor activity (log p=-7.20), ligand-activated transcription factor activity (log p=-7.20), hormone binding (log p=-6.25) showed high significant level, suggesting that drug-target interaction may be related to these factors. In the study, enrichment of the potential targets was quantified as Reach Frequency (RF). The term insulin binding (RF=0.75), acetylcholinesterase activity (RF=1), insulin-activated receptor activity (RF=0.67) showed high enrichment, suggesting that PHE may participate in the regulation of DM through these molecular functions. The term PI3K complex, class IA (RF=0.67) and insulin receptor complex (RF=0.5) showed high enrichment and ficolin-1-rich granule lumen (log p=- 5.66) showed high significant level in the enrichment analysis of CC.
KEGG database showed that 77 signaling pathways matched with intersection targets, among which pathway IL-17 signaling pathway (map04657) had the highest significant level (log p=-7.23) and 11 binding targets, MMP9, MMP1, MAPK14, GSK3B, HSP90AA1, S100A9, LCN2, MMP3, MAPK8, MAPK1 and CASP3 were included (fig. 3). Additionally, KEGG database revealed that 11 PHE target molecules were related with Coronavirus Disease-19 (COVID-19), such as PIK3CA, MMP1, MAPK14, F2, PIK3R1, MMP3, MAPK8, MAPK1, IL2, EGFR and CASP1, the significant level was log p=-3.12.
PPI network (fig. 4) shows that the six nodes with the highest degree, including PIK3R1, PIK3CA, MAPK1, HSP90AA1, INSR, ESR1 and NR3C1, play a core role in PHE against DM, target networks were formed around them.
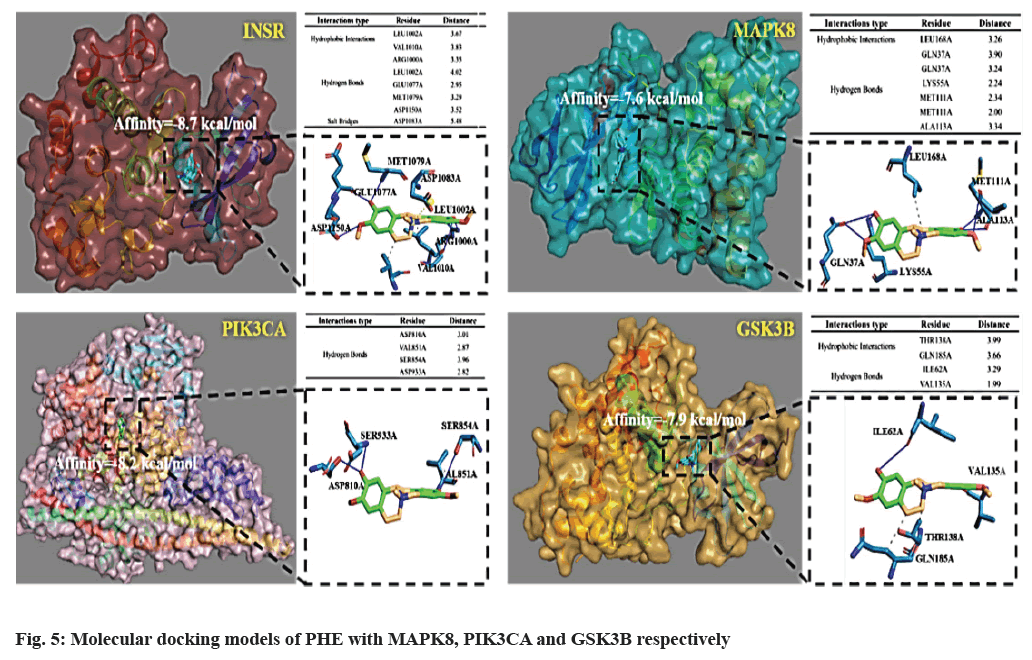
Molecular docking was performed to evaluate the binding capacity of PHE to key targets (fig. 5), INSR, MAPK8, PI3KCA, GSK3B were selected. The binding energies of the small molecules to the six proteins were -8.7, -7.6, -8.2 and -7.9 kcal/mol, respectively, which were all less than -7 kcal/mol, indicating that PHE could form stable complexes with these target proteins. The interaction between PHE and these targets mainly include hydrophobic interaction and hydrogen bond. In the case of INSR docking, leucine 1002, valine 1010 of INSR form hydrophobic interaction with PHE. Moreover, arginine 1000, leucine 1002, glutamic acid 1077, methionine 1079 and aspartic acid 1150 of INSR form hydrogen bonds with PHE and aspartic acid 1083 of INSR forms salt bridges with PHE.
DM, especially Type 2 DM (T2DM) is characterized by both resistance to the action of insulin and defects in insulin secretion[20]. The former has become an important mechanism to explore drug targets in diabetes. Pathological Insulin Resistance (IR) refers to the impaired physiological response of target organs to insulin, including the regulation of glucose and lipid metabolism, protein synthesis, growth and proliferation[21]. Based on the KEGG database, among 84 intersection targets, 9 are associated with IR, include INSR, PIK3CA, PTPN1, GSK3B, PCK1, MAPK8, PIK3R1, NOS3 and PPARA. IR develops through a complex interaction of genes and environment. It mainly acts on liver, adipose tissue and muscle tissue. In liver tissue, IR is mainly manifested that insulin cannot synthesize glycogen, inhibiting gluconeogenesis and leading to increased glucose production and lipid synthesis, finally leading to glucose and lipid metabolism disorder. In adipose tissue, insulin’s inhibitory effect on lipolysis was declined by IR, resulting that peripheral glucose utilization rate decreases and free fatty acid increases. IR induces decrease of glucose transport capacity of cell membrane and glucose utilization rate in muscle tissue[22]. Some severe IR has been proved to be related to the abnormality of insulin before binding to the receptor, defect of insulin receptor itself and the disturbance of signal transmission into the cell after binding. Current researches focus more on the latter two. Defect at the receptor level can be divided into functional and structural abnormalities of INSR. INSR is expressed in all cells, but INSR messenger Ribonucleic Acid (mRNA) levels across multiple human tissue samples show a large range in abundance, spleen, ovary and uterus have the highest INSR expression and blood has the lowest INSR expression[23]. The response to insulin can be influenced by the availability of INSR on the plasma membrane, INSR trafficking processes and the balance between these processes[24]. Reduced INSR expression has been found in adipose tissue of both obese animals and patients, which is associated with IR[25]. In this study, PHE was proved to achieve stable docking with INSR (affinity=-8.7 kcal/mol), suggesting that PHE may participate in the influence of diabetes by regulating the structure of INSR. Insulin binds to α subunit of INSR and changes the configuration of β subunit, then intracellular tyrosine kinases are activated, which can phosphorylate a dozen tyrosine residues on Insulin Receptor Substrates (IRS) and the phosphorylated IRSs bind and activate downstream effectors. IRS proteins are involved in activating two downstream signaling pathways, PI3K/AKT pathway, which is important for insulin’s metabolic activity, and MAPK pathway, which is responsible for cell growth and development[26].
PIK3CA and PIK3R1 respectively encode the p110 catalytic subunit (PI3Kp110a) and p85 regulatory subunit (PI3Kp85a) of class I PI3Ks. The downstream effects of PI3K are mainly reflected in the regulation of PIP3, which eventually leads to partial activation of AKT that stimulates the movement of the glucose membrane transporter towards the cell membrane, thereby increasing glucose absorption in the blood. GSK-3β, a downstream gene of PI3K/AKT signaling pathway, is an evolutionarily conserved serine/threonine kinase that regulates glycogen synthase activity and regulates cell differentiation, proliferation and apoptosis[27,28]. Insulin signaling was the first pathway found to increase the inhibitory serine-phosphorylation of GSK3 by activation of PI3K and AKT. Insulin receptors in most peripheral tissues stimulate a signaling cascade leading to activation of AKT which phosphorylates and inhibits GSK-3[29]. In previous studies, diabetic resistance has been shown to be associated with the over activation of GSK-3 in peripheral tissues, and as a pro-inflammatory enzyme, GSK-3 promotes the production of IL-6, which is well documented to cause IR in liver and adipose tissue[30]. Therefore, inhibiting the hyper phosphorylation of GSK-3 can effectively control IR, such as reducing the expression of PI3K and AKT or inhibiting the expression of GSK-3. It has been proven that high concentrations of PHE can activate the PI3K/AKT pathway by significantly increasing AKT1 gene expression level in zebrafish embryos[31]. In the results above, the PI3K/AKT pathway appears in the BP enrichment, KEGG enrichment and PPI network, indicating that it is highly likely to be the intermediate mechanism of PHE against DM. After screening and molecular docking of proteins belonging to the PI3K/AKT/GSK-3β pathway in 84 targets, PIK3CA and GSK-3β are selected for high affinity; PHE may achieve the hypoglycemic effect by binding to these targets. In the other pathway of IR, MAPK pathway, MAPK8 is in the target intersection, which is also known as JNK1, a member of MAPK family. It can be triggered by various stress stimuli. JNK1 and JNK2 express in all cells and tissues throughout the body and JNK3 expresses in heart, brain and testicles[32]. MAPK8 plays an important biological role in affecting cell proliferation, apoptosis, differentiation, cell death and inflammation, in particular, it is one of the most studied molecules in the mechanism of IR. MAPK8 has been proposed to drive IR in obesity through four different mechanisms, among which direct inhibition of phosphorylation of IRS1 and IRS2 and negative regulation of the PPARα-Fibroblast Growth Factor 21 (FGF21) axis are associated with PHE[33]. In the former one MAPK8 decreases recruitment of the PI3K/AKT signaling pathway in response to insulin. In hepatocyte, MAPK8 acts as a negative regulator of the transcription factor PPARα, leading to reduction of FGF21, fatty acid oxidation and ketogenesis, promoting fatty liver and IR[34]. By molecule docking, MAPK8 shows high binding energy with PHE (affinity=-7.6 kcal/mol), PHE may be involved in the regulation of IR by directly affecting the phosphorylation of MAPK8 on IRS or the effect of the PPARα-FGF21 axis.
In obesity state, lipid accumulation is significantly increased, and the secretion of inflammatory cell promoting factor is increased, which leads to relevant inflammatory response and induces IR[35]. The main inflammatory mediators include C-Reactive Protein (CRP), Tumour Necrosis Factor α (TNF-α), IL- 6, etc. Among all mediators, IL-17 appears to be implicated in the pathogenesis of many auto-immune and inflammatory diseases including DM[36]. In T2DM, IL-17 activates the NF-κB pathway through up-regulation of proinflammatory cytokine gene expression, eventually stimulates production of IL-1β, IL-6 and TNF-α, which contributes to the induction of IR[37]. In this study, the IL-17 signaling pathway was observed a high correlation with the intersection targets (log p=-7.23). Based on the KEGG database, among 84 intersection targets, 11 are associated with IL-17 signaling pathway, include MAPK14, MMP1, MMP9, GSK3B, HSP90AA1, S100A9, LCN2, MMP3, MAPK8, MAPK1 and CASP3. Except CASP3, the other 10 targets were involved in anti-microbial and tissue remodeling mediated by IL-17A/IL-17F, entirely through MAPK pathway. Although this pathway is different from the involvement of IL-17 in T2DM, it provides insights into the mechanism by which PHE may be involved in diabetic complications such as Diabetic Foot Ulcer (DFU).
Currently, researches on the endocrine function of adipocytes have made continuous progress. Adipocytokines such as leptin, resistin, adiponectin, Free Fatty Acid (FFA) are closely related to IR[38- 41]. Additionally, mitochondrial dysfunction[42] and Endoplasmic Reticulum Stress (ERS)[43] have been shown to be associated with IR, but these mechanisms did not appear in the results and will not be detailed.
Another pathogenic mechanism of DM is defects of insulin secretion. Insulin secretion by islet β-cells is an extremely complex process, at present, it can be regulated by regulating ion channels, protecting and repairing islet β-cells, anti-apoptosis and regulating signaling pathways[44]. It has been confirmed that the ability of β-cells to secrete insulin is closely related to intracellular Calcium ions (Ca2+) concentration and cell membrane potential, involving the Potassium ion (K+) and Sodium ion (Na+) channels[45]. The complete structure and function of islet β-cells is a necessary prerequisite for insulin secretion. Studies have shown that the protection and repair of islet β-cells mainly through alleviating oxidative stress and anti-inflammation pathway[46,47]. Promoting cell proliferation and inhibiting apoptosis is an important link in maintaining normal physiological function of islet β-cells, including the inhibition of ERS[48], apoptosis pathway[49], etc. Signaling pathways associated with insulin secretion generally include PI3K/Akt signaling pathway, cAMP-PKA signaling pathway, MAPK signaling pathway, G-Protein- Coupled Receptor (GPCR) signaling pathway, Arf6- Cdc42-Rac1 signaling pathway and so on, studies have found that many plant active ingredients affect insulin secretion through these pathways[46]. Although it is unclear for us whether PHE performs drug effects on insulin secretion via regulating these four ways, it can be predicted that PHE may be involved.
Type 1 DM (T1DM) accounts for around 10 % of DM, which is mainly caused by immune-mediated selective destruction of islet β-cells, and is also related to genetic, environmental factors and viral infection. In this study, the target pathway associated with T1DM is mainly IL-17. As mentioned above, IL-17 is involved in T lymphocyte-mediated islet β-cell damage. T cells play a role mainly by secreting cytokines, mediating cellular immunity and inducing inflammation, damaging β-cells and causing T1DM[50].
It’s worth pointing out that in the era of the COVID-19 pandemic, diabetics are at greater risk than ever before. The lack of physical activity and diet caused by the lockdown and infection due to COVID-19 are two situations that may worsen glycemic control[51]. In a summary report from the Chinese Center for Disease Control of 44 672 confirmed cases showed that those with diabetes have a higher mortality rate[52]. Available evidence suggests that hyperglycemia can increase glucose levels in airway secretion and viral replication. It also causes respiratory dysfunction and functional lung changes through increased vasculature permeability and alveolar dysfunction[53]. On the contrary, infection with COVID-19 may lead to increased blood hypercoagulability, increased inflammation and poor blood sugar control in diabetics[54]. In the fight against COVID-19, TCM is proven to be effective in preventing[55]. PHE is one of the most important ingredients in the Phellodendri Cortex, one of the 50 fundamental herbs, its antidiabetic activity has attracted great attention[12]. In the KEGG analysis, “COVID-19 Homo sapiens” pathway has high significant level (Log p=-3.12) and 11 binding targets, PIK3CA, MMP1, MAPK14, F2, PIK3R1, MMP3, MAPK8, MAPK1, IL- 2, EGFR and CASP1. These targets mainly focus on the inflammatory cytokines, cytokine storm. Besides, PHE’s potential effects on IR and insulin secretion may help control blood sugar.
Conflict of interests:
The authors declared no conflict of interest.
References
- Sun Y, Lenon GB, Yang AWH. Phellodendri cortex: A phytochemical, pharmacological, and pharmacokinetic review. Evid Based Compl Alt Med 2019;7621929.
- Li Y, Liu XG, Wang HY, Dong X, Gao W, Xu XJ, et al. Pharmacokinetic studies of phellodendrine in rat plasma and tissues after intravenous administration using ultra-high performance liquid chromatography–tandem mass spectrometry. J Chromatogr B 2016;1029(1030):95-101.
- Hattori T, Furuta K, Hayashi K, Nagamatsu T, Ito M, Suzuki Y. Antinephritic effects and mechanisms of phellodendrine (OB-5) on crescentic-type anti-GBM nephritis in rats. Jpn J Pharmacol 1992;60(3):187-95.
[Crossref] [Google Scholar] [Pub Med]
- Mori H, Fuchigami M, Inoue N, Nagai H, Koda A, Nishioka I. Principle of the bark of Phellodendron amurense to suppress the cellular immune response. Planta Med 1994;60(5):445-9.
[Crossref] [Google Scholar] [Pub Med]
- Mori H, Fuchigami M, Inoue N, Nagai H, Koda A, Nishioka I, et al. Principle of the bark of Phellodendron amurense to suppress the cellular immune response: Effect of phellodendrine on cellular and humoral immune responses. Planta Med 1995;61(1):45-9.
[Crossref] [Google Scholar] [Pub Med]
- Bonté F, Dumas JM, Saunois A, Meybeck A. Phospholipase A2 inhibition by alkaloid compounds from Phellodendron amurense bark. Pharm Biol 1999;37(1):77-9.
- Zhang DM, Li HJ, Zhou Q. Protective effect of phellodendrine on bacterial vaginosis mice. Chin Trad Herbal Drug 2018;49(24):5849-53.
- Jiang YY, Lin Y. Effect and mechanism of phellodendrine on acute kidney injury in sepsis rats. Chin J Surg Int Trad West Med 2019;25(6):882-7.
- Xu MQ, Lu YW, Yi HM, Yu GR. Study on the inhibitory effect of phellodendrine on LPS-induced microglia activation and its mechanism. Guiding J Trad Chin Med Pharm 2015;27(7):10-5.
- Li L, Huang T, Tian C, Xiao Y, Kou S, Zhou X, et al. The defensive effect of phellodendrine against AAPH-induced oxidative stress through regulating the AKT/NF-κB pathway in zebrafish embryos. Life Sci 2016;157:97-106.
[Crossref] [Google Scholar] [Pub Med]
- Zheng LT, Zhou H, Liu YM, Lin AH. Inhibitory effect of phellodendrine on α-glucosidase in vitro. J Nanjing Univ Trad Chin Med 2020;36(6):853-8.
- Tian X, Xu Z, Hu P, Yu Y, Li Z, Ma Y, et al. Determination of the antidiabetic chemical basis of Phellodendri Chinensis Cortex by integrating hepatic disposition in vivo and hepatic gluconeogenese inhibition in vitro. J Ethnopharmcol 2020;263:11325.
[Crossref] [Google Scholar] [Pub Med]
- Kim YJ, Lim HS, Kim Y, Lee J, Kim BY, Jeong SJ. Phytochemical quantification and the in vitro acetylcholinesterase inhibitory activity of Phellodendron chinense and its components. Molecules 2017;22(6):925.
[Crossref] [Google Scholar] [Pub Med]
- Su S, Wang X, Xi X, Zhu L, Chen Q, Zhang H, et al. Phellodendrine promotes autophagy by regulating the AMPK/mTOR pathway and treats ulcerative colitis. J Cell Mol Med 2021;25(12):5707-20.
[Crossref] [Google Scholar] [Pub Med]
- Thu PM, Zheng ZG, Zhou YP, Wang YY, Zhang X, Jing D, et al. Phellodendrine chloride suppresses proliferation of KRAS mutated pancreatic cancer cells through inhibition of nutrients uptake via micropinocytosis. Eur J Pharm 2019;850:23-34.
[Crossref] [Google Scholar] [Pub Med]
- International Diabetes Federation. IDF Diabetes Atlas. 9th ed. Diabetes Atlas; 2019.
- Behl T, Kaur I, Kotwani A. Implication of oxidative stress in progression of diabetic retinopathy. Surv Ophthalmol 2016;61:187-96.
[Crossref] [Google Scholar] [Pub Med]
- Yildirim MA, Goh KI, Cusick ME, Barabasi AL, Vidal M. Drug-target network. Nat Biotechnol 2007;25(10):1119-26.
[Crossref] [Google Scholar] [Pub Med]
- Fu JY, Zhang Y. Mechanism of Astragali Radix-Coptis Rhizoma pairin treating type 2 diabetes mellitus based on network pharmacology. Zhongguo Zhong Yao Za Zhi 2021; 46(18):4808-15.
[Crossref] [Google Scholar] [Pub Med]
- Posner BI. Insulin signaling: The inside story. Can J Diabetes 2017;41(1):108-13.
[Crossref] [Google Scholar] [Pub Med]
- American Diabetes Association. Consensus Development Conference on Insulin Resistance. Diabetes Care 1998;21(2):310-14.
- Jung SH, Jung CH, Reaven GM, Kim SH. Adapting to insulin resistance in obesity: Role of insulin secretion and clearance. Diabetologia 2018;61(3):681-7.
[Crossref] [Google Scholar] [Pub Med]
- Payankaulam S, Raicu AM, Arnosti DN. Transcriptional regulation of INSR, the insulin receptor gene. Genes 2019;10(12):984.
[Crossref] [Google Scholar] [Pub Med]
- Chen Y, Huang L, Qi X, Chen C. Insulin receptor trafficking: Consequences for insulin sensitivity and diabetes. Int J Mol Sci 2019;20(20):5007.
[Crossref] [Google Scholar] [Pub Med]
- Zhou L, Zhang J, Fang Q, Liu M, Liu X, Jia W, et al. Autophagy-mediated insulin receptor down-regulation contributes to endoplasmic reticulum stress-induced insulin resistance. Mol Pharmacol 2009;76(3):596-603.
[Crossref] [Google Scholar] [Pub Med]
- Haeusler RA, McGraw TE, Accili D. Biochemical and cellular properties of insulin receptor signaling. Nat Rev Mol Cell Biol 2018;19(1):31-44.
[Crossref] [Google Scholar] [Pub Med]
- Cervello M, Augello G, Cusimano A, Emma MR, Balasus D, Azzolina A, et al. Pivotal roles of glycogen synthase-3 in hepatocellular carcinoma. Adv Biol Regul 2017;65:59-76.
[Crossref] [Google Scholar] [Pub Med]
- Beurel E, Grieco SF, Jope RS. Glycogen synthase kinase-3 (GSK3): Regulation, actions and diseases. Pharmacol Ther 2015;148:114-31.
[Crossref] [Google Scholar] [Pub Med]
- Cross DAE, Alessi DR, Cohen P, Andjelkovich M. Inhibition of glycogen synthase kinase-3 by insulin mediated by protein kinase B. Nature 1995;378(6559):785-9.
[Crossref] [Google Scholar] [Pub Med]
- Amar S, Belmaker RH, Agam G. The possible involvement of glycogen synthase kinase-3 (GSK-3) in diabetes, cancer and central nervous system diseases. Curr Pharm Des 2011;17(22):2264-77.
[Crossref] [Google Scholar] [Pub Med]
- Li L. Study on the relative mechanism of phellodendrine protective effect on ROS-mediated oxidative stress. J Cent South Univ 2017.
- Anfinogenova ND, Quinn MT, Schepetkin IA, Atochin DN. Alarmins and c-Jun N-terminal kinase (JNK) signaling in neuroinflammation. Cells 2020;9(11):9112350.
[Crossref] [Google Scholar] [Pub Med]
- Solinas G, Becattini B. JNK at the crossroad of obesity, insulin resistance and cell stress response. Mol Metab 2017;6(2):174-84.
[Crossref] [Google Scholar] [Pub Med]
- Vernia S, Cavanagh Kyros J, Garcia Haro L, Sabio G, Barrett T, Jung DY, et al. The PPARalpha-FGF21 hormone axis contributes to metabolic regulation by the hepatic JNK signaling pathway. Cell Metab 2014;20(3):512-25.
[Crossref] [Google Scholar] [Pub Med]
- Rehman K, Akash MSH. Mechanisms of inflammatory responses and development of insulin resistance: How are they interlinked? J Biomed Sci 2016;23(1):87.
[Crossref] [Google Scholar] [Pub Med]
- Roohi A, Tabrizi M, Abbasi F, Ataie Jafari A, Nikbin B, Larijani B, et al. Serum IL-17, IL-23 and TGF-beta levels in type 1 and type 2 diabetic patients and age-matched healthy controls. Biomed Res Int 2014;2014:718946.
[Crossref] [Google Scholar] [Pub Med]
- Abdel Moneima A, Bakerya HH, Allam G. The potential pathogenic role of IL-17/Th17 cells in both type 1 and type 2 diabetes mellitus. Biomed Pharmacother 2018;101:287-92.
[Crossref] [Google Scholar] [Pub Med]
- Blüher M. Adipokines-removing road blocks to obesity and diabetes therapy. Mol Metab 2014;3(3):230-40.
[Crossref] [Google Scholar] [Pub Med]
- Benomar Y, Amine H, Crépin D, Rifai SA, Riffault L, Gertler A, et al. Central resistin/TLR4 impairs adiponectin signaling, contributing to insulin and FGF21 resistance. Diabetes 2016;65(4):913-26.
[Crossref] [Google Scholar] [Pub Med]
- Luo J, Qi J, Wang W, Luo Z, Liu L, Zhang G, et al. Antiobesity effect of flaxseed polysaccharide via inducing satiety due to leptin resistance removal and promoting lipid metabolism through the AMP-activated protein kinase (AMPK) signaling pathway. J Agric Food Chem 2019;67(25):7040-9.
[Crossref] [Google Scholar] [Pub Med]
- Huang F, Liu K, Du H, Kou J, Liu B. Puerarin attenuates endothelial insulin resistance through inhibition of inflammatory response in an IKKβ/IRS-1-dependent manner. Biochimie 2012;94(5):1143-50.
[Crossref] [Google Scholar] [Pub Med]
- Fang P, Zhang L, Yu M, Sheng Z, Shi M, Zhu Y, et al. Activiated galanin receptor 2 attenuates insulin resistance in skeletal muscle of obese mice. Peptides 2018;99:92-8.
[Crossref] [Google Scholar] [Pub Med]
- Guerrero Hernández A, Leon Aparicio D, Chavez Reyes J, Olivares Reyes JA, de Jesus S. Endoplasmic reticulum stress in insulin resistance and diabetes. Cell Calcium 2014;56(5):311-22.
[Crossref] [Google Scholar] [Pub Med]
- Zhang XY, Zhang XQ, Mei XH. Research progress on the regulation mechanisms of insulin secretion by plant active ingredients. Sci Technol Food Ind 2021;42(16):412-20.
[Crossref]
- Rorsman P, Braun M. Regulation of insulin secretion in human pancreatic islets. Annu Rev Physiol 2013;75:155-79.
[Crossref] [Google Schoalr] [Pub Med]
- Girgis CM, Cheng K, Scott CH, Gunton JE. Novel links between HIFs, type 2 diabetes and metabolic syndrome. Trends Endocrinol Metab 2012;23(8):372-80.
[Crossref] [Google Scholar] [Pub Med]
- Dinarello CA, Donath MY, Mandrup Poulsen T. Role of IL-1beta in type 2 diabetes. Curr Opin Endocrinol Diabetes Obes 2010;17(4):314-21.
[Crossref] [Google Scholar] [Pub Med]
- He YM, Zhang Q, Zheng M, Fan ZH, Li YH, Zhang D, et al. Protective effects of a G.lucidum proteoglycan on INS-1 cells against IAPP-induced apoptosis via attenuating endoplasmic reticulum stress and modulating CHOP/JNK pathways. Int J Biol Macromol 2018;106:893-900.
[Crossref] [Google Scholar] [Pub Med]
- Wang Y, Peng GQ, Jiang XQ, Song WG, Zhang QL, Wang QB, et al. The protective mechanism of polygona-polysaccharose on rat model of diabetes. Guiding J Trad Chin Med Pharm 2017;23(2):8-16.
- Zhao SF, Fang PH, Shi MY, Zhang ZW. Research progress on the mechanisms of autoimmunity and type 1 diabetes mellitus. Med Recapitulate 2014;20(16):2980-2.
- Bouhanicka B, Cracowskib JL, Failliec JL. Diabetes and COVID-19. Therapies 2020;75:327-33.
- Wu Z, McGoogan JM. Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: Summary of a report of 72 314 cases from the Chinese center for disease control and prevention. JAMA 2020;323(13):239-42.
[Crossref] [Google Scholar] [Pub Med]
- Moradi marjaneh R, Asgharzadeh F, Khordad E, M. Marjaneh M. Diabetes and COVID-19: A review of possible mechanisms. Curr Pharm Des 2021;27(21):2522-7.
[Crossref] [Google Scholar] [Pub Med]
- Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical characteristics of 138 hospitalized patients with 2019 novel coronavirus-infected pneumonia in Wuhan, China. JAMA 2020;323(11):1061-9.
[Crossref] [Google Scholar] [Pub Med]
- Zhao Z, Li Y, Zhou L, Zhou X, Xie B, Zhang W, et al. Prevention and treatment of COVID-19 using Traditional Chinese Medicine: A review. Phytomedicine 2021;85:153308.
[Crossref] [Google Scholar] [Pub Med]
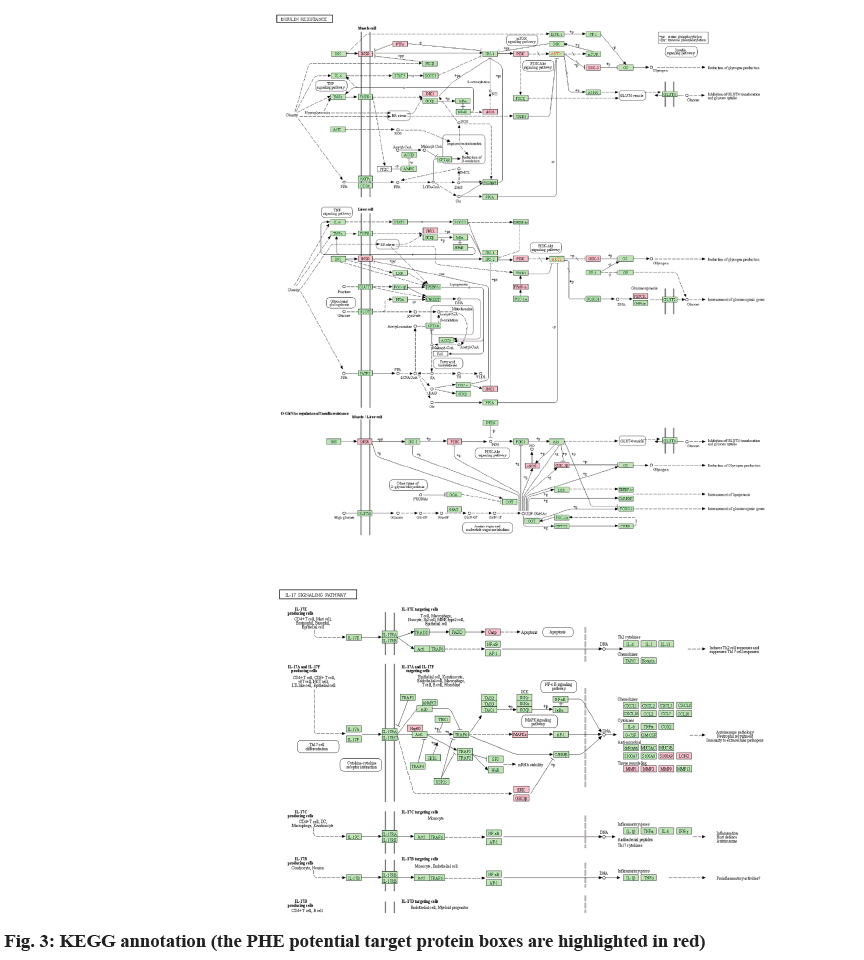
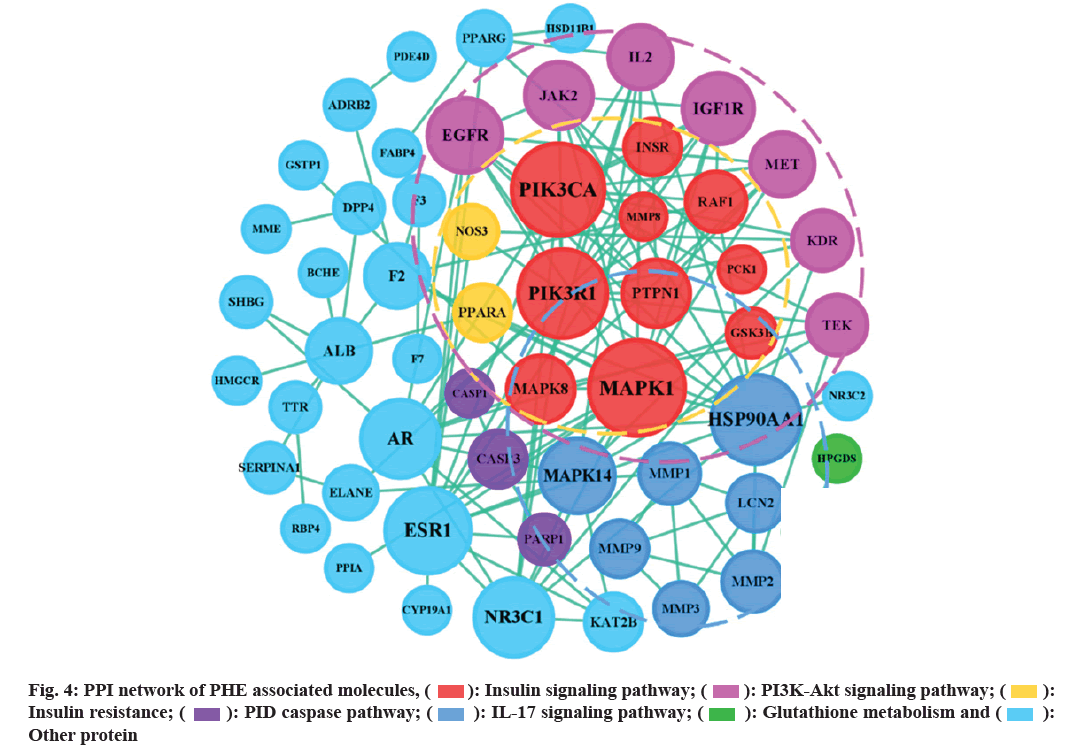
 ): Insulin signaling pathway; (
): Insulin signaling pathway; ( ): PI3K-Akt signaling pathway; (
): PI3K-Akt signaling pathway; ( ):
Insulin resistance; (
):
Insulin resistance; ( ): PID caspase pathway; (
): PID caspase pathway; ( ): IL-17 signaling pathway; (
): IL-17 signaling pathway; ( ): Glutathione metabolism and (
): Glutathione metabolism and ( ):
Other protein
):
Other protein