- *Corresponding Author:
- Yong Shao
Department of Urology, The Second Affiliated Hospital of Harbin Medical University, Nangang, Harbin 150001, China
E-mail: pshy7481@163.com
| This article was originally published in a special issue, “Advanced Targeted Therapies in Biomedical and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2023:85(1) Spl Issue “226-234” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
RAB42, member rat sarcoma oncogene family was proved to play a key role in cancer-promoting progression. However, its function in the progression of bladder cancer was still uncertain. We explored the role of RAB42, member rat sarcoma oncogene family in bladder cancer and pan-cancer using bioinformatics analysis, including expression, prognostic, genetic alteration, functional pathways, tumor microenvironment and drug resistance. Our results indicated that RAB42, member rat sarcoma oncogene family expression was increased in most cancers, including bladder cancer and that its expression generally indicated poorer patient prognosis. In addition, RAB42, member rat sarcoma oncogene family expression was positively associated with most immune cells, such as tumor-associated macrophages and Tregs. Elevated expression of RAB42, member rat sarcoma oncogene family predicted resistance to several anti-tumor drugs. In summary, our study showed that RAB42, member rat sarcoma oncogene family is a potential oncogene and prognostic marker in bladder cancer and pan-cancer. Elevated RAB42, member rat sarcoma oncogene family expression indicated tumor immunosuppressive microenvironment.
Keywords
RAB42, member rat sarcoma oncogene family, bladder cancer, pan-cancer, prognostic marker
RAB42, member of the Rat Sarcoma (RAS) oncogene family (RAB42) was reported to involve in metabolism-related cancer-promoting pathways[1]. A recent study showed that RAB42 is a prognostic risk factor for gliomas and could serve as a potential target for the treatment of glioma patients[2]. Furthermore, RAB42 is associated with multiple malignant clinical features and can promote glioma proliferation, migration and invasion by activating the Vascular Endothelial Growth Factor (VEGF) signaling pathway[3]. However, the roles and mechanisms of RAB42 in prognosis, progression, genetic alterations and Tumor Microenvironment (TME) of Bladder Cancer (BLCA) have not been investigated.
BLCA is a worldwide tumor, which may lead to more than 170 000 death each year[4]. The main treatment for BLCA is transurethral resection of bladder tumor. The 5 y survival rate after first-line therapy for patients with non-metastatic BLCA is only 36 %-48 %, while the 5 y relative survival rate for metastatic BLCA is 5 %-36 %[5]. Therefore, new prognostic markers and therapeutic targets are required to elevate the treatment and survival of BLCA patients.
In this work, we assessed the expression pattern of RAB42 and its relationship with BLCA prognosis and its possible biological functions through bioinformatics analysis. The association of RAB42 and TME were deeply explored. Our work revealed that RAB42 may be a potential target and prognostic marker in BLCA.
Materials and Methods
Data sources:
The messenger Ribonucleic Acid (mRNA) expression profiles and clinical data of 33 cancer and normal tissues were obtained from the UCSC Xena (https://xenabrowser.net/datapages/) website. The genetic information of RAB42 was obtained from cBioPortal database (http://www.cbioportal.org/).
RAB42 expression:
To evaluate the expression of RAB42 in pan-cancer, we collected tumor tissues of 33 cancers from The Cancer Genome Atlas (TCGA) database and the normal tissues from the TCGA and Genotype-Tissue Expression (GTEx) database.
Prognostic analysis of RAB42:
Univariate Cox regression (uniCox) analysis of RAB42 in pan-cancer was conducted and visualized using R package “survminer” and “survival”.
Functional enrichment analysis:
Gene Set Variation Analysis (GSVA) and Gene Set Enrichment Analysis (GSEA) analysis was performed using the R package “clusterProfiler” to explore the potential biological functions of RAB42 in BLCA, including results based on Hallmark, Gene Ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) and Reactome databases.
TME analysis:
We downloaded the immune infiltration data from ImmuCellAI and Tumor Immune Estimation Resource 2 (TIMER2) database. The ImmuCellAI database uses the “single sample GSEA (ssGSEA)” method to assess the content of immune cells in tumors. TIMER2 database collected the immune infiltration results calculated using 6 algorithms, including TIMER, CIBERSORT, quanTIseq, xCell, Microenvironment Cell Populations-counter (MCP-counter) and EPIC methods.
Statistical analysis:
R software (version 4.1.1) was used to perform bioinformatics analysis, statistics and visualization. Correlations were calculated using the Pearson test. The comparison between the two groups of samples was performed using the t-test, p<0.05 was considered statistically significant.
Results and Discussion
RAB42 expression analysis was shown in fig. 1 and fig. 2. We explored the differential expression of RAB42 in 33 cancers. Results indicated that the expression of RAB42 in tumor tissues was higher than that in normal tissues in 24 tumors, including BLCA (fig. 1A). For tumor tissues, RAB42 expression was highest in Thymoma (THYM) and lowest in Pheochromocytoma and Paraganglioma (PCPG) (fig. 1B). For normal tissues, RAB42 expression was highest in spleen and lowest in muscle (fig. 1C). We next evaluated the expression of RAB42 in paired tumor and adjacent normal tissues and in different tumor stage using TCGA data. We found that RAB42 expression was higher in 8 tumors, such as BLCA, Cholangiocarcinoma (CHOL), Breast Cancer gene (BRCA), Head and Neck Squamous Cell Carcinoma (HNSC), Kidney Renal Clear Cell Carcinoma (KIRC), Kidney Renal Papillary Cell Carcinoma (KIRP), Lung Adenocarcinoma (LUAD) and Stomach Adenocarcinoma (STAD), and lower in Colon Adenocarcinoma (COAD), Rectum Adenocarcinoma (READ) and Thyroid Carcinoma (THCA) (fig. 2A). In addition, RAB42 expression was higher in stage III or stage IV than that in stage I or stage II in BLCA, Liver Hepatocellular Carcinoma (LIHC), Lung Squamous Cell Carcinoma (LUSC), Uterine Corpus Endometrial Carcinoma (UCEC) and Uterine Carcinosarcoma (UCS). These results suggested that RAB42 may play an important role in the BLCA and other tumors (fig. 2B).
Genetic alteration of RAB42 was shown in fig. 3. We further assessed the genetic alteration of RAB42 and found that the genetic alteration frequency of RAB42 was low in BLCA, in which “Amplification” occupies the largest proportion (fig. 3A). We also calculated the correlation of RAB42 with Copy Number Alteration (CNA) and methylation level in pan-cancer. We found that RAB42 expression was positively linked to its CNA level (fig. 3B) and negatively linked to methylation level in the promoter of RAB42 (fig. 3C).
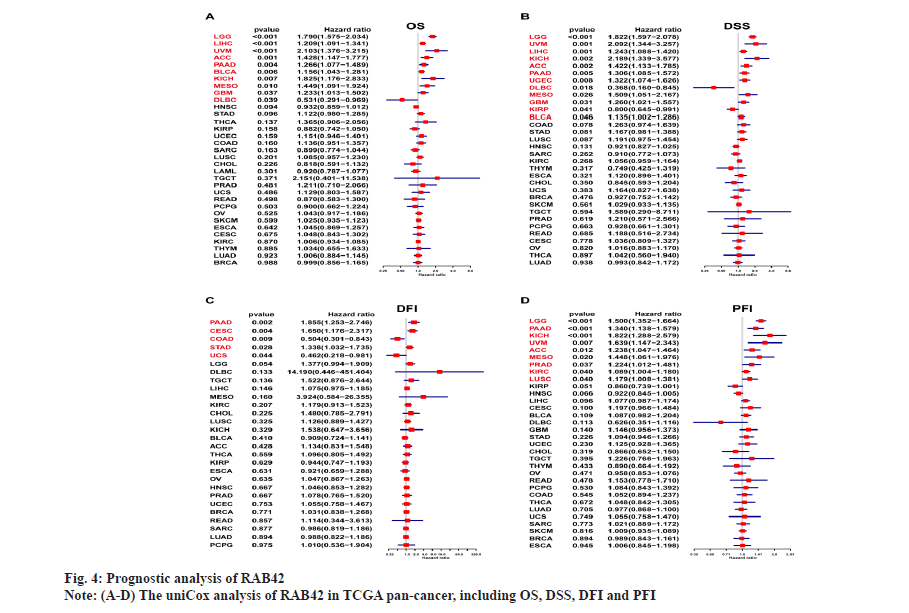
The prognostic role of RAB42 in pan-cancer was shown in fig. 4. To further study the prognostic role of RAB42 in pan-cancer, we used forest plot to display the results of uniCox analysis of RAB42 in pan-cancer, including Overall Survival (OS), Disease-Specific Survival (DSS), Disease-Free Interval (DFI) and Progression-Free Interval (PFI). Our results showed that RAB42 is a risk factor of OS in Low-Grade Glioma (LGG), LIHC, Uveal Melanoma (UVM), Adrenocortical Carcinoma (ACC), Pancreatic Adenocarcinoma (PAAD), BLCA, Kidney Chromophobe (KICH), Mesothelioma (MESO) and Glioblastoma Multiforme (GBM), and a favorable factor of OS in Diffuse Large B Cell Lymphoma (DLBCL) (fig. 4A). For DSS results, RAB42 is a risk factor LGG, UVM, LIHC, KICH, ACC, PAAD, UCEC, MESO, GBM, KIRP and BLCA, and a favorable factor in DLBC (fig. 4B). RAB42 is an unfavorable factor for DFI in PAAD, Cervical Squamous Cell Carcinoma (CESC) and STAD, and a favorable factor in COAD and UCS (fig. 4C). For PFI results, RAB42 act as a risk factor in LGG, PAAD, KICH, UVM, ACC, MESO, PRAD, KIRC and LUSC (fig. 4D).
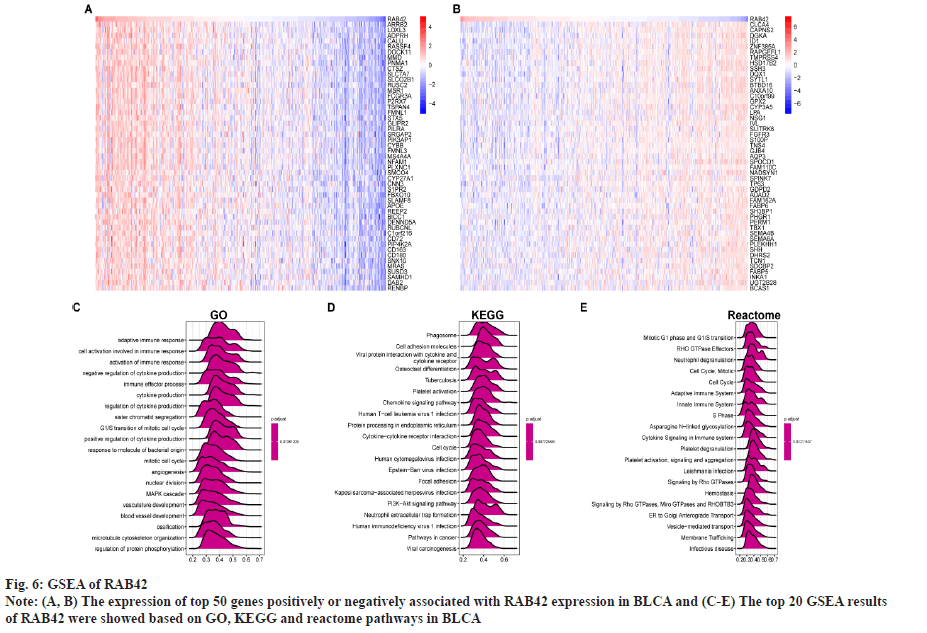
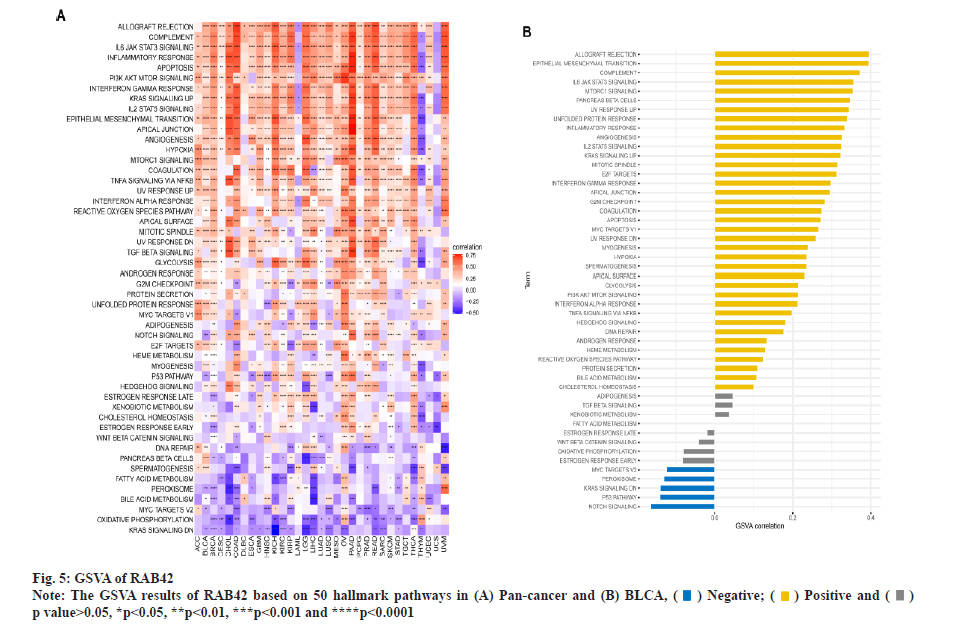
Functional analysis of RAB42 was shown here. To evaluate the possible functional pathways of RAB42 in BLCA, we performed GSVA and GSEA of RAB42 in BLCA and pan-cancer. GSVA results indicated that RAB42 was positively correlated with immune-related cancer-promoting malignant pathways, such as epithelial mesenchymal transition, Interleukin (IL)-6/Janus Kinase (JAK)/Signal Transducer and Activator of Transcription (STAT) 3 signaling, mammalian Target of Rapamycin Complex 1 (mTORC1) signaling, inflammatory response, angiogenesis, IL-2/STAT5 signaling and interferon gamma response pathways in BLCA and pan-cancer (fig. 5A and fig. 5B). Next, we conducted correlation analysis of RAB42 with all mRNAs in BLCA and performed GSEA based on correlation analysis. Fig. 6A and fig. 6B displayed the expression of top 50 genes which were positively or negatively associated with RAB42. GO-based GSEA analysis showed that RAB42 was mainly associated with immune-related biological processes, such as adaptive immune response and immune effector process (fig. 6C). GSEA based on the KEGG pathway showed that RAB42 was mainly associated with phagosome, cell adhesion molecules, chemokine signaling pathway and Phosphoinositide-3-Kinase (PI3)-Protein Kinase B (Akt) signaling pathway (fig. 6D). Reactome pathway-based GSEA indicated that RAB42 was mainly associated with adaptive immune system and innate immune system. These results suggested that RAB42 have close association with tumor immune microenvironment.
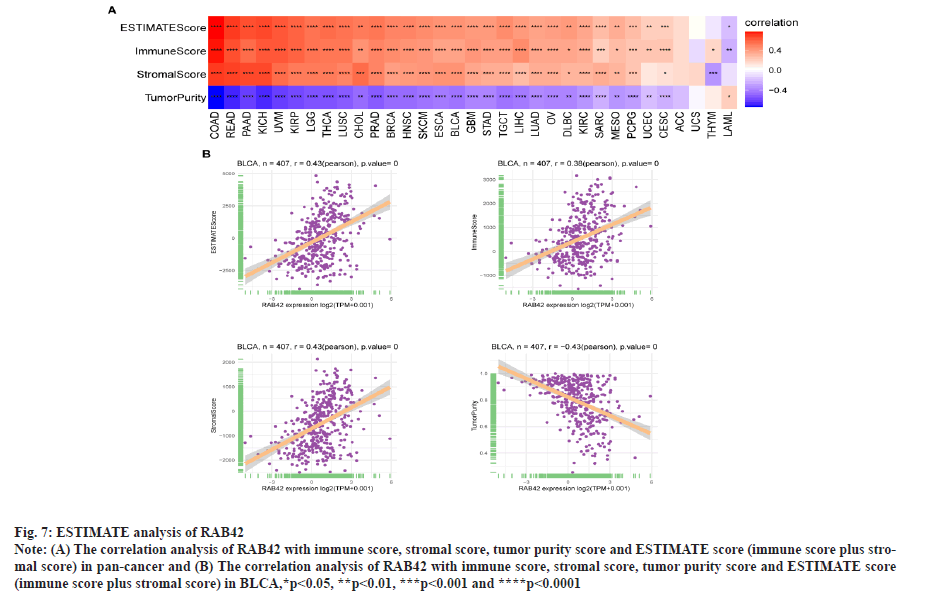
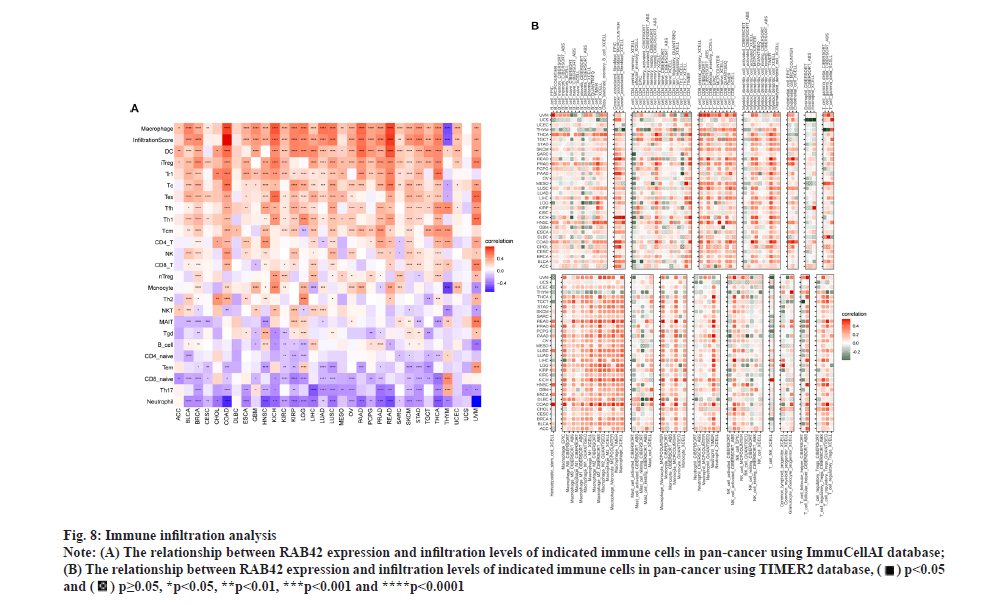
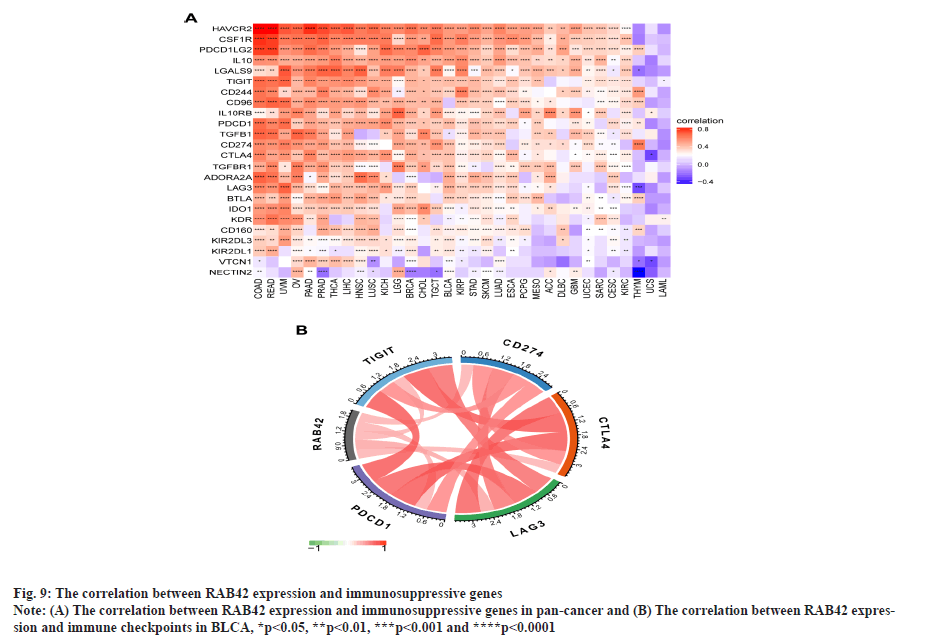
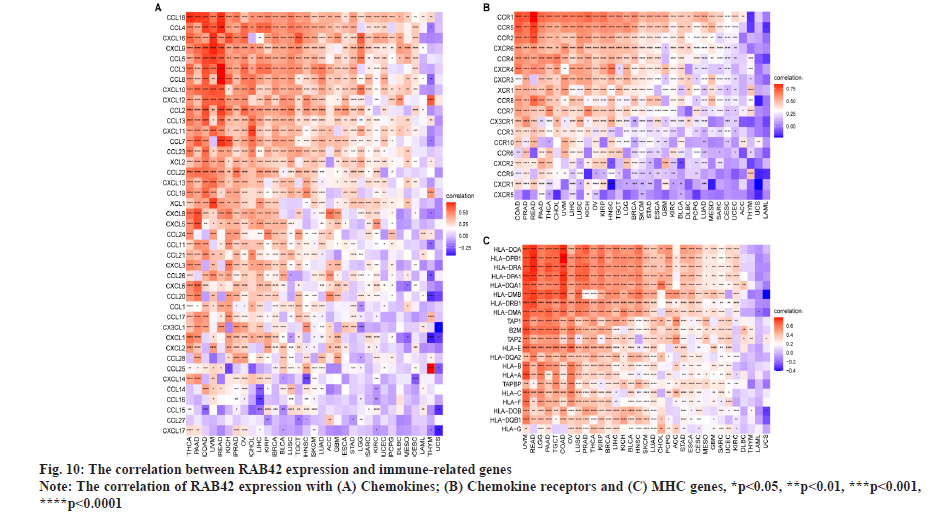
RAB42 is associated with immune cell infiltration in BLCA. Given that RAB42 is associated with immune-related pathways in BLCA, we further explore the relationship between RAB42 and TME. We calculated the TME score using R package “Estimation of Stromal and Immune cells in Malignant Tumours using Expression data’ (ESTIMATE)”, including immune score, stromal score and tumor purity score. We found that RAB42 have positive correlation with immune and stromal score and negative correlation with tumor purity score in BLCA and pan-cancer (fig. 7A and fig. 7B). We then explored the relationship between RAB42 and 24 types of immune cell infiltration in each cancer using immune infiltration data from the ImmuCellAI database. The results showed that RAB42 was positively correlated with most immune cells, such as macrophages and regulatory T cells (Tregs) (fig. 8A). We further verified the results that RAB42 was positively correlated with most immune cells using immune infiltration data from TIMER2 database (fig. 8B). We have proved the association of RAB42 with most immune cells. We further assessed the potential relationship of RAB42 with immune-related genes. We found that RAB42 expression was positively related with immunosuppressive genes (fig. 9A), immune checkpoints (fig. 9B), chemokines (fig. 10A), chemokine receptors (fig. 10B) and Major Histocompatibility Complex (MHC) genes (fig. 10C) in BLCA and pan-cancer. Finally, our results demonstrated that RAB42 is a potential regulator of tumor immune microenvironment.
Fig. 7: ESTIMATE analysis of RAB42.
Note: (A) The correlation analysis of RAB42 with immune score, stromal score, tumor purity score and ESTIMATE score (immune score plus stromal
score) in pan-cancer and (B) The correlation analysis of RAB42 with immune score, stromal score, tumor purity score and ESTIMATE score
(immune score plus stromal score) in BLCA,*p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001.
Fig. 8: Immune infiltration analysis.
Note: (A) The relationship between RAB42 expression and infiltration levels of indicated immune cells in pan-cancer using ImmuCellAI database;
(B) The relationship between RAB42 expression and infiltration levels of indicated immune cells in pan-cancer using TIMER2 database, 
 *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001.
*p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001.
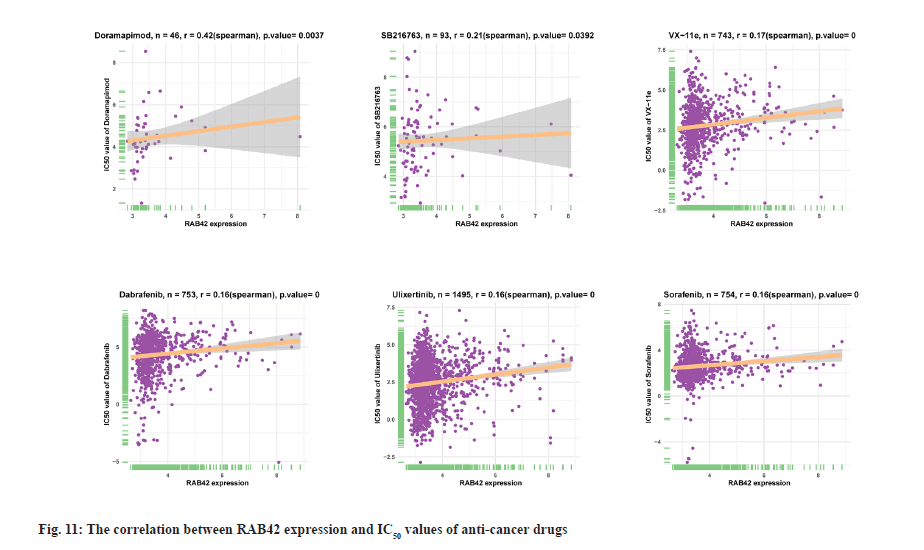
Drug resistance analysis of RAB42 was shown here. In addition, we conducted the association analysis of RAB42 with Half-Maximal Inhibitory Concentration (IC50) of anti-cancer drugs from Genomics of Drug Sensitivity in Cancer (GDSC) database. Among the 192 anti-cancer drugs, RAB42 expression was positively correlated with IC50 of 109 anti-cancer drugs, such as doramapimod, SB216763, VX-11e, dabrafenib, ulixertinib and sorafenib (fig. 11), indicating that patients with high expression of RAB42 may be resistant to the treatment of these anti-cancer drugs.
BLCA is the most common malignant tumor in women and the fourth most common malignant tumor in men. It represents a range of diseases ranging from chronically treated recurrent non-aggressive tumors, to aggressive or advanced disease requiring multimodal and aggressive treatment. Advances in the understanding of the underlying biology of BLCA have fundamentally changed how the disease is diagnosed and treated[5]. RAB42 is a member of the RAS oncogene family, which has been reported to be involved in metabolism-related cancer-promoting pathways[1]. Recent studies showed that RAB42 is a prognostic risk factor and a potential treatment target in hepatocellular carcinoma and glioblastoma[6,7]. However, the roles and mechanisms of RAB42 in prognosis, progression, genetic alterations and TME of BLCA have not been investigated. Our results show that RAB42 is over-expressed in 24 cancers. Increased expression in BLCA predicted worse tumor stages and poor survival of BLCA patients, suggesting that RAB42 may be a potential biomarker for predicting cancer progression. Our study is the first to reveal that RAB42 is a potential oncogene and biomarker in BLCA. GSVA and GSEA results indicated that RAB42 was positively correlated with immune-related cancer-promoting malignant pathways, such as epithelial mesenchymal transition, IL-6/JAK/STAT3 signaling, mTORC1 signaling, inflammatory response, angiogenesis, IL-2/STAT5 signaling and interferon gamma response pathways in BLCA and pan-cancer, suggesting that RAB42 have close association with tumor immune microenvironment. Immune cells in the TME have an essential role in tumor progression[8,9]. On the one hand, some infiltrating immune cells such as cytotoxic T cells and Natural Killer (NK) cells can play a role in killing tumor cells. On the other hand, some immunosuppressive cells such as macrophage and Treg cells can play a role in promoting tumor progression[10,11]. Given that our enrichment analysis results showed that RAB42 is associated with immune processes, we used two different approaches to investigate the role of RAB42 in immune cell infiltration in TME of BLCA. Our results suggested that RAB42 expression is positively related to most immune cells, such as macrophages and Tregs based on immune cell infiltration data from ImmuCellAI and TIMER2 database.
In addition, we found that RAB42 expression was positively related with immunosuppressive genes, immune checkpoints, chemokines, chemokine receptors and MHC genes in BLCA and pan-cancer. Additionally, we also revealed that patients with high expression of RAB42 may be resistant for the treatment of anti-cancer drugs, such as doramapimod, SB216763, VX-11e, dabrafenib, ulixertinib and sorafenib. In conclusion, our results demonstrated that RAB42 is a potential prognostic marker associated with tumor immune microenvironment in BLCA.
Author’s contributions:
Yong Shao conceived and designed the study. Yunhui Chan and Rong Zhao contributed to the writing of the manuscript. Yuxin Zhu supervised the study. All authors have reviewed the manuscript.
Conflict of interests:
The authors declared no conflict of interest.
References
- He X, Xu C. Immune checkpoint signaling and cancer immunotherapy. Cell Res 2020;30(8):660-9.
[Crossref] [Google scholar] [PubMed]
- Zhang GH, Zhong QY, Gou XX, Fan EX, Shuai Y, Wu MN, et al. Seven genes for the prognostic prediction in patients with glioma. Clin Transl Oncol 2019;21(10):1327-35.
[Crossref] [Google scholar] [PubMed]
- Liu B, Su Q, Xiao B, Zheng G, Zhang L, Yin J, et al. RAB42 promotes glioma pathogenesis via the VEGF signaling pathway. Front Oncol 2021;11:1-13.
[Crossref] [Google scholar] [PubMed]
- Wang G, Gao Y, Chen Y, Wang K, Zhang S, Li G. Identification of novel tumor antigens and the immune landscapes of bladder cancer patients for mrna vaccine development. Front Oncol 2022;12:1-16.
[Crossref] [Google scholar] [PubMed]
- Lenis AT, Lec PM, Chamie K. Bladder cancer: A review. JAMA 2020;324(19):1980-91.
[Crossref] [Google scholar] [PubMed]
- Peng H, Du X, Zhang Y. RAB42 is a potential biomarker that correlates with immune infiltration in hepatocellular carcinoma. Front Mol Biosci 2022;9:1-21.
[Crossref] [Google scholar] [PubMed]
- Sun L, Yan T, Yang B. The progression related gene RAB42 affects the prognosis of glioblastoma patients. Brain Sci 2022;12(6):1-12.
[Crossref] [Google scholar] [PubMed]
- Magrini E, Minute L, Dambra M, Garlanda C. Complement activation in cancer: Effects on tumor-associated myeloid cells and immunosuppression. Semin Immunol 2022;60:101642.
[Crossref] [Google scholar] [PubMed]
- Liu C, Zhou X, Long Q, Zeng H, Sun Q, Chen Y, et al. Small extracellular vesicles containing miR-30a-3p attenuate the migration and invasion of hepatocellular carcinoma by targeting SNAP23 gene. Oncogene 2021;40(2):233-45.
[Crossref] [Google scholar] [PubMed]
- Payandeh Z, Khalili S, Somi MH, Mard‐Soltani M, Baghbanzadeh A, Hajiasgharzadeh K, et al. PD‐1/PD‐L1‐dependent immune response in colorectal cancer. J Cell Physiol 2020;235(7):5461-75.
[Crossref] [Google scholar] [PubMed]
- Malka D, Lièvre A, André T, Taïeb J, Ducreux M, Bibeau F. Immune scores in colorectal cancer: Where are we? Eur J Cancer 2020;140:105-18.
[Crossref] [Google scholar] [PubMed]
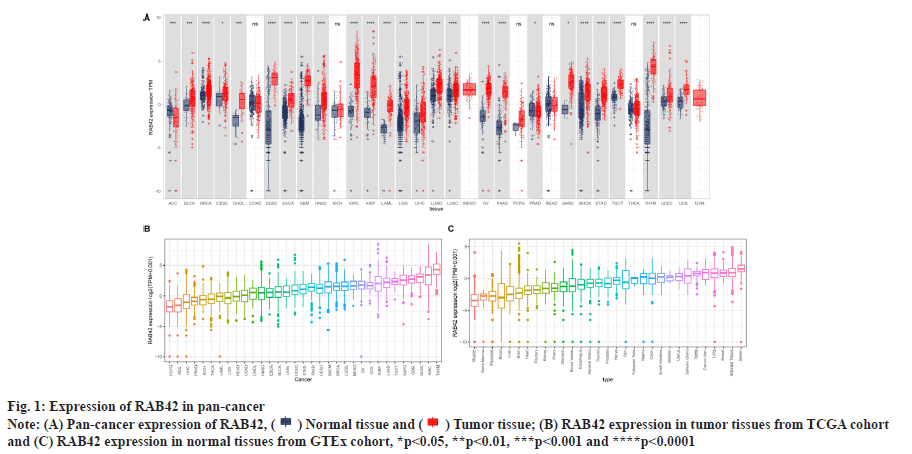
 (B) RAB42 expression in tumor tissues from TCGA cohort
and (C) RAB42 expression in normal tissues from GTEx cohort, *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001.
(B) RAB42 expression in tumor tissues from TCGA cohort
and (C) RAB42 expression in normal tissues from GTEx cohort, *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001.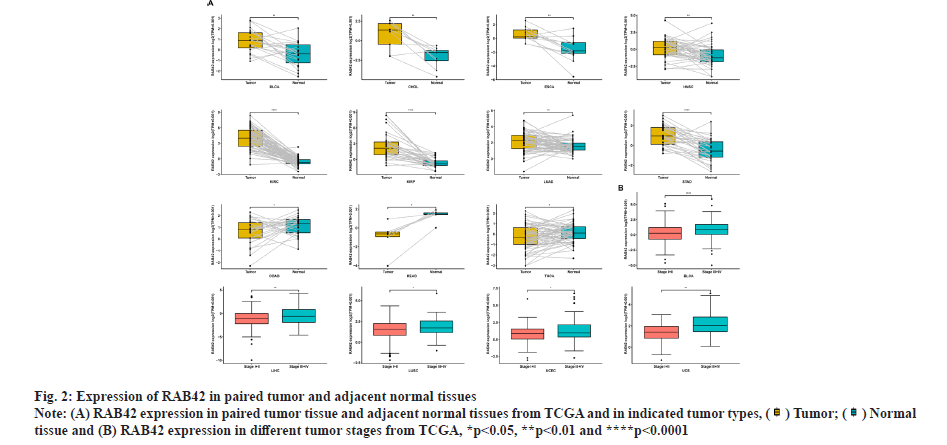
 tissue and (B) RAB42 expression in different tumor stages from TCGA, *p<0.05, **p<0.01 and ****p<0.0001.
tissue and (B) RAB42 expression in different tumor stages from TCGA, *p<0.05, **p<0.01 and ****p<0.0001.
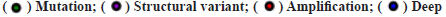 deletion; (B) The correlation of RAB42 expression and CNA level in pan-cancer,
deletion; (B) The correlation of RAB42 expression and CNA level in pan-cancer,  and (C) The
correlation of RAB42 expression with Deoxyribonucleic Acid (DNA) methylation level in pan-cancer
and (C) The
correlation of RAB42 expression with Deoxyribonucleic Acid (DNA) methylation level in pan-cancer  .
.
 p value>0.05, *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001.
p value>0.05, *p<0.05, **p<0.01, ***p<0.001 and ****p<0.0001.