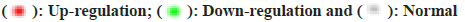- *Corresponding Author:
- Yinghui Liu
Department of Traditional Chinese Medicine, Changchun University of Traditional Chinese Medicine, Changchun, Jilin Province 130117, China
E-mail: ntxingjl@163.com
| This article was originally published in a special issue, “Recent Progression in Pharmacological and Health Sciences” |
| Indian J Pharm Sci 2024:86(2) Spl Issue “202-209” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Aortic dissection is a serious cardiovascular emergency with a very high early mortality rate, so it is particularly important to develop new targeted therapies to reduce prevalence and improve survival, and we hope to explore potential biomarkers involved in the development of aortic dissection and available pharmaceutical components sensitive to therapeutic targets through bioinformatics technology. First, download the gene expression omnibus dataset (GSE190635), use the affy software package of the R software for data standardization, the limma software package to screen the differential genes, and use the relevant software package to draw the volcano map and heat map of the differential gene; then database for annotation, visualization, and integrated discovery software was used for gene ontology and Kyoto encyclopedia of genes and genomes pathway enrichment analysis. Next, the protein-protein interaction network was further constructed, and the STRING exported data was visualized using Cytoscape software, and finally the hub gene was extracted using cytoHubba and MCODE plugins. Finally, the Coremine medical information retrieval platform was used to screen for possible therapeutic drugs. Through differentially expressed genes analysis, 45 up-regulated differentially expressed genes and 132 down-regulated differentially expressed genes were successfully distinguished from the GSE79768. They are mainly enriched in positive regulation of urine volume, cell activation, extracellular region, lipopeptide binding and nucleotide oligomerization domain-like receptor signaling pathway, etc. Through protein-protein interaction, the top 7 hub genes were ranked, including early growth response protein 1, C-X-C motif chemokine ligand 8, fructo-oligosaccharides, filamin C, toll-like receptor 2, identified integrin alpha 7, myosin light chain-2. Coremine medical was used to retrieve a variety of related drugs, among which recombinant interleukin-6, recombinant tumor necrosis factor family protein, lipopolysaccharide and tyrosine co-regulated the above genes. In this study, 7 differentially expressed genes were identified as potential biomarkers for aortic dissection patients, which may provide new targets for aortic dissection therapy. But this is just the beginning, and we need to carry out experiments to verify the accuracy and feasibility of these results in the future.
Keywords
Aortic dissection, bioinformatics, biomarkers, differentially expressed gene, protein-protein interaction
Acute Aortic Dissection (AD), in which blood from the lumen of the aorta enters the lining of the aorta from a tear in the intima of the aorta and may expand along the long axis of the aorta, resulting in separation of the true and false lumen of the aorta, a cardiovascular emergency with a high risk of death[1]. Most patients present with acute onset of symptoms, including sudden onset of severe pain, shock, and hematoma compression of the corresponding aortic branch, and are easily misdiagnosed due to the complexity of the clinical presentation[2]. The early mortality rate of the disease is extremely high, many patients have died before admission to the hospital, so the epidemiological data is not sufficient, studies show that the incidence in the American population is 4.6/100 000/y [3], from 2012 to 2019, the mortality rate of AD has increased year by year[4], studies have shown that the occurrence of the disease is related to genetics, ancestry, pregnancy and adverse cardiovascular events, and modifiable risk factors include hypertension, dyslipidemia, aortitis, obstructive sleep apnea, diabetes and cocaine[5]. As the world's demographics change, the incidence of AD may increase, because the risk of morbidity is generally higher in the elderly, so the social medical burden it brings will become heavier[6]. There is growing evidence acknowledging the severity of the disease[7].
The risk of death from drug therapy alone remains high in treatment, and surgical treatment and endovascular repair can significantly reduce the incidence of the disease, but the improvement of longterm quality of life still needs to be improved[8-10], so the intervention has to be further advanced, from early treatment to early diagnosis, over the years, doctor’s diagnostic tools and management methods for the disease have continued to evolve, from the initial electrocardiogram, chest X-ray to echocardiography to computed tomography[11,12], Although it effectively reduces the missed diagnosis rate, it cannot identify and accurately prevent the potential risk of AD in advance, and the risk of death with AD increases by 1 % to 3 % for each additional hour[13]. Therefore, a high degree of clinical suspicion is required for early diagnosis and targeted therapy to control disease progression in time and improve the chance of longterm survival.
Medical researchers need to explore more reliable biomarkers for diagnosis and treatment, and bioinformatics technology has opened the door to several long noncoding Ribonucleic Acid (RNAs) and circular RNAs that have been found to be expressed differently in AD samples, suggesting their potential role in vascular physiology and disease[13]. One untargeted metabolomics study identified succinic acid as a biomarker and therapeutic target for AD[14], and another used a multiomics approach to collect data and attempt to use a systems biology approach to dissect the underlying molecular mechanisms of AD for low-budget, accessible, highly sensitive, and specific biomarkers[15]. It can be seen that there is still a lot of room for mining of bioinformatics data related to AD, so we carried out this study in the hope of helping to explore reliable molecular biomarkers and potential therapeutic targets for AD.
Materials and Methods
Ethical statement:
Ethical approval is not applicable for this study because the datasets used are all from open databases. The Institute’s application network software and data can be collected as shown in Table 1.
| S. No | Database/software | Website | Feature comments |
|---|---|---|---|
| 1 | GEO | https://www.ncbi.nlm.nih.gov/geo/ | Experiment set, verification set chip download |
| 2 | R 4.0.3 | https://www.r-project.org/ | Statistical analysis and differential gene screening |
| 3 | DAVID | https://david.ncifcrf.gov/ | GO and KEGG pathway enrichment |
| 4 | STRING 11.0 | https://string-db.org | PPI network construction |
| 5 | Cytoscape 3.7.2 | https://cytoscape.org/ | Key gene screening and visualization |
| 6 | Coremine medical | https://www. coremine. com/medical/ | Targeted drug/ingredient screening |
Table 1: Databases/Software and Their Websites As Well As Functions
Microarray data source and information:
The entire microarray data used in this study was sourced from the Gene Expression Omnibus (GEO) database, a public functional genomics data repository. The search terms were atrial fibrillation, Homo sapiens, and expression profiling by array. After screening, the gene expression profile data provided by Wang and Fan from Fuwai Hospital were selected for GSE190635. The GSE190635 data series included 8 samples (4 AD samples and 4 healthy control samples). Affymetrix HG-U133 plus 2.0 GeneChip was used to determine genome-wide expression profiles of aortic tissue samples from patients with AD and healthy controls.
Screening of Differentially Expressed Genes (DEGs) and correlation analysis:
First, use the Affy package in R 4.2.2 software for data standardization preprocessing and normalization (fig. 1a and fig. 1b), and according to the platform annotation information, first convert the probe id into the gene serial number gene id, and then convert the gene id to gene symbol and save it. The Limma package was then applied to log2 transformation and DEGs filtering of the GSE190635 matrix data[16,17]. The Benjamini and Hochberg false discovery rate methods were used to correct for false positives. Set the statistically significant difference threshold to |log2FC|>1 and p<0.01 as the statistical significance threshold level for the DEGs sample[18,19].
Gene Ontology (GO) and Kyoto Encyclopedia of Genes Genomes (KEGG) pathway enrichment analysis:
To explore the function of differential genes, the online bioinformatics tools Database for Annotation, Visualization, and Integrated Discovery (DAVID) were used to annotate the functions of GO and KEGG pathway enrichment analysis[20-22].
Protein-Protein Interaction (PPI) network and module analysis:
To further understand the interaction of differential genes, a PPI analysis was performed using the online database String 11.5 to set a high confidence >0.700 and species limited to Homo sapiens, after removing discrete nodes, we chose Cytoscape 3.8.2 software as the visualization tool for PPI networks. Next, we used Cytoscape Software's Molecular Complex Detection (MCODE) to screen key modules of the PPI network (degree cut-off=2, node score cut-off=0.2, k-core=2, and maximum depth=100)[23].
Determination of hub genes:
We chose Cytoscape’s cytoHubba plugin to identify key genes in the network named hub gene. The top hub genes were selected and sorted according to the topological algorithm[24].
Analysis of drugs for early prevention and treatment of AD:
Medical Ontology Information Retrieval Platform (https://www. coremine. com/medical/)-Coremine medical is an input into human gene/protein to retrieve diseases, biological processes, cellular composition, molecular function, possible therapeutic drugs, etc. The core genes screened in the early stage were entered into the Coremine medical search box and screened for potential therapeutic drugs with p<0.05 as the criterion[25]. Finally, drugs suitable for AD prevention and treatment targets were screened.
Results and Discussion
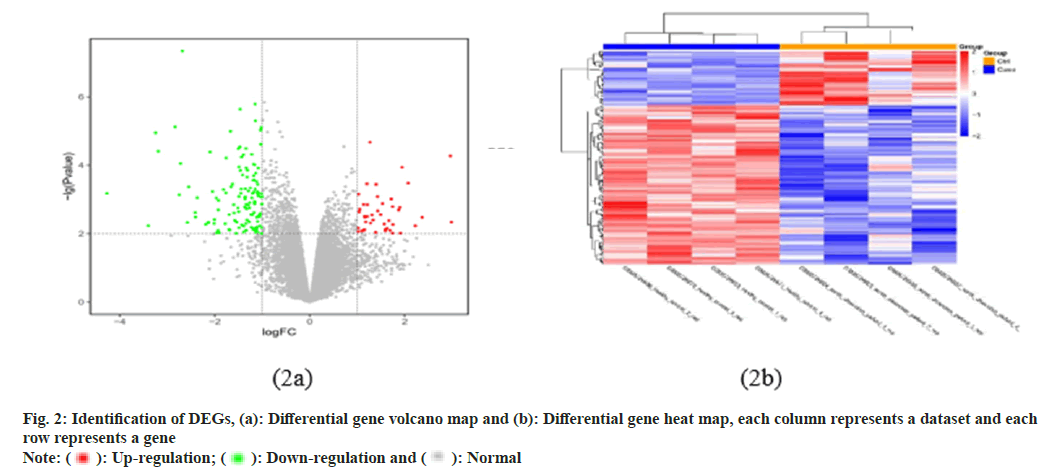
In this study, a total of 54 675 probes were identified, corresponding to 20 824 genes. There were 177 genes that differed between samples from patients with AD and healthy controls, including 45 upregulated DEGs and 132 down-regulated DEGs. We plotted volcano maps (fig. 2a) and heat maps (fig. 2b) to show differential genes, where blue indicate down-regulated gene expression, red indicates upregulation, and gray indicates the rest of the DEG. In addition, expression levels and aggregation states of all DEGs are shown in the heat map (fig. 2b).
Through analysis we obtained the first 10 GO molecular functional terms, cellular component terms, bioprocess terms, and KEGG pathways (fig. 3a and fig. 3b). According to the p value, the regulation of biological processes by DEGs is mainly reflected in the positive regulation of urine output, cell activation, cardiac muscle hypertrophy, cardiac muscle tissue morphogenesis, commissural neuron axon guidance and cellular response to parathyroid hormone stimulus, in terms of cellular components, DEGs significantly enriched Z disc, extracellular region, actin filament bundle, sarcoplasmic reticulum lumen and cell projection, and for MF analysis, significantly enriched terms are ion binding, lipopeptide binding, cytokine activity, calcium-release channel activity and identical protein binding. In addition, as shown in fig. 3b, the KEGG enrichment pathway for DEGs includes; Gonadotropin-Releasing Hormone (GnRH) secretion, legionellosis, Nucleotide Oligomerization Domain (NOD)-like receptor signaling pathway, Transforming Growth Factor Beta (TGF-β) signaling pathway and parathyroid hormone synthesis, secretion and action.
Integrate rich terminology into the network based on gene ID and p value. We use a circle map to show the results (fig. 3a and fig. 3b). The circle map is composed of 4 concentric circles, the outermost circle reflects the classification of enrichment, and the coordinate bar outside the circle represents the number of genes, yellow part represents molecular function, pink part represents cellular component, purple part represents biological process; the longer the bars in the second circle indicate more background genes, the redder the color indicates a smaller p value, and the third circle is the total number of foreground genes. The innermost circles represent their respective rich factor values, and the cells on a guide line represent 0.1
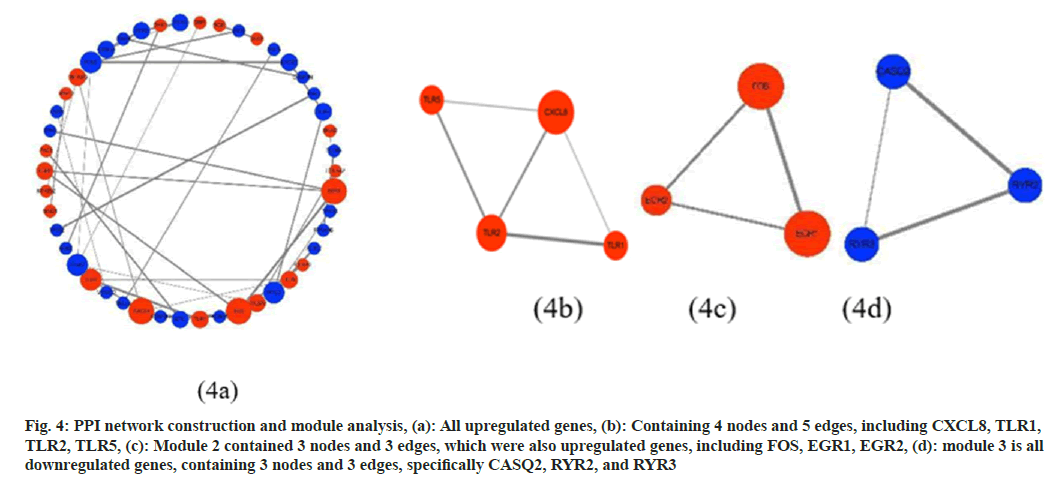
To investigate the association between DEGs, PPI networks were established using the STRING database. Using Cytoscape software to visualize the network (fig. 4a), the network with a total of 170 nodes and 53 edges, and after removing discrete nodes, fig. 4a retains a high degree of 46 nodes and 37 edges, including 20 upregulated genes and 26 downregulated genes. In addition, the analysis of the Cytoscape molecular complex detection plugin showed that in the PPI network of DEG, a total of 3 key modules were found, all of which were upregulated genes (fig. 4b-fig. 4d).
Fig. 4: PPI network construction and module analysis, (a): All upregulated genes, (b): Containing 4 nodes and 5 edges, including CXCL8, TLR1, TLR2, TLR5, (c): Module 2 contained 3 nodes and 3 edges, which were also upregulated genes, including FOS, EGR1, EGR2, (d): module 3 is all downregulated genes, containing 3 nodes and 3 edges, specifically CASQ2, RYR2, and RYR3
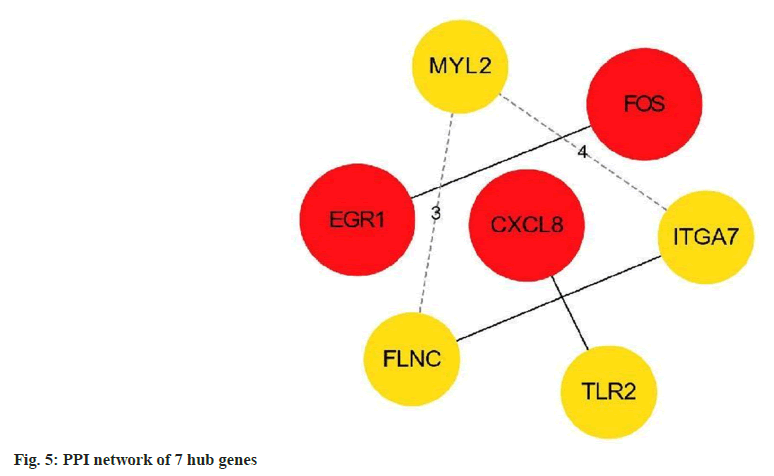
Finally, using the Cytoscape version 3.8.2 plugin cytoHubba, according to the degree value, filter out the top 7 hub DEGs and plot the interaction network between these 7 hub DEGs (fig. 5).
The results showed that Early Growth Response 1 (EGR1), C-X-C Motif Chemokine Ligand 8 (CXCL8) and Fructo-Oligosaccharides (FOS) were the most critical genes, with the highest degree=4, and the other genes Filamin C (FLNC), Toll Like Receptor 2 (TLR2), Integrin Alpha 7 (ITGA7), and Myosin Light Chain 2 (MYL2) had equal degrees (degree=3) as shown in Table 2.
| Gene name | Protein name | Expression level | Degree | Enriched significant modules |
|---|---|---|---|---|
| EGR1 | Early growth response 1 | Up | 4 | Module 2 |
| CXCL8 | Interleukin-8 | Up | 4 | Module 1 |
| FOS | Proto-oncogene | Up | 4 | Module 2 |
| FLNC | Filamin C | Down | 3 | None |
| TLR2 | Toll like receptor 2 (TLR2) | Up | 3 | Module 1 |
| ITGA7 | Integrin subunit alpha 7 | Down | 3 | None |
| MYL2 | Myosin light chain 2 | Down | 3 | None |
Table 2: The Information of Top 7 Hub Genes Based on Their Degree Value
By entering human gene into Coremine medical, the system showed all potential therapeutic drugs (drug and TCM) associated with that gene, and the search results for 7 genes are shown in Table 3.
| Gene name | Drugs | Co-acting drugs |
|---|---|---|
| EGR1 | Golnerminogene Pradenovec, Brivoligide, MEK inhibitor PD-98059, phorbol | Recombinant interleukin-6 |
| CXCL8 | Recombinant Interleukin-6, recombinant tumor necrosis factor family protein, recombinant tumor necrosis factor-α, recombinant interleukin-1β | |
| FOS | c-FOS Antisense, tetradecanoy iphorbol acetate, phorbol | |
| FLNC | Itraconazole, Omalizumab, Fulacimstat, Mepolizumab, Bacillus anthracis strain V770-NP1-R antigen | Recombinant tumor necrosis factor family protein and lipopolysaccharide tyrosine |
| TLR2 | lipopolysaccharide, macrophage stimulator lipopeptide 2, recombinant interleukin-6, Pam2CSK4 | |
| ITGA7 | integrin alphabeta1, Adenosine Diphosphate (ADP)-ribosylarginine | |
| MYL2 | Y 27632 |
Table 3: Effective Drugs Corresponding to All Hub Genes Obtained Through Retrieval
Through DEG analysis, we learned that there were 177 differential genes between samples from patients with AD and healthy control samples, including 45 up-regulated DEGs and 132 down-regulated DEGs. (|log2FC|>1 and p<0.05). To further understand molecular function, biological processes, cell composition, and related signaling pathways, we performed GO annotation and KEGG pathway enrichment assays. According to the PPI data, the top 7 hub genes were ranked, including EGR1, CXCL8, FOS, FLNC, TLR2, ITGA7, MYL2, of which CXCL8 and TLR2 were found in key module 1, EGR1 and FOS, which were enriched in significant module 2, however, FLNC, ITGA7 and MYL2 were not shown in key modules. GO analysis showed that positive regulation of urine volume and cell activation were the most significant biological process that enriches. However, in the KEGG pathway note, we see that DEGs are enriched in GnRH secretion, legionellosis, NOD-like receptor signaling pathway, TGF-β signaling pathway and parathyroid hormone synthesis, secretion and action. Both hub genes in module one were enriched in legionellosis.
First of all, through the above results, we are seeing a lot of results related to inflammation and immunity, which has been recognized by scientists[26-30], i.e., from the pathophysiological point of view, inflammation of the aortic wall is closely related to the development of AD, from the etiological point of view, AD is often considered to be a special form of atherosclerosis, caused by chronic inflammation. In our study, CXCL8 and TLR2 in the hub gene, NODlike receptor signaling pathway and TGF-β signaling pathway in the KEGG-enriched pathway are all inextricably linked to inflammation. We know that CXC chemokines are considered to be key factors in the inflammatory response, and that it can mediate the recruitment of inflammatory leukocytes[31-33], CXCL8 promotes the formation of new blood vessels by mediating the recruitment and activation of neutrophils, leading to exogenous inflammation associated with IL-6, and ultimately forcing the aorta to dilate and rupture[34].
The TGF-β signaling pathway is significant for the maintenance of normal structure and function of the aorta[35], and genetic evidence suggests that TGF-β signaling-related genes play an important role in AD, such as mutations in genetic specimens from patients with AD, often accompanied by paradoxically enhanced TGF-β signaling[36]. Chen et al.[37] then performed a target sequencing that showed that activation of the TGF-β signaling pathway may be related to the pathogenesis of AD.
In other respects, we see that seramine protein (FLN) is actin cross-linked protein, as a scaffold protein, FLN is closely related to cytoskeletal stability[38], studies have shown that FOS can regulate Filamin A (FLNA) high expression in aortic Smooth Muscle Cells (SMCs) in healthy people, while both FOS and FLNA are significantly downregulated in aortic smooth muscle cell SMCs in AD samples, which may be related to the pathological mechanism of AD occurrence[39]. GnRH secretion, as gonadotropinreleasing hormone, may disrupt the intracellular concentration and contractile process of calcium ions with agonist therapy and may lead to pathological remodeling of the heart[40]. On the other hand, cardiac involvement is one of the most common complications of Legionella pneumonia, which can manifest as myocarditis, pericarditis, and prosthetic valve endocarditis, and even further and permanent myocardial damage[41]. TLR2 also offers therapeutic potential for limiting AD formation[42]. In addition, the role of parathyroid glands has been seen in both GO function notes and KEGG enrichment, and then there are no relevant studies showing the relationship between the two, which also provides more space for scholars to reverie.
There were also some limitations to this study, such as the small sample size of the microarray dataset we used. Secondly, and we did not perform richer survival analysis, correlation analysis, etc., we were able to determine very few enrichment pathways and core DEGs, so a larger sample was needed for research, and all the results of the study were discussed in detail and further experimental studies were done.
AD is an acute severe disease caused by hematoma dissection of the aortic intima. Through analysis, this study determined that EGR1, CXCL8, FOS, FLNC, TLR2, ITGA7, MYL2 are the key genes in the pathogenesis of AD, and the prediction of targeted drugs was carried out according to the results of the study, which provided a scientific basis for the pathogenesis study of AD and the exploration of drug treatment, and we also need a series of related experiments to confirm these genes and, MF, CC, BP is intrinsically associated with AD, and further research can be carried out to predict the therapeutic effect of drugs on AD.
Acknowledgments:
This work was supported by the Jilin Provincial Science and Technology Department Project (20200201369JC).
Conflict of interests:
The authors declared no conflict of interests.
References
- Hiratzka LF, Bakris GL, Beckman JA, Bersin RM, Carr VF, Casey DE, et al. ACCF/AHA/AATS/ACR/ASA/SCA/SCAI/SIR/STS/SVM guidelines for the diagnosis and management of patients with thoracic aortic disease. Circulation 2010;121(13):e266-369.
- Rodrigues Bento J, Meester J, Luyckx I, Peeters S, Verstraeten A, Loeys B. The genetics and typical traits of thoracic aortic aneurysm and dissection. Ann Rev Genom Hum Genet 2022;23:223-53.
[Crossref] [Google Scholar] [PubMed]
- Sen I, Erben YM, Franco-Mesa C, deMartino RR. Epidemiology of aortic dissection. Semin Vasc Surg 2021:4(1);1-17.
[Crossref] [Google Scholar] [PubMed]
- Nazir S, Ariss RW, Minhas AM, Issa R, Michos ED, Birnbaum Y, et al. Demographic and regional trends of mortality in patients with aortic dissection in the United States, 1999 to 2019. J Am Heart Assoc 2022;11(7):e024533.
[Crossref] [Google Scholar] [PubMed]
- Zhou Z, Cecchi AC, Prakash SK, Milewicz DM. Risk factors for thoracic aortic dissection. Genes 2022;13(10):1814.
[Crossref] [Google Scholar] [PubMed]
- Howard DP, Banerjee A, Fairhead JF, Perkins J, Silver LE, Rothwell PM. Population-based study of incidence and outcome of acute aortic dissection and premorbid risk factor control: 10-year results from the oxford vascular study. Circulation 2013;127(20):2031-7.
[Crossref] [Google Scholar] [PubMed]
- Meszaros I, Morocz J, Szlavi J, Schmidt J, Tornoci L, Nagy L, et al. Epidemiology and clinicopathology of aortic dissection. Chest 2000;117(5):1271-8.
[Crossref] [Google Scholar] [PubMed]
- Hameed I, Cifu AS, Vallabhajosyula P. Management of thoracic aortic dissection. JAMA 2023;329(9):756-7.
- Zhu Y, Lingala B, Baiocchi M, Tao JJ, Toro Arana V, Khoo JW, et al. Type A aortic dissection-experience over 5 decades: JACC historical breakthroughs in perspective. J Am Coll Cardiol 2020;76(14):1703-13.
[Crossref] [Google Scholar] [PubMed]
- Ahmad W, Liakopoulos OJ. Commentary: A tailored strategy for repair of acute type a aortic dissection: Balancing risk vs. benefit. J Thor Cardiovasc Surg 2022;164(6):1710-1.
[Crossref] [Google Scholar] [PubMed]
- Augoustides JG. Commentary: Acute type B aortic dissection: Navigating new horizons. J Thorac Cardiovasc Sur 2022;164(4):1066-7.
[Crossref] [Google Scholar] [PubMed]
- Baman JR, Malaisrie SC. What is aortic dissection? JAMA 2023;330(2):198.
- Yin ZQ, Han H, Yan X, Zheng QJ. Research progress on the pathogenesis of aortic dissection. Curr Probl Cardiol 2022;48(8):101249.
[Crossref] [Google Scholar] [PubMed]
- Cheng M, Yang Y, Xin H, Li M, Zong T, He X, et al. Non-coding RNAs in aortic dissection: From biomarkers to therapeutic targets. J Cell Mol Med 2020;24(20):11622-37.
[Crossref] [Google Scholar] [PubMed]
- Cui H, Chen Y, Li K, Zhan R, Zhao M, Xu Y, et al. Untargeted metabolomics identifies succinate as a biomarker and therapeutic target in aortic aneurysm and dissection. Eur Heart J 2021;42(42):4373-85.
[Crossref] [Google Scholar] [PubMed]
- Hao X, Cheng S, Jiang B, Xin S. Applying multi-omics techniques to the discovery of biomarkers for acute aortic dissection. Front Cardiovasc Med 2022;9:961991.
[Crossref] [Google Scholar] [PubMed]
- Liu S, Wang Z, Zhu R, Wang F, Cheng Y, Liu Y. Three differential expression analysis methods for RNA sequencing: Limma, EdgeR, DESeq2. J Vis Exp 2021;18(175):e62528.
[Crossref] [Google Scholar] [PubMed]
- Guo S, Wu J, Zhou W, Liu X, Liu Y, Zhang J, et al. Identification and analysis of key genes associated with acute myocardial infarction by integrated bioinformatics methods. Medicine 2021;100(15):e25553.
[Crossref] [Google Scholar] [PubMed]
- Ding Q, Xing J, Bai F, Shao W, Hou K, Zhang S, et al. C1QC, VSIG4, and CFD as potential peripheral blood biomarkers in atrial fibrillation-related cardioembolic stroke. Oxid Med Cell Longev 2023;2023:5199810.
[Crossref] [Google Scholar] [PubMed]
- Yu S, Yu J, Guo Y, Chu Y, Zhang H. Bioinformatic analysis for the identification of potential gene interactions and therapeutic targets in atrial fibrillation. Eur Rev Med Pharmacol Sci 2021;25(5):2281-90.
[Crossref] [Google Scholar] [PubMed]
- Aleksander SA, Balhoff J, Carbon S, Cherry JM, Drabkin HJ, Ebert D, et al. The gene ontology knowledgebase in 2023. Genetics 2023;224(1):31.
[Crossref] [Google Scholar] [PubMed]
- Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucl Acids Res 2000;28(1):27-30.
[Crossref] [Google Scholar] [PubMed]
- Nashiry A, Sarmin Sumi S, Islam S, Quinn JM, Moni MA. Bioinformatics and system biology approach to identify the influences of COVID-19 on cardiovascular and hypertensive comorbidities. Brief Bioinform 2021;22(2):1387-401.
[Crossref] [Google Scholar] [PubMed]
- Peng C, Zhang Y, Lang X, Zhang Y. Role of mitochondrial metabolic disorder and immune infiltration in diabetic cardiomyopathy: New insights from bioinformatics analysis. J Transl Med 2023;21(1):1-8.
[Crossref] [Google Scholar] [PubMed]
- Kong Q, Ma Y, Yu J, Chen X. Predicted molecular targets and pathways for germacrone, curdione, and furanodiene in the treatment of breast cancer using a bioinformatics approach. Sci Rep 2017;7(1):15543.
[Crossref] [Google Scholar] [PubMed]
- Gawinecka J, Schönrath F, von Eckardstein A. Acute aortic dissection: Pathogenesis, risk factors and diagnosis. Swiss Med Wkly 2017;147(3334):w14489.
[Crossref] [Google Scholar] [PubMed]
- Melo RG, Mourao M, Caldeira D, Alves M, Lopes A, Duarte A, et al. A systematic review and meta-analysis of the incidence of acute aortic dissections in population-based studies. J Vasc Surg 2022;75(2):709-20.
[Crossref] [Google Scholar] [PubMed]
- Elsayed RS, Cohen RG, Fleischman F, Bowdish ME. Acute type a aortic dissection. Cardiol Clin 2017;35(3):331-45.
[Crossref] [Google Scholar] [PubMed]
- Zhang X, Hong S, Qi S, Liu W, Zhang X, Shi Z, et al. NLRP3 inflammasome is involved in calcium-sensing receptor-induced aortic remodeling in SHRs. Mediators Inflamm 2019;2019:6847087.
[Crossref] [Google Scholar] [PubMed]
- Wortmann M, Peters AS, Erhart P, Korfer D, Bockler D, Dihlmann S. Inflammasomes in the pathophysiology of aortic disease. Cells 2021;10(9):2433.
[Crossref] [Google Scholar] [PubMed]
- Muñoz A, Costa M. Nutritionally mediated oxidative stress and inflammation. Oxidat Med Cell Longev 2013;2013:610950.
[Crossref] [Google Scholar] [PubMed]
- Paudel S, Baral P, Ghimire L, Bergeron S, Jin L, deCorte JA, et al. CXCL1 regulates neutrophil homeostasis in pneumonia-derived sepsis caused by Streptococcus pneumoniae serotype 3. Blood 2019;133(12):1335-45.
[Crossref] [Google Scholar] [PubMed]
- Frangogiannis NG. Chemokines in ischemia and reperfusion. Thromb Haemost 2007;97(5):738-47.
[Google Scholar] [PubMed]
- Anzai A, Shimoda M, Endo J, Kohno T, Katsumata Y, Matsuhashi T, et al. Adventitial CXCL1/G-CSF expression in response to acute aortic dissection triggers local neutrophil recruitment and activation leading to aortic rupture. Circ Res 2015;116(4):612-23.
[Crossref] [Google Scholar] [PubMed]
- Takeda N, Hara H, Fujiwara T, Kanaya T, Maemura S, Komuro I. TGF-β signaling-related genes and thoracic aortic aneurysms and dissections. Int J Mol Sci 2018;19(7):2125.
[Crossref] [Google Scholar] [PubMed]
- Chen J, Chang R. Association of TGF-β canonical signaling-related core genes with aortic aneurysms and aortic dissections. Front Pharmacol 2022;13:888563.
[Crossref] [Google Scholar] [PubMed]
- Chen P, Yu B, Li Z, Chen Y, Sun Y, Wang DW. COL5A1 Variants cause aortic dissection by activating TGF-β-signaling pathway. J Am Heart Assoc 2021;10(11):e019276.
[Crossref] [Google Scholar] [PubMed]
- Bandaru S, Ala C, Zhou AX, Akyürek LM. Filamin A regulates cardiovascular remodeling. Int J Mol Sci 2021;22(12):6555.
[Crossref] [Google Scholar] [PubMed]
- Chen Y, Wei X, Zhang Z, He Y, Huo B, Guo X, et al. Downregulation of filamin a expression in the aorta is correlated with aortic dissection. Front Cardiovasc Med 2021;8:690846.
[Crossref] [Google Scholar] [PubMed]
- Poljak Z, Hulin I, Maruscakova L, Carter A, Mladosievicova B. Androgen deprivation therapy and cardiovascular complications. Bratisl Lek Listy 2016;117(10):557-61.
- Irigaray Sierra P, Matute-Blanco L, Pueo Crespo E, Salas Lorente C, Viles Bertrán MD. Behind Legionella pneumonia: Not just lung involvement. Eur Heart J Cardiovasc Imag 2022;23(8):e291.
[Crossref] [Google Scholar] [PubMed]
- Shi X, Ma W, Li Y, Wang H, Pan S, Tian Y, et al. miR-144-5p limits experimental abdominal aortic aneurysm formation by mitigating M1 macrophage-associated inflammation: Suppression of TLR2 and OLR1. J Mol Cell Cardiol 2020;143:1-4.
[Crossref] [Google Scholar] [PubMed]