- *Corresponding Author:
- Z. Li
School of Electronics and Information Engineering, Tianjin Vocational Institute, Tianjin 300410, China
E-mail:13920210252@163.com
| This article was originally published in a special issue, “Trends in Therapeutic Management of Various Clinical Conditions II” |
| Indian J Pharm Sci 2021:83(2)Spl issue;204-212 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
In order to better study the biofilm formation process, image structure analyzer software was used to quantitatively analyze the biofilm formation process of Pseudomonas aeruginosa under different concentrations of glucose and allicin. Glucose solution (5.6, 7.0, 11.1, 16.7, 20 and 30 mmol/L) was added to the culture medium of non repetitive Pseudomonas aeruginosa and 6 groups were formed. Minimum inhibitory concentration of allicin to Pseudomonas aeruginosa PAO1 was determined by Tryptic Soy Broth micro dilution method. Single colony was inoculated into Tryptic Soy Broth according to garlic concentration (10; 128 μg/mL). Concomitantly, 3 groups were divided into 1, 3 and 7 d study. The groups of experimental materials were placed in confocal laser scanning microscope for observation and the obtained pictures were imported into the image structure analyzer software, through which the quantitative analysis of the biofilm structure was performed. As main findings we found that as glucose concentration increases, the thickness of Pseudomonas aeruginosa biofilm gradually increased. After glucose (30 mmol/L) treatment for 3 d, areal porosity decreased from 0.93±0.10 to 0.62±0.02 and the biofilm became thicker; textural entropy changed from 6.67±0.99 to 7.88±0.21, meaning an increase in biofilm heterogeneity and thickness; the average diffusion distance decreased from 2.44±0.28 to 1.72±0.36, the biofilm became thicker and the average diffusion distance between colonies also decreased. Under the intervention of allicin 10 μg/mL, the thickness of biomembrane was significantly reduced, the structure of biomembrane was sparse and the numbers of dead and living bacteria were significantly reduced. Under the action of allicin 128 μg/mL, the thickness of biomembrane was further reduced and the dead bacteria were clustered. After the action of allicin 128 μg/mL for 7 d, areal porosity increased from 0.68±0.10 to 0.92±0.02, the biomembrane became sparser; textural entropy increased from 6.67±0.93 changed to 5.52±0.548, the membrane heterogeneity decreased. Image structure analyzer software showed to be suitable for quantitative analysis of biofilm formation process. It is concluded that with the increase of glucose concentration, the maturation speed of Pseudomonas aeruginosa biofilm is accelerated, and the thickness of biofilm is gradually increased; allicin can destroy the self structure of Pseudomonas aeruginosa biofilm.
Keywords
Adhesion, allicin, biofilm, glucose, ISA software, Pseudomonas aeruginosa
As other microorganisms, bacteria are widespread in the environment, where they live irreversibly attached to the surface of inert or active entities, allowing them to reproduce, differentiate and secrete polysaccharide matrix substances, that culminates in the formation of a bacterial collective membrane formed by wrapping of the bacterial community (biofilm)[1]. Indeed, biofilms are characteristically viewed as the aggregation of microbial tissues, and it may be composed of one single biofilm or more mixed biofilm microorganisms[1].
A large number of studies have reported that microorganisms play a dominant role in solid surface colonization, being also gradually recognized that the formation of such biofilms involves both complex physical and chemical processes, as well as the interaction of biological communities. The existence of such biomembrane not only creates a stable internal environment that confers protection and ensures cell activities, and mediates the connection between cells and matrix, but also undertakes functions of material transport, information transfer and energy conversion, closely determined by the biomembrane’s structure. Briefly, bacterial biofilm formation is a dynamic process that can be divided into 4 stages: colonization stage of bacterial reversible adhesion; aggregation stage of irreversible adhesion; maturity stage of biofilm and bacterial abscission and re-colonization[2].
In general and as referred above, biofilm is a type of bacterial aggregation with a certain spatial structure, irreversibly attached to the surface and encapsulated by extracellular polymeric substances (EPS). EPS is mainly composed of polysaccharides, proteins, lipids and nucleic acids[3]. Various substances in EPS interact to form biofilms with certain mechanical properties. In fact, bacteria are encapsulated in the biofilm by extracellular polymers in order to resist various environmental pressures. Almost all kinds of bacteria can form biofilms on any surface (i.e. plastic, metal, glass, soil, wood, surfaces of various medical devices, living tissues, food), although the structure and morphology of biofilms formed in different environments are quite different. For example, data obtained from scanning atomic microscope show that the biofilms formed by Proteus mirabilis are flat in structure and low in bacterial density, having extracellular polymers tightly wrapped with bacteria, being able to reach 40 μM in thickness. Biofilms cultured in the laboratory flow cell have the shape of a mill and occasionally there are parts on the top layer; they have a high density of bacteria and a thickness of hundreds of microns. At present, there is much research on biofilm formation, but the specific mechanism is unclear. Many studies have shown that the process of bacterial biofilm formation can be divided into the following steps[4,5]: bacteria use their own adhesion factors, such as extracellular protein, polysaccharide or pili to help bacteria to initially adhere to the surface of the medium; more EPS is secreted to achieve reversible adhesion of bacteria to non-adhesion conversion; bacterial division and expansion on the surface to form smaller agglomerations; after the local density reaches a certain degree, the group perception signal system is opened, bacterial biofilm is gradually formed, internal bacterial differentiation is completed, gene expression changes, a large number of EPS are secreted and the bacterial metabolic level and even gene mutation occurs at the same time; finally, biofilm diffuses and suffers local disintegration, the mature biofilm can be partially shed under the internal regulation mechanism.
In terms of mechanical strength, bacteria can stick firmly to the substrate surface and the biofilm gene expression is different. Some bacteria secrete extracellular hydrolases that hydrolyze local extracellular polymers, ultimately changing the adhesion to the biofilm substrate. It can also use pili or flagella to fall off the biofilm and thus expand and infect other surfaces. Several studies have used comparative proteomics and immunoproteomics to get the expression differences of bacterial internal proteins in different periods, so as to analyze the role of each protein in the biofilm formation. For instance, Karin Sauer[6] analyzed the proteomics of biofilm in different periods and found that the difference of protein regulation between planktonic bacteria and mature biofilm bacteria was as high as 50 %, with more than 800 proteins having more than 6 times the difference of expression level, the difference protein mainly focused on motion regulation aspects, Nguyen alginate expression and group perception[6]. The great difference on protein expression indicated that P. aeruginosa has different phenotypes at different stages of biofilm formation[6].
Thus considering that these different phenotypes plays an important role in understanding the process of biofilm formation and prevention, in this study, P. aeruginosa, one of the three major pathogens of acquired infection[7], was selected as research object, and the influence of different glucose concentrations and the effect of allicin on P. aeruginosa biofilm formation was assessed by image structure analyzer (ISA) software.
Materials and Methods
Strains
Non repetitive strains of P. aeruginosa were collected from the pulmonology, oncology, hematology and orthopedy wards of Tianjin Medical University General Hospital (China).
Antibacterials, Reagents and Equipments
Several drugs in different concentrations were used: allicin at 30 mg/2 mL (ChuzhouLaien Pharmaceutical Co., Ltd.); ceftazidime at 10 mg/2 mL (Ruiyang Pharmaceutical Co., Ltd.) and levofloxacin oxonate at 20 mg/2 mL (Hunan Wuzhoutong Pharmaceutical Co., Ltd.). Agar and tryptone soybean broth (TBS) were purchased from Oxford Company (UK), phosphate buffer solution (PBS, 10 mmol/L, pH 7.4) and glucose powder were from Guangxi Wuzhou Pharmaceutical Group Co., Ltd.
24-well plates from Corning (USA) and 10 mm×10 mm slide (PVC material) from Shanghai Tiancheng Technology Co., Ltd, and ISA software was kindly provided by Professor Halukbeyenal, Montana State University, USA.
Preparation of Culture Medium
In this study, Tryptic Soy Broth (TSB), diluted twice, without glucose was considered as the control group (Group 1), while culture medium with different glucose concentrations, i.e. 5.6, 7.0, 11.1, 16.7, 20, and 30 mmol/L as the experimental groups (Group 2 to 7 respectively). Briefly, a single pure colony was selected with the bacterial inoculation ring and placed into the 0.9 % saline tube with high pressure sterilization. Five groups of parallel experiments were established, each group with a blank. The control pore (control group) and the 6 bacterial pores (experimental groups) were composed[8] and they were cultured for 1, 2, 3 and 4 d respectively. A cover glass was placed into all the bacteria holes to be tested and the blank control holes of the 24-well plates. Then, 2100 μL sugar free TSB culture solution was added to each hole of the blank control hole (control group). To the experimental groups, 100 μL bacteria solution and 2 mL TSB culture solution with the 6 different concentrations of sugar containing TSB culture solution were added to each hole to be tested (Experiment 2-7 groups). Afterwards, cultures were incubated at 37º and the culture solution was changed every 48 h.
Fluorescein isothiocyanate (FITC) labeled Concanavalin A (FITC-Con A) (Xi'an Qiyue Biological Technology Co., Ltd.) and Immunoprecipitation(PI) (Shanghai Jiandun Biological Technology Co., Ltd.) were used for double fluorescence staining [9]. Briefly after 1, 2, 3 and 4 d of culture, the slides were taken out, washed repeatedly with PBS and the remaining water was absorbed by the filter paper, after the non-adherent bacteria were washed away. Then, 2 % glutaraldehyde 2 mL was fixed for 1.5 h and washed with PBS; a small amount of 50 μg/mL FITC-Con A was taken and dyed in the refrigerator at 4º for 30 min in dark. After Phosphate-buffered saline (PBS) washing, 10 μg/mL Propidium iodide (PI) was stained in the refrigerator at 4º for 15 min in dark, then washed and dried. In the presence of excess moisture, the slide was sealed with 40 % glycerin.
Minimal inhibitory concentration (MIC) of allicin to P. aeruginosa PAO1 was determined by the microdilution method and the parallel determination of P. aeruginosa quality control bacteria ATCC 27853 was performed[9]. Briefly, fresh P. aeruginosa colonies were inoculated in TSB and then the solution was inoculated in 24-well plate, 1 mL per well. Each period was divided into 3 groups: saline control group, allicin high concentration group (128 μg/mL) and allicin low concentration group (10 μg/mL) (clinical concentration). PI was stained in a refrigerator at 4º for 15 min and then washed with PBS.
Confocal laser scanning microscope Observation
Confocal laser scanning microscope (CLSM) was from Leica (Germany), argon laser (488 nm) was used to excite, the green channel to receive the green signal, red channel to receive the red signal, and the biomembrane remained free to scan layer-by-layer (scanning along Z axis) towards the attachment surface of the biomembrane and slide. Five random fields were obtained from each specimen, and 8 to 16 layers were scanned for each field[10].
Quantitative Analysis of Biofilm Structure by ISA Software
Image data obtained from the observation was transferred to the ISA software, the parameters were defined and ISA and ISA3d command was executed[11]. Operation data included: structural parameters, such as textural entropy (TE), structural energy (E), homogeneity (H); and plane parameters, such as area porosity (AP), average diffusion distance (ADD) and surface area (SA).
Statistical Analysis
The Statistical Package for the Social Sciences (SPSS, Chicago, USA) software, version 26.0 was used for the analysis. All results obtained were tested for normal and non normal distribution and were analyzed for homogeneity of variance after logarithmic conversion. Mean±standard deviation (x±s) was used to represent the comparison between groups. The difference was statistically significant when p<0.05.
Results and Discussion
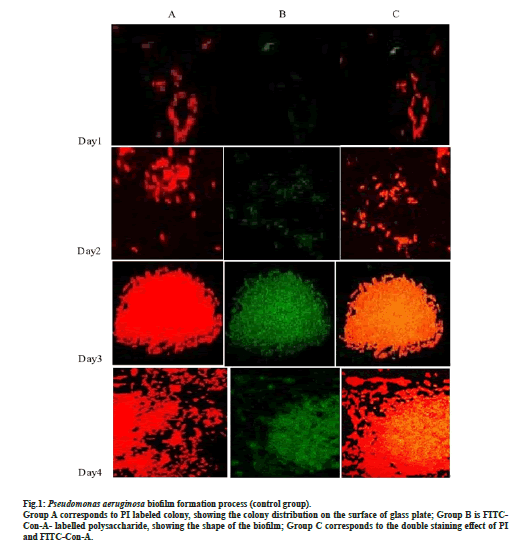
After double immunofluorescence staining of P. aeruginosa, PI combined with DNA in the body of P. aeruginosa gave out red fluorescence, while FITC-ConA combined with the biofilm polysaccharide led to a green fluorescence, which combined with CLSM allowed to observe the biofilm formation. Fig. 1 shows P. aeruginosa biofilm formation at days 1, 2, 3 and 4 (control group) after being cultured in sugar free broth.
On d 1, a small number of bacteria gather and adhere and the biofilm began to form; the green fluorescence is significantly weaker than the red one, indicating that the biofilm is still in the initial formation stage, with weak adhesion and easy to be remove (fig. 1); on d 2, both the red and green fluorescence increase and enhance, with more colonies gather, more polysaccharide matrix of biofilm and enhance adhesion (fig. 1); on d 3, a large number of bacteria gather and integrate (fig. 1). In dense colonies, the fluorescence intensity of polysaccharide matrix of biofilm was stronger, which indicate that the biofilm had entered the maturity stage. On the 4th d, the bacteria on the edge of the biofilm began to spread everywhere, but the fluorescence intensity of the polysaccharide matrix of the biofilm did not decrease, indicating that the biofilm enter the understanding polymerization stage.
Figure 1: Pseudomonas aeruginosa biofilm formation process (control group). Group A corresponds to PI labeled colony, showing the colony distribution on the surface of glass plate; Group B is FITCCon- A- labelled polysaccharide, showing the shape of the biofilm; Group C corresponds to the double staining effect of PI and FITC-Con-A.
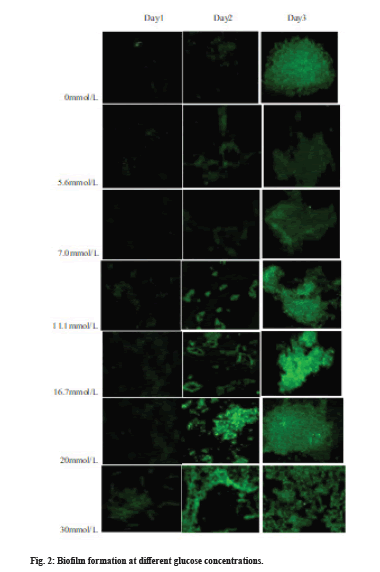
In order to clarify the impact of glucose on the biofilm formation process, the green fluorescence of the FITC-ConA combined with the polysaccharide components of the biofilm was assessed to compare the effect of different glucose concentrations on the biofilm formation at different time periods, as shown in fig. 2.
With the increase in glucose concentration in the culture medium, the biofilm maturation speed is gradually accelerated and the maturity time is gradually decreased; the fluorescence intensity of the biofilm polysaccharide matrix is increasingly strong, indicating that the adhesion ability increases with the increase in glucose concentration. In addition, when the glucose concentration in the culture medium was greater than or equal to 11.1 mmol/L, both the biofilm maturity and stability were significantly higher than that of the control group (sugar free culture medium) and the glucose concentrations were 5.6 and 7.0 mmol/L.
When P. aeruginosa biofilm began to depolymerize at d 4, the effect of different glucose concentrations on biofilm thickness was analyzed (d 3). The analysis of variance (SNK-q test) was used for comparison between groups and the results are shown in Table 1.
| Glucose concentration (mmol/L) | 0 | 6.7 | 7 | 11.1 | 16.7 | 20 | 30 |
|---|---|---|---|---|---|---|---|
| Average biofilm thickness (µm) | 7.74 | 7.04 | 7.86 | 9.54 | 10.66 | 12.08 | 17.24 |
| Standard deviation (µm) | 0.22 | 0.44 | 0.37 | 0.53 | 0.28 | 0.18 | 0.48 |
| p | d,f* | d,f* | d,f* | a,c,e* | a,c,e* | b,c,d* | b,c,d* |
*(a) p<0.05; (b) p<0.01 vs. no glucose; (c) p<0.05 vs. 6.7 and 7.0 mmol/L glucose group; (d) p<0.05 vs. 11.1 and 16.7 mmol/L glucose group; (e) p<0.05; (f) p<0.01 vs. 20 and 30 mmol/L glucose group.
Table 1: Effect of Different of Glucose Concentration of Pseudomonas Aeruginosa Biofilm Thickness At D 3
With the increase of glucose concentration, the biofilm thickness gradually increased. At a glucose concentration of ≤11.1 mmol/L, the biofilm thickness was 7.74-7.86 µm and there were no significant differences (p>0.05) when compared to the glucose free group. On the other side, at a glucose concentration ≥11.1 mmol/L, the biofilm thickness was statistically different (p<0.05) from that of the glucose free group and the thickness was greater than or equal to 9.54 µm. Finally, with a glucose concentration ≥20 mmol/L, the biofilm thickness was statistically different (p<0.01) when comparing with the control group and the thickness was >12.08 µm.
Data obtained from the quantitative analysis of biofilm structure under different glucose concentrations are shown in Table 2. As can be seen, the biofilm becomes thicker with the increase in glucose concentrations. The changes in thickness, AP, ADD and TE at d 1 and d 2 groups were the same as those in the d 3 group, indicating that glucose interference was feasible for each stage of biofilm infection. After glucose increased from 0 to 30 mmol/L for 3 d, AP decreased from 0.93±0.10 to 0.62±0.02, showing that glucose contributed to a thicker biofilm, and to a decrease of the pore and gap between colonies. Also, TE changed from 6.67±0.99 (before the action) to 7.88±0.21 (after the action), indicating that both biofilm heterogeneity and thickness increased after the action of glucose. ADD decreased from 2.44±0.28 to 1.72±0.36, which confirmed that glucose made the biofilm thicker and decreased the average diffusion distance between colonies.
| Glucose (mmol/L) | Date | Biofilm thickness (µm) | AP | ADD | TE |
|---|---|---|---|---|---|
| 0 mmol/L | D 1 | 6.72±0.31a | 0.99±0.01 | 1.58±0.11 | 5.58±0.875 |
| D 2 | 10.28±0.53a | 0.97±0.02 | 1.73±0.19 | 6.09±0.76 | |
| D 3 | 12.16±0.46a | 0.93±0.10 | 2.44±0.28 | 6.67±0.99 | |
| 5.6 mmol/L | D 1 | 7.25±0.39a | 0.97±0.01a | 1.49±0.10a | 5.73±0.57a |
| D 2 | 11.59±0.65a | 0.94±0.01a | 1.66±0.08a | 6.21±0.74a | |
| D 3 | 13.46±0.46a | 0.90±0.01a | 2.37±0.18a | 6.82±0.28a | |
| 7.0 mmol/L | D 1 | 8.72±0.39a | 0.93±0.01a | 1.41±0.03a | 5.89±0.79a |
| D 2 | 12.97±0.64a | 0.89±0.01a | 1.57±0.17a | 6.32±0.31a | |
| D 3 | 15.68±0.43a | 0.87±0.02a | 2.25±0.43a | 6.98±0.75a | |
| 11.1 mmol/L | D 1 | 9.62±0.38a | 0.88±0.01a | 1.39±0.03a | 5.97±0.63a |
| D 2 | 13.74±0.65a | 0.83±0.01a | 1.52±0.27a | 7.23±0.45a | |
| D 3 | 16.82±0.46a | 0.81±0.02a | 2.17±0.38a | 7.42±0.65a | |
| 16.7 mmol/L | D 1 | 10.48±0.33a | 0.85±0.01a | 1.27±0.23a | 6.21±0.65a |
| D 2 | 14.65±0.68a | 0.77±0.01a | 1.48±0.47a | 7.59±0.14a | |
| D 3 | 18.37±0.86a | 0.74±0.02a | 1.97±0.39a | 7.62±0.64a | |
| 20 mmol/L | D 1 | 11.98±0.68a | 0.76±0.01a | 1.22±0.52a | 6.33±0.27a |
| D 2 | 16.33±2.26a | 0.71±0.01a | 1.32±0.48a | 7.64±0.48a | |
| D 3 | 20.76±0.71a | 0.69±0.02a | 1.86±0.33a | 7.72±0.53a | |
| 30 mmol/L | D 1 | 13.32±0.47a | 0.71±0.01a | 1.17±0.29a | 6.47±0.52a |
| D 2 | 18.65±0.73a | 0.68±0.01a | 1.21±0.31a | 7.78±0.92a | |
| D 3 | 22.73±0.49a | 0.62±0.02a | 1.72±0.36a | 7.88±0.21a |
*AP-regional pore rate; ADD-average diffusion distance; TE-textural entropy.
A: p<0.05 (compared with the normal saline control group)
Table 2: Quantitative Analysis of Biofilm Structure Under Different Glucose Concentration (X̄±S)
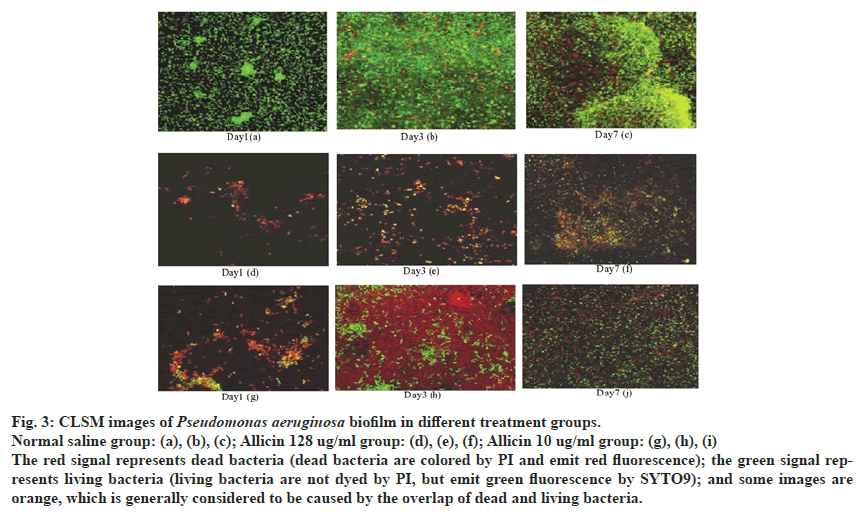
MIC value of allicin against P. aeruginosa PAO1 was 128 μg/mL. Fig. 3 shows 3 specimens in each group of CLSM scanning photos, each specimen with 3 random fields and each field with 8 layers of green channel scanning.
Fig. 3: CLSM images of Pseudomonas aeruginosa biofilm in different treatment groups.
Normal saline group: (a), (b), (c); Allicin 128 ug/ml group: (d), (e), (f); Allicin 10 ug/ml group: (g), (h), (i)
The red signal represents dead bacteria (dead bacteria are colored by PI and emit red fluorescence); the green signal represents
living bacteria (living bacteria are not dyed by PI, but emit green fluorescence by SYTO9); and some images are
orange, which is generally considered to be caused by the overlap of dead and living bacteria.
Without drug intervention, bacteria were dense and there were black areas in the form of tubes or bubbles that could not be stained by SYTO9/PI, in the biofilm group on d 7. At the same time, there were more dead bacteria and the thickness of biofilm was about 29 μm (compared with d 1 and d 3 models). The surface of biofilm was rough and uneven with ridge like protuberances when dead and living bacteria were mixed.
Under the intervention of allicin (10 μg/mL), the biofilm thickness was significantly reduced (compared to the saline group) in about 20 μm. Many areas in the biofilm structure could not be stained by SYTO9/PI, seeming that the biofilm structure was sparse and the number of dead and living bacteria is significantly reduced, compared with the saline group at d 7. Under the action of allicin (128 μg/mL), the biofilm thickness was further reduced, in about 16 μm and the dead bacteria (red signal) significantly increased, while dead bacteria were clustered. After allicin intervention, the thickness of biofilm decreased, the structure was sparse and the resistance of mature biofilm structure to the outside world was further enhanced. Data obtained from the quantitative analysis of biofilm structure is shown in Table 3.
| Group | Date | Biofilm thickness (µm) | AP | ADD | TE |
|---|---|---|---|---|---|
| Normal saline | D 1 | 11.82±0.548 | 0.77±0.01 | 1.58±0.11 | 5.58±0.875 |
| D 3 | 22.88±0.88 | 0.75±0.02 | 1.73±0.19 | 6.09±0.76 | |
| D 7 | 27.73±0.58 | 0.68±0.10 | 2.44±0.28 | 6.67±0.988 | |
| Allicin (10 μg/mL) | D 1 | 9.98±0.33a | 0.89±0.00a | 1.58±0.05 | 5.04±0.42a |
| D 3 | 16.33±2.26a | 0.94±0.01a | 1.66±0.08a | 5.46±0.74a | |
| D 7 | 19.86±0.77a | 0.89±0.01a | 1.73±0.18a | 5.75±0.28a | |
| Allicin (128 μg/mL) | D 1 | 6.88±0.40a | 0.97±0.01a | 1.39±0.03a | 4.83±0.69a |
| D 3 | 13.28±0.76a | 0.96±0.01a | 1.52±0.18a | 5.28±0.38a | |
| D 7 | 15.47±0.57a | 0.92±0.02a | 1.66±0.439a | 5.52±0.48a |
A: p<0.05 (compared with the normal saline control group)
Table 3: Result of Biofilm Model Parameters Analysis (x̄±s, n=9).
As to other microorganisms, P. aeruginosa biofilm formation is a dynamic process[12-14]. In this study, we addressed P. aeruginosa biofilm formation, the impact of different glucose concentrations on P. aeruginosa growth and the effect of allicin (10 and 128 μg/mL) on P. aeruginosa biofilm formation. Double immunofluorescence technique was used to observe the polysaccharide formation and bacterial distribution during P. aeruginosa biofilm formation, with collected images being analyzed by ISA software. It was noticed that, at d 1, the biofilm (initial formation stage) had a weak adhesion; at d 2, the number of colonies, the matrix polysaccharides and the adhesion ability increased. D 3 showed a large number of bacteria gathered into dense colonies, and the biofilm reached the mature stage. At d 4, the bacteria on the biofilm edge began to spread around and the biofilm entered the depolymerization stage. Furthermore, as the glucose concentration increased in the culture medium, the biofilm matures faster and adhered more effectively. Compared with other studies, this study uses ISA software to analyze that it is easy to use, can provide advanced protection for the network and has better expansion performance. Thus, it is feasible to suppose that the use of glucose interferes in all stages of biofilm formation. In our study, when glucose was applied at 30 mmol/L, AP and ADD decreased, TE changed from before to after the action and both biofilm heterogeneity and thickness increased.
When the effect of allicin (10 and 128 μg/mL) on theP. aeruginosa biofilm formation process was addressed, it was stated that, with the increase in culture days, the biofilm thickness gradually increased, indicating that in the later stage of biofilm formation, due to the limitation of nutrients and the sensing system of population density, the biofilm density reached the domain value and sent out signals to trigger biofilm formation[15,16]. In order to reflect the changes of gap channels and nutrient supply distance during the development of the biofilm structure, AP and ADD parameters were analyzed by ISA software[17]. First of all, AP decreased with the increase in biofilm thickness[18]. The increase of ADD indicated that the biofilm became more and more dense, the pores and gaps of biofilm decreased, the supply of oxygen and nutrients was limited and the growth rate of biofilm slowed down to a stable state, while reflected the heterogeneity of biofilm[19].
Indeed, there are significant differences between bacteria in the biofilm and that floating in the logarithmic growth period, or between surface and inner bacteria in biofilm in both morphology and physiological features, as well as the sensitivity to antibiotics[20]. Heterogeneity is an important strategy for bacterial survival[21]. The increasing heterogeneity of the biofilm indicates that the structure of the mature biofilm is more and more complex and its resistance is stronger[22]. In terms of the d 7 model, the biofilm thickness using allicin (128 μg/mL) decreased 6 h later, which indicated that allicin destroyed the intrinsic structure of the biofilm[23]. Allicin also was able to inhibit extracellular polysaccharide complex formation. Moreover, AP increased, meaning that allicin makes the biofilm sparser and the pore and gap between the colonies increase, which could increase the antibiotics penetration[24]. TE also changed from before to after the allicin action, further indicating that the biomembrane heterogeneity decreased and the structure tended to be simple after the action of allicin, which led to the mature biomembrane returning to the active state of metabolism, also weakening its resistance[25]. Moreover, allicin (10 μg/mL) thinned of the biofilm thickness and structure, although the effect was not obvious at high dose[26]. Likewise, compared with the existing similar studies of allicin, the change trend of thickness, AP, ADD and TE in the d 1 and d 3 groups was similar to that of the d 7 group, it shows that it is feasible to use allicin to intervene in various stages of the biofilm formation process using the research method in this paper[27]. In summary, these data highlight again the importance of early intervention due to the initial formation of biofilms with sparse and even structures and vigorous bacterial metabolism[28]. In addition, allicin showed to destroy the self structure of biofilm and help other antibiotics to penetrate and kill bacteria, ultimately improving its efficacy at low doses[29,30].
P. aeruginosa is one of the most common bacteria causing infection and its ability to acquire resistance to antibiotics has always brought difficulties for clinical research[31]. Here, we assessed the process of its biofilm formation and used the ISA software to quantitatively analyze the influence of different glucose concentrations and allicin on its ability to form biofilms. As main findings, we state that, with the increase of glucose concentration, the rate of biofilm maturation increases, while allicin exerted an effective inhibitory effect on biofilm formation. Nonetheless, despite these advances, this study did not detect and analyze other structural biomembrane parameters, where there is still a notable lack of experimental data. Thus, in future research, the biomembrane morphology will be further elucidated from the perspective of different bacterial interactions.
Conflict of Interests:
The authors declare no conflict of interests.
Acknowledgement:
NM acknowledges the Portuguese Foundation for Science and Technology under the Horizon 2020 Program (PTDC/PSI‐GER/28076/2017). CFR acknowledges the UID/EQU/00511/2020 Project—Laboratory of Process Engineering, Environment, Biotechnology and Energy— LEPABE financed by national funds through FCT/ MCTES (PIDDAC).
References
- Learman DR, Voelker BM, Madden AS, Hansel CM. Constraints on superoxide mediated formation of manganese oxides. Front Microbiol 2013;4:262.
- Bardiau M, Caplin J, Detilleux J, Graber H, Moroni P, Taminiau B, et al. Existence of two groups of Staphylococcus aureus strains isolated from bovine mastitis based on biofilm formation, intracellular survival, capsular profile and agr-typing. Vet Microbiol 2016;185:1-6.
- Sachit DE, Veenstra JN. Analysis of reverse osmosis membrane performance during desalination of simulated brackish surface waters. J Membr Sci 2014;453:136-54.
- Jamali NS, Jahim JM, Isahak WN. Biofilm formation on granular activated carbon in xylose and glucose mixture for thermophilic biohydrogen production. Int J Hydrogen Energy 2016;41(46):21617-27.
- Schmidt KE, Auschill TM, Heumann C, Frankenberger R, Eick S, Sculean A, et al. Influence of different instrumentation modalities on the surface characteristics and biofilm formation on dental implant neck, in vitro. Clin Oral Implants 2017;28(4):483-90.
- Gomes LC, Mergulhão FJ. SEM analysis of surface impact on biofilm antibiotic treatment. Scanning 2017;2960194:1-8.
- Sugawara G, Yokoyama Y, Ebata T, Igami T, Yamaguchi J, Mizuno T, et al. Preoperative biliary colonization/infection caused by multidrug-resistant (MDR) pathogens in patients undergoing major hepatectomy with extrahepatic bile duct resection. Surgery 2018;163(5):1106-13.
- Cazelles B, Boudjema G. The Moran effect and phase synchronization in complex spatial community dynamics. Am Nat 2001;157(6):670-6.
- Li N, Nan XY, Chen JN. Research on ORP Prediction of Gold Ore Biological Oxidation Pretreatment Process. Comput Simul 2016;6:70.
- Liu CM, Xu B, Liu JX. Research progress of electrochemistry and biological technologies in microbial fuel cells. Chin J Power Source 2016;1:225-28.
- Ning C, Xiang DW, Sun B. On-site measurement of igbt module lossesin power converter system by discharging DC capacitor. J Power Supply 2016;14:40-45.
- Bodelón G, Montes-García V, López-Puente V, Hill EH, Hamon C, Sanz-Ortiz MN, et al. Detection and imaging of quorum sensing in Pseudomonas aeruginosa biofilm communities by surface-enhanced resonance Raman scattering. Nat Mater 2016;15(11):1203-11.
- Kim LH, Chong TH. Physiological responses of salinity-stressed Vibrio sp. and the effect on the biofilm formation on a nanofiltration membrane. Environ Sci Technol 2017;51(3):1249-58.
- Sommer R, Wagner S, Rox K, Varrot A, Hauck D, Wamhoff EC, et al. Glycomimetic, orally bioavailable LecB inhibitors block biofilm formation of Pseudomonas aeruginosa. J Am Chem Soc 2018;140(7):2537-45.
- Xu Z, Zhang H, Yu H, Dai Q, Xiong J, Sheng H, et al. Allicin inhibits Pseudomonas aeruginosa virulence by suppressing the rhl and pqs quorum-sensing systems. Can J Microbiol 2019;65(8):563-74.
- Yang X, Sha K, Xu G, Tian H, Wang X, Chen S, et al. Subinhibitory concentrations of allicin decrease uropathogenic Escherichia coli (UPEC) biofilm formation, adhesion ability and swimming motility. Int J Mol Sci 2016;17(7):979.
- Yang F, Yang F, Wang G, Kong T, Wang H, Zhang C. Effects of water temperature on tissue depletion of florfenicol and its metabolite florfenicol amine in crucian carp (Carassius auratus gibelio) following multiple oral doses. Aquaculture 2020;515:734542.
- Wu Z, Liu Y, Jia X. A Novel Hierarchical Secret Image Sharing Scheme with Multi-Group Joint Management. Mathematics 2020;8(3):448.
- Cheng H, Liu Y. An improved RSU-based authentication scheme for VANET. J Internet Technol 2020;21(4):1137-50.
- Gao N, Cheng B, Hou H, Zhang R. Mesophase pitch based carbon foams as sound absorbers. Mater Lett 2018;212:243-6.
- Gao NS, Guo XY, Cheng BZ, Zhang YN, Wei ZY, Hou H. Elastic wave modulation in hollow metamaterial beam with acoustic black hole. IEEE Access 2019;7:124141-6.
- Gao N, Hou H, Wu JH. A composite and deformable honeycomb acoustic metamaterial. Int J Mod Phys B 2018;32(20):1850204.
- Gao N, Hou H, Cheng B, Zhang R. A hollow inclusion self-similarity phononic crystal with an ultra-low-frequency bandgap. Int J Mod Phys B 2018;32(02):1850005.
- Gao N, Hou H, Zhang Y, Wu JH. Sound absorption of a new oblique-section acoustic metamaterial with nested resonator. Mod Phys Lett B 2018;32(04):1850040.
- Gao N, Wei Z, Hou H, Krushynska AO. Design and experimental investigation of V-folded beams with acoustic black hole indentations. J Acoust Soc Am 2019;145(1):EL79-83.
- Gao N, Wei Z, Zhang R, Hou H. Low-frequency elastic wave attenuation in a composite acoustic black hole beam. App Acoust 2019;154:68-76.
- Gao N, Wu JH, Yu L, Hou H. Ultralow frequency acoustic bandgap and vibration energy recovery in tetragonal folding beam phononic crystal. Int J Mod Phys B 2016;30(18):1650111.
- Gao N, Zhang Y. A low frequency underwater metastructure composed by helix metal and viscoelastic damping rubber. J Vib Control 2019;25(3):538-48.
- Brian M. Structure and antioxidant activity of extracellular polysaccharides from antarctic marine filamentous fungi. Ccamlr Sci 2018;25(1):49-56.
- Kazemi E, Zargooshi J, Kaboudi M, Heidari P, Kahrizi D, Mahaki B, et al. A genome-wide association study to identify candidate genes for erectile dysfunction. Brief Bioinform 2020.
- Lei Z, Jihao C, Zhang L, Huibin H, Yusu W, Yonghui L. Preparation of soybean oil factory sludge catalyst and its application in selective catalytic oxidation denitration process. J Clean Prod 2019;225:220-6.