- *Corresponding Author:
- H. Sun
Department of Neurology,
Shandong Changyi People’s Hospital,
636 Limin Street,
Changyi City,
China
E-mail: qiff0536@yeah.net
| This article was originally published in a special issue: Special issue on “Trends in therapeutic Management of Various Conditions” |
| Indian J Pharm Sci 2020:82(3)Spl issue 6: 16-23 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
In order to explore the anticancer mechanism of bile acid receptor TGR5 in lung cancer mouse models, healthy C57BL/6J mice of the wild-type and the TGR5 gene knockout type and human lung adenocarcinoma cell line A549 were selected as test system to construct animal models. The mouse models were used to observe the inhibitory effects of bile acid receptor TGR5 on lung cancer and related indicators, inflammatory mediators and mRNA expression levels. Through comparisons between the genes of wild-type mice and TGR5 knockout mice, it was found that the expression levels of IP-10, IL-1β, IFN-γ, CCL4, CXCL2, and CCL3 were significantly higher in the TGR5 knockout model group than those in the wild-type control group. Analysis of the effects of TGR5 on the proliferation of lung adenocarcinoma cells, A549, revealed that the RGT5 agonist could effectively inhibit proliferation of lung cancer cells. Analysis of the effects of TGR5 on apoptosis of lung adenocarcinoma cells A549, indicated that after adding DY240 or INT777 agonist, the level of cell apoptosis was significantly increased compared to the control group. Analysis of the effects of TGR5 on NF-κB signalling pathway showed that inactivating TGR5 could effectively inhibit the expression and phosphorylation of inflammatory factors related to signalling pathways in lung cancer cells. Therefore, it was found that the intervention with TGR5 agonist could effectively inhibit the proliferation of lung cancer cells, which achieved the expected results. Despite the deficiencies in the research process, it has provided a certain basis and ideas for subsequent research on exploring the treatments of lung cancer more accurately. Thus, the study is of great significance.
Keywords
TGR5, lung cancer, Western blotting, RT-PCR
Introduction
Lung cancer is one of the most common malignant tumours in the world, about 80 % of lung cancer is non-small cell lung cancer (NSCLC); in addition, since the onset of NSCLC is latent, more than half of the lung cancer patients are already in the middle or late stage when they are diagnosed, where the surgical treatments are impossible to cure the disease [1]. The cause of lung cancer has not been clearly identified yet. A large number of studies have indicated that the incidence rate of lung cancer in people who smoke for a long time is 10 times higher than that of non-smokers and patients who started smoking at younger ages are more likely to suffer from lung cancer; the younger the age is, the higher the incidence rate of lung cancer. The general clinical manifestations of lung cancer patients are relatively complicated with individual differences. The discovery time and symptoms of each patient are different [2]. The manifestations of lung cancer in the early stage is mild and often irritate no discomforts among patients; therefore, lung cancer is often discovered during physical examinations. In order to better understand the mechanism of lung cancer, domestic and foreign scholars have attracted more and more attention.
The bile acid receptor on cell membrane, TGR5 mainly regulates processes such as bile acid metabolism, energy metabolism, and carbohydrate metabolism in humans [3]. TGR5 is a member of the G-protein coupled receptor family, which contains 7 structural domains spanning the cell membrane and transduces the extracellular signals to the intracellular environment via the G protein in vivo [4]. A study reported that TGR5 can inhibit inflammation in vivo in a variety of cells and tissues and during the course of action, the nuclear factor kappa B (NF-κB) pathway gets activated, allowing inflammatory factors to pass through, leading to inflammation. For example,lipopolysaccharides (LPS) and tumour necrosis factor (TNF-α) are inflammatory mechanisms that mediate the activation of inhibitor of kappa B kinases (IKKs) [5]. TNF-α can induce phosphorylation of proteins such as IκBα, which leads to a series of degradations and the transfer of p50 and p65 proteins from the NF-κB pathway to the nuclei; thus, the expression levels of a variety of pro-inflammatory factors are increased, and the inflammatory factors in the cells are activated and the body shows the inflammatory reactions [6,7]. TGR5, as a membrane receptor, binds to the corresponding ligand on the NF-κB pathway and upregulates cAMP to inhibit the phosphorylation of IκBα, thereby exhibiting the inflammatory effects in humans; in addition, it also upregulates protein expression levels on which protein kinases are dependent, resulting in a reduction in the production of a series of proinflammatory factors. Moreover, TGR5 also regulates other organs of the human body, such as stomach tissue and liver, and participates in inflammatory response process [8]. A large number of studies reported that bile acids are involved in the onset of cancers; however, the active mechanism of bile acid TGR5 on lung cancer has been rarely reported. Therefore, it is necessary to explore the possible mechanism of bile acid TGR5 on lung cancer.
In summary, despite relevant studies on the biological treatments of lung cancer, drugs that can completely cure lung cancer have not been discovered yet. Therefore, the healthy C57BL/6J mice of the wild-type and the gene knockout type and human lung adenocarcinoma cell line A549 were selected as test systems to construct animal models. The mouse models were used to observe the inhibitory effects of bile acid receptor TGR5 on lung cancer by studying related indicators, inflammatory mediators and mRNA expression levels, with an aim to provide new ideas for the clinical treatments of lung cancer in the future.
In order to clearly understand the bile acid TGR5, many studies have been reported. Carino et al. studied the association between TGR5 activation and advanced gastric cancer. It was found that TGR5 is expressed in advanced gastric cancer and its expression is associated with EMT markers, which suggested that TGR5 antagonist could play a role in the treatment of gastric cancer [9]. Su et al. studied the correlation between TGR5, gastric cancer and kidneys in mice; it was found that TGR5 can inhibit the proliferation and migration of renal cancer cells, TGR5 is resistant to the renal inflammation and the proliferation and migration of renal cancer cells passing through the NF-κB and STAT3 signalling pathways partially, which provided the experimental basis for the research on human kidney inflammation [10]. Deutschmann et al. studied that whether TGR5 can promote the invasiveness and metastasis of CCA and it was found that the BAsactivated TGR5 can promote the development of CCA through not only increasing the cell proliferation but also promoting the metastasis, thereby promoting the tumor proliferation [11]. Zhuo et al. studied whether TGR5 can be used as an important pathophysiological feature of osteoarthritis (OA) and it was found that INT777 treatment could inhibit the phosphorylation of p38 and IκB kinase activation/inhibition of the κBα/NF- κB (IKK/IκBα/NF-κB) signalling pathway and TGR5 may be a potential therapeutic target for OA therapy [12].
Materials and Methods
C57BL/6J mice, including the gene knockout mice and the wild-type (WT) mice (18-22 g) were selected as the test system, which were purchased from Shanghai Guandao Bioengineering Co., Ltd., China. The study was performed after the mice were allowed to acclimate for 1 w in the Experimental Animal Center. All experimental protocols were conducted in accordance to the Chinese Experimental Animal Protection and Management Regulations. All animal experiments were approved by the Ethic Committee of Shandong Changyi People’s Hospital. Human lung adenocarcinoma cell line A549 was purchased from Shanghai Sixin Biotechnology Co., Ltd., China.
Animal model construction:
The purchased gene knockout purebred mice were hybridized with the WT mice, the reproduced heterozygous offspring mice were hybridized again, and the reproduced offspring mice were submitted for gene identification. The mice were marked on the ear) as they grew to 4-6 w, including gender and other information. Generally, the probability of occurrence of heterozygous offspring mice in the early stage of gene identification was relatively low. Therefore, only one PCR was performed in the early stage to identify the gene knockout mice (both heterozygous and homozygous) then, the identified mice were hybridized again and this step was repeated for about 6 mo. With the increase of hybridized offspring, the probability of the band appearing after the first PCR was gradually increased; thus, the PCR was performed twice at this time. In terms of the second round of gene identification, the initial PCR was not stable; through continuous improvement of the method, finally, after 18 mo, the first pair of homozygous mice were obtained, which were then coupled and their progenies were found to be homozygous by the first and second PCR tests, indicating successful construction of the model, and thus the TGR5 mouse models were obtained.
DNA extraction:
Mice of the same gender that had similar experimental dates were selected from the constructed mouse models. Then, 1/5 of the mouse ear without ear tags was cut and placed in an Eppendorf (EP) tube (1.5 ml) and each EP tube was labeled. Next, lysis buffer was added to the tubes directly to lyse the ear tissue. The well-lysed ear tissue was centrifuged in a centrifuge for 10 min at 14 000 rpm. After the centrifugation was completed, 200 μl of supernatant was pipetted from each sample tube and was placed in an empty EP tube that was labeled correspondingly. Saturated NaCl solution (100 μl/tube) was added for salting out, mixed well and centrifuged according to the procedure described above. The supernatant was pipetted, the processes were repeated. Subsequently, since the sedimented DNA contained a large number of salt ions, it was washed with ethanol. The centrifugation was repeated twice to obtain the relatively pure DNA, which was dissolved by adding double distilled water and stored at -20°.
Cell passage and culture:
The A549 cells were taken out from the liquid nitrogen tank and quickly placed in a lunch box that contained liquid nitrogen. The box was then placed in a 37° water bath and carefully observed. Once the cells were thawed, they were transferred to a biological cabinet, and then transferred into a 15 ml centrifuge tube that contained 1640 complete medium; the centrifuge tube was centrifuged at 800 rpm for 5 min. Then, the supernatant was discarded, the sedimentation was added with 10 ml of 1640 complete medium, mixed evenly and was cultured in an incubator. The experiment could be started once the culture of cells was stable in the state. Generally, the passage required at least 3 generations.
In the process of cell culture, the supernatant was first pipetted, pancreatin added thereto after the end of the cell fusion growth and once the cells were observed to be separated, they were submitted to centrifugation (800 rpm, 5 min). The supernatant was discarded, and 6 ml of 1640 complete medium was added through a pipette. The cellular fluid in the centrifuge tube was gently mixed with a pipette, and the cells were counted by a cytometer. The concentration of A549 cells was 1.6×105/ml based on the needed volume of cytometer, the required volume of cellular fluid was calculated. A new 50 ml centrifuge tube was added with the mother cellular fluid and 1640 complete medium, mixed evenly, and incubated respectively in a 5 % CO2 incubator and a 2 % O2 incubator at 37°.
Processing of cells by adding TGR5:
The water bath was set to 37°; then, the 1640 incomplete medium (without fetal bovine serum and double antibody) was taken out from the 4° refrigerator, placed into the water bath, preheated for 30 min, and placed into the ultra-clean workbench. The TGR5 agonist (the concentration of INT777 was 50 μl/mol and the concentration of DY240 was 30 μl/mol) was formulated. Then, the culture medium was pipetted out from the medium, added with TGR5 agonist, and added with TNF-α continuously for induction. In term of the control group, DMSO solvent was added when the TGR5 agonist was added, and the other procedures were the same.
Real time (RT) PCR:
Cells in the logarithmic growth phase were washed thrice with pre-cooled long buffer; after being added with RNA extraction agent Trizol (Shanghai Beyotime Biotechnology, China), the cells were repeatedly blown and beaten until the cell membrane was completely lysed to extract the RNA. After being stood for 5 min, 100 μl of chloroform was added; then, after being oscillated, the DNA was allowed to stand for 10 min, centrifuged at 12 000 rpm (Beijing Sihuan Scientific Instrument Factory, China) for 15 min; next, 150 μl of the supernatant was removed, and isopropanol was added thereto at the ratio of 1:1, after being mixed, the DNA was allowed to stand for 10 min, centrifuged at 12 000 rpm for 10 min, the supernatant was discarded, DNA was added with 1 ml of 75 % ethanol, mixed well, centrifuged at 12 000 rpm for 5 min, and the supernatant was discarded; once the samples were dried, 20 μl DEPC (Pierce, USA) water was added to dissolve the samples. The quantitative machine (Zhejiang Lecheng Electric Appliance Factory, China) was first cleaned and zeroed, and the absorbance values of RNA at 260 nm and 280 nm were measured to calculate the concentration of RNA and evaluate the purity of RNA, the total RNA=500/sample concentration. The reverse transcription system was prepared, as shown in Table 1 and was reacted at 42° for 2 min. After the reverse transcription was completed, the obtained cRNA was added to 80 μl double distilled water and stored at -20° or directly subjected to PCR amplification (ABI, USA).
| Components | Volume | |
|---|---|---|
| A | Total RNA | 2 µl |
| 5×first-strand Buffer | 4 µl | |
| M-Mu LV Reverse Transcriptase (200 U/?l) | 0.8 µl | |
| Oligo dT Primer (50 ?M) | 2 µl | |
| dNTP Mixture (10 mM each) | 2 µl | |
| RNase Inhibitor (40 U/?l) | 0.5 µl | |
| RNase Free dH2O | up to 50 µl | |
| B | 2×Super SYBR premixture | 9 µl |
| PCR Forward Primer (10 µM) | 0.5 µl | |
| PCR Reverse Primer (10 µM) | 0.5µl | |
| cDNA | 4 µl | |
| RNase Free dH2O | up to 20 µl |
A. reverse transcription, B. quantitative RT-PCR
Table 1: The Reaction System
Detection of target protein expression by Western blotting:
The volume of the supernatant sample per pore was calculated from the protein concentration measured by the BCA protein concentration test, and the total protein amount of each supernatant sample was equal. The loading buffer was added to the supernatant sample protein in each hole, and the samples were boiled; then, they were submitted to the SDS-polyacrylamide gel electrophoresis (SDS-PAGE); the conditions of SDSPAGE were 80 V for about 0.5 hours and then 110 V for about 1 hour. After the electrophoresis was terminated, the membrane transfer of supernatant sample protein was carried out through the wet method under the condition of 100 V; the protein on the polyvinylidene fluoride (PVDF) membrane was transferred to the nitrocellulose (NC) membrane, and the process lasted for 1.5 hours. Next, the membrane was washed with Tris-buffered saline and Tween (TBST) buffer and soaked in the container. After the membrane transfer of supernatant protein samples was completed, the membrane was blocked by using the blocking solution (20 ml of TBST, 0.75 g of bovine serum albumin, and 3 scoops of skimmed milk powder) at room temperature for 60 min. After the membrane was blocked, the fresh 1 % skim milk powder was used to diluent the antibody according to the recommendations given by the instructions; the PLDL1 antibody was diluted at 1:500, the β-actin was diluted at 1:3000, and stored at 4°; the transferred PVDF membrane and the appropriate amount of the first antibody were added into the antibody incubation box, with the PVDF membrane was completely soaked, and stored in a 4° shaker overnight. In the morning of the next day, the first antibody was pipetted and discarded; the PVDF membrane was gently washed thrice with TBST for 10min each on a horizontal shaker at room temperature. The HRP-labelled second antibody was diluted with fresh 1 % skim milk powder in the incubation of secondary antibody. The antimouse second antibody was 1:3000 and the anti-rabbit second antibody was 1:2000. The operations were the same as the incubation of the first antibody at room temperature for 1 hour. After the above operations were successfully completed, the image analysis software processing system (Image pro plus) (BIO-RAD, USA) was used to analyze the protein band of the NC membrane specimen. The gray value of the target protein band on the NC membrane (BIO-RAD, the USA) was compared with the gray value of the β-actin protein band to obtain the ratio, and the ratio of the bands was compared; after 3 times of repetition, the mean number and standard deviation of the ratio were calculated.
MTT assay:
Cells in the well-growth state (the cell fusion degree was about 80 % and the cell morphology was normal) were used for the MTT assay in a 96 well plate. The plate was incubated for 12-24 h; once the cell fusion state was observed to be about 80 %, drug treatment was given. A total of 3 duplicate wells in the same treatment group were prepared. Immediately after the addition of the drug, the MTT solution was added to one row of each treatment group and the plate was placed in a cell culture incubator. After 4 h, the culture plate was taken out. A tractor was used to remove the upper liquid from the wells were added with MTT. Then, 100 μl of DMSO solution was added to the liquid, and the liquid was extracted for 10 min, the extracted liquid was pipetted and placed into a transparent 96-well plate and assayed at the wavelength of 450 nm on an enzyme mark instrument. The MTT was added at the same time for the next 4 d and the same operation was performed afterward, and the readings were recorded.
Detection of cell apoptosis:
The cell plate was taken out, and a pipette was used to transfer the medium into a 15 ml centrifuge tube labelled with the corresponding treatments. The supernatant was washed with PBS and collected in the corresponding tube. All cells that were digested by pancreatin were collected. The tube was placed in a centrifuge and centrifuged at 1500 rpm for 5 min at room temperature. During the centrifugal process, the double-distilled water was used to dilute the 10× sample loading buffer to 1×, which was then placed on ice. After the centrifugation was terminated, the supernatant was discarded, and the cells were gently resuspended by adding 1 ml of pre-cooled PBS, transferred into a 1.5 ml EP tube at 1500 rpm for 5 min. The supernatant was discarded, and the cells were resuspended by adding sample loading buffer; then, the PI and AnnexinVFITC dye solutions were added under the light-proof environment. The cells were allowed to react for 15 min under the light-proof environment at room temperature and were submitted to the detection of flow cytometry.
Statistics analysis:
SPSS 22.0 statistics software was used to analyse the data and the measurement data were expressed as mean±SD. The pairwise comparisons of the mean number between samples of the groups were submitted to t-test, the multiple comparisons of the mean number between samples of the groups were submitted to the One-way ANOVA test, P<0.05 indicated the statistical significance of the difference.
Results and Discussion.
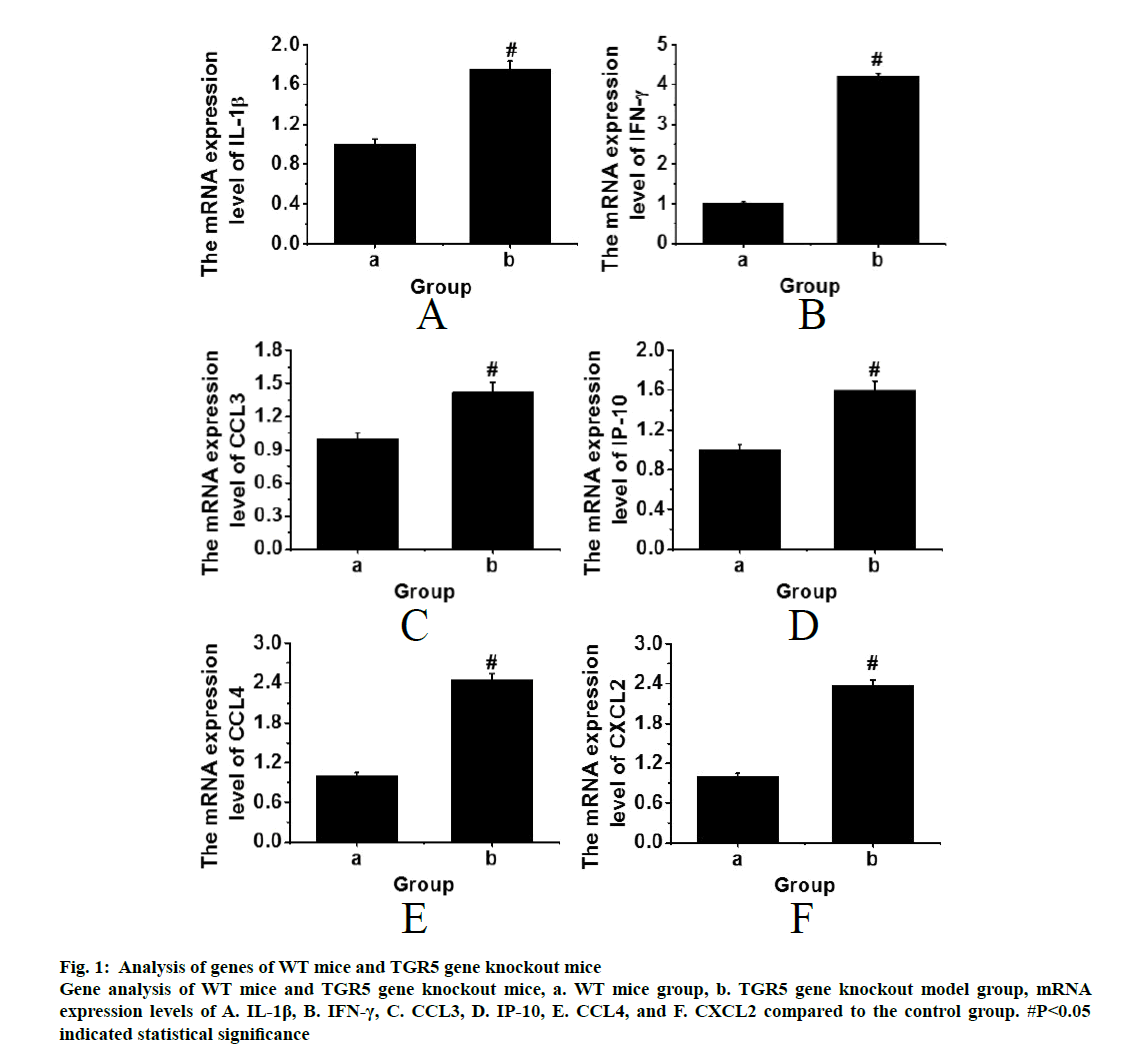
The WT mice and the TGR5 gene knockout mice with the same physiological status were processed by cutting the spinal cords. After euthanasia, the lungs were subjected to RT-PCR for genetic analysis. The results were shown in fig. 1. It was found that the expression levels of IP-10, IL-1β, IFN-γ, CCL4, CXCL2 and CCL3 were significantly higher in the TGR5 gene knockout model group than those in the WT control group (p<0.05). Therefore, it can be inferred that the bile acid receptor, TGR5 could have an inhibitory effect on pulmonary inflammation through the NF-κB signalling pathway, thereby achieving a therapeutic effect.
Figure 1: Analysis of genes of WT mice and TGR5 gene knockout mice Gene analysis of WT mice and TGR5 gene knockout mice, a. WT mice group, b. TGR5 gene knockout model group, mRNA expression levels of A. IL-1β, B. IFN-γ, C. CCL3, D. IP-10, E. CCL4, and F. CXCL2 compared to the control group. #P<0.05 indicated statistical significance
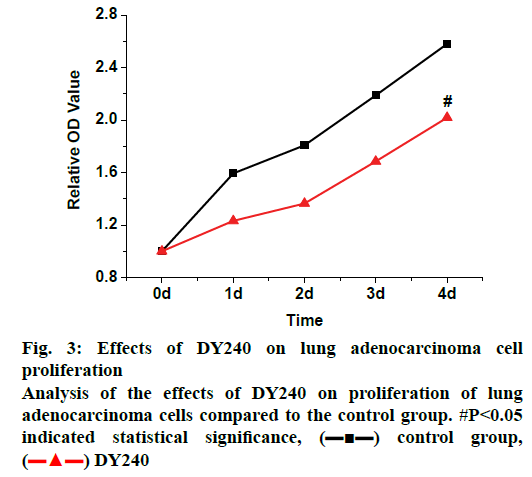
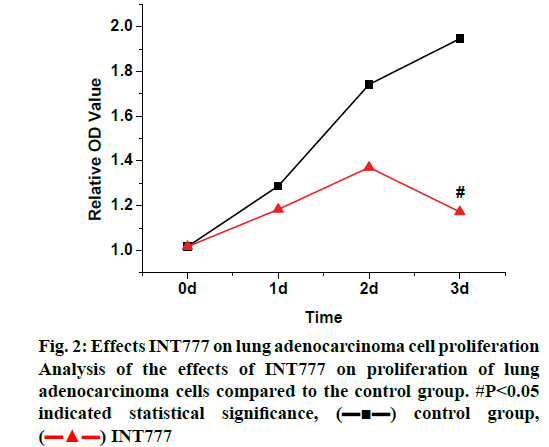
In the study of the effects of TGR5 on the proliferation of lung adenocarcinoma cells, the A549 cells in good growth status were selected. The concentration of cells was first adjusted to 7000 per well, INT777 and DY240 agonists were added to each well to process the cells. The time of drug addition was set as the initial time node, and the cell growth status was recorded by MTT assay. The results were shown in the following fig. 2. When INT777 agonist was added to the cells, it was found that the inhibitory effects on lung adenocarcinoma cells showed since the 2nd d, the inhibition observed was significantly greater than that of the WT control group (p<0.05). It can be observed from fig. 3 that when the DY240 agonist was added to the cells, the inhibitory effects on lung adenocarcinoma cells developed slowly from the beginning and after 4 d of observation, DY240 was found significantly inhibit proliferation of lung adenocarcinoma cells (p<0.05). Therefore, it can be seen that RGT5 agonist can effectively inhibit the proliferation of lung cancer cells, which can be an effective target for the treatment of lung cancer.
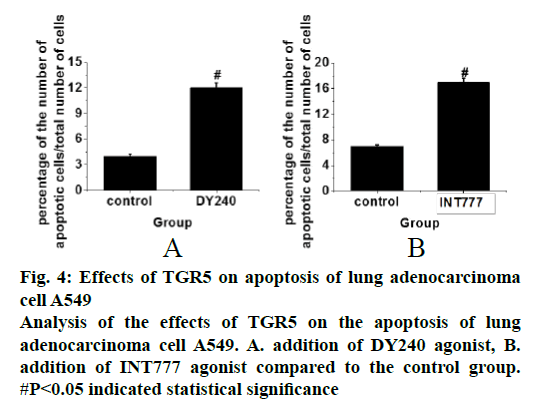
In the cell apoptosis study flow cytometry analysis was performed on cells that were in good growth states and were treated with DY240 and INT777 and the obtained data were recorded. After the addition of the DY240 agonist, the results in fig. 4A showed that the apoptosis level of cells was significantly increased compared to the control group (p<0.05). After the addition of the INT777 agonist. The results in fig. 4B showed that the apoptosis level of the cells was significantly increased, compared to the control group (p<0.05). Therefore, the results of the analysis indicated that TGR5 promoted apoptosis of lung adenocarcinoma A549 cells, which could reduce lung cancer cells and thereby inhibit lung cancer.
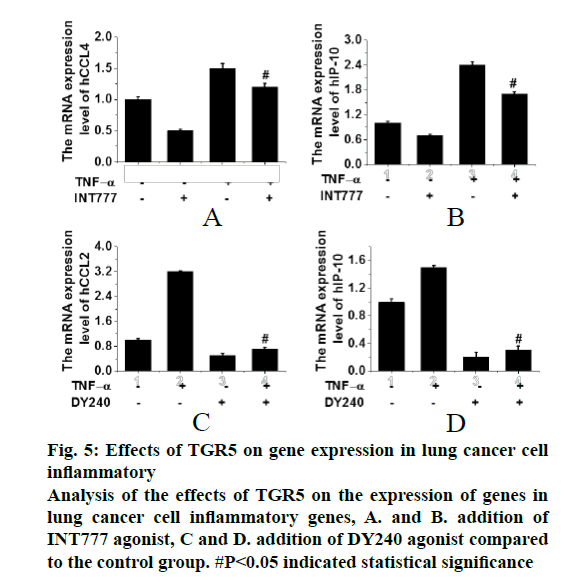
In the analysis of the effects TGR5 on the NF-κB signalling pathway, cells that were in good growth states were selected and treated with DY240 and INT777. TNFα was added to induce the signalling pathway and RT-PCR was used to detect the gene expression. The results were shown in fig. 5. It can be seen that the addition of TNFα can induce the activation of NF-κB signalling pathway; in addition, once the agonists of TGR5 (DY240 or INT777) were added activation of the signalling pathway was inhibited. Therefore, it can be speculated that inflammatory factors such as CCL2 and IP-10 were related to the proliferation of lung cancer and the activation of TGR5 can inhibit the expression of inflammatory factors in lung cancer cells. Therefore, it is speculated that TGR5 may be a tumour suppressor of lung cancer.
Figure 5: Effects of TGR5 on gene expression in lung cancer cell inflammatory Analysis of the effects of TGR5 on the expression of genes in lung cancer cell inflammatory genes, A. and B. addition of INT777 agonist, C and D. addition of DY240 agonist compared to the control group. #P<0.05 indicated statistical significance
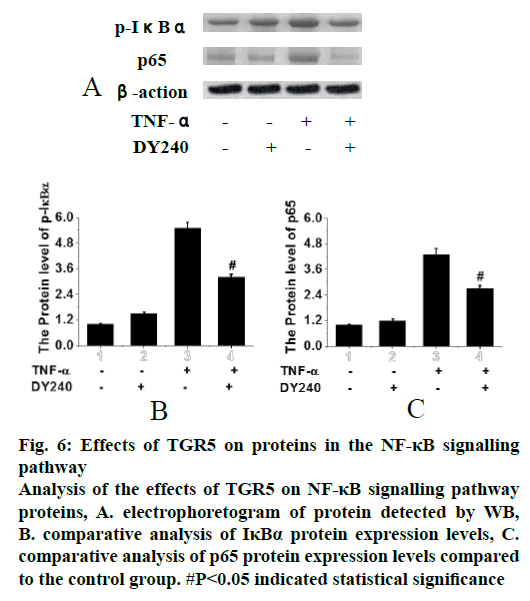
In the analysis of the effects of TGR5 on the proteins in the NF-κB signalling pathway, cells that were in good growth states and treated with TGR5 DY240 agonist were selected, TNFα was added to induce the signalling pathway and Western blotting (WB) method was used to detect the protein expression. Results were shown in fig. 6. It can be seen that the phosphorylation of IκBα was most obvious when only TNFα was added, while the addition of TGR5 agonist DY240 significantly inhibited phosphorylation of IκBα (p<0.05). The nuclei transfer of p65 protein was most obvious when only TNFα was added, while the addition of TGR5 agonist DY240 significantly inhibited the nuclei transfer of p65 protein (p<0.05). Therefore, it was speculated that TGR5 agonist could inhibit the phosphorylation of IκBα and the nucleus transfer of p65 protein, and it was speculated that TGR5 could inhibit the occurrence of lung cancer.
Figure 6:Effects of TGR5 on proteins in the NF-κB signalling pathway Analysis of the effects of TGR5 on NF-κB signalling pathway proteins, A. electrophoretogram of protein detected by WB, B. comparative analysis of IκBα protein expression levels, C. comparative analysis of p65 protein expression levels compared to the control group. #P<0.05 indicated statistical significance
In order to explore the anticancer mechanism of bile acid receptor TGR5 in lung cancer mouse models, healthy C57BL/6J mice of the wild-type and the gene knockout type and human lung adenocarcinoma cell line A549 were selected as experimental models. The mouse models were used to observe the inhibitory effects of bile acid receptor TGR5 on lung cancer by observing the related indicators, inflammatory mediators and related mRNA expression levels. Through the comparisons between the genes of WT mice and TGR5gene knockout mice, it was found that the expression levels of IP-10, IL-1β, IFN-γ, CCL4, CXCL2, and CCL3 were significantly higher in the TGR5 gene knockout model group compared to those in the WT control group. Through the analysis of the effects of TGR5 on the proliferation of lung adenocarcinoma cell A549, it was found that the RGT5 agonist could effectively inhibit the proliferation of lung cancer cells. Through the analysis of the effects of TGR5 on the apoptosis of lung adenocarcinoma cell A549, it was found that after adding the DY240 or INT777 agonist, the level of cell apoptosis was significantly increased. Through the analysis of the effects of TGR5 on the NF-κB signalling pathway, it was found that inactivating TGR5 could effectively inhibit the expression and phosphorylation of inflammatory factors related to signalling pathways in lung cancer cells.
Therefore, through the exploration of the correlation of TGR5 to lung cancer, it was found that the intervention of TGR5 agonist administration could effectively inhibit the proliferation of lung cancer cells, which achieved the expected results. The experiments and analysis had provided experimental data for the clinical treatments of lung cancer in the future. There are certain deficiencies in the research progress; for example, the collected sample capacity is not large enough in the experiment to accurately explore the mechanism. Therefore, it is necessary to increase the sample capacity to conduct a more detailed exploration of the characteristics.
References
- Chen M J, Chen Y L, Wang T W. Membrane bile acid receptor TGR5 predicts good prognosis in ampullary adenocarcinoma patients with hyperbilirubinemia. Oncol Rep 2016;36(4):1997-2008.
- Cariello M, Peres C, Zerlotin R. Long-term Administration of Nuclear Bile Acid Receptor FXR Agonist Prevents Spontaneous Hepatocarcinogenesis in Abcb4−/− Mice. Sci Rep 2017;7(1):11203.
- Li D, Cao W. Bile acid receptor TGR5, NADPH Oxidase NOX5-S and CREB mediate bile acid-induced DNA damage in Barrett’s esophageal adenocarcinoma cells. Sci Rep 2016;6:31538.
- Jin Q, Noel O, Nguyen M. Bile acids upregulate BRCA1 and downregulate estrogen receptor 1 gene expression in ovarian cancer cells. Eur J Cancer Prev 2018;27(6):553-56.
- Wang Y, Aoki H, Yang J. The role of sphingosine 1-phosphate receptor 2 in bile-acid–induced cholangiocyte proliferation and cholestasis-induced liver injury in mice. Hepatol 2017;65(6):2005-18.
- Liu X, Chen B, You W. The membrane bile acid receptor TGR5 drives cell growth and migration via activation of the JAK2/STAT3 signalling pathway in non-small cell lung cancer. Cancer Lett 2018;412:194-207.
- Deutschmann K, Reich M, Klindt C. Bile acid receptors in the biliary tree: TGR5 in physiology and disease. Biochim Biophys Acta (BBA) 2018;1864(4):1319-25.
- Pathak P, Xie C, Nichols RG. Intestine farnesoid X receptor agonist and the gut microbiota activate G?protein bile acid receptor-1 signaling to improve metabolism. Hepatol 2018;68(4):1574-88.
- Carino A, Graziosi L, D'Amore C, The bile acid receptor GPBAR1 (TGR5) is expressed in human gastric cancers and promotes epithelial-mesenchymal transition in gastric cancer cell lines. Oncotarget 2016;7(38):61021.
- Su J, Zhang Q, Qi H. The G-protein-coupled bile acid receptor Gpbar1 (TGR5) protects against renal inflammation and renal cancer cell proliferation and migration through antagonizing NF-κB and STAT3 signaling pathways. Oncotarget 2017;8(33):54378.
- Deutschmann K, Reich M, Lang A. Role of the bile acid receptor TGR5 (GPBAR1) in cholangiocarcinoma (CCA). Z Gastroenterol 2018;56(01):A4. 26.
- Zhuo W, Li B, Zhang D. Activation of G-protein-coupled bile acid receptor Gpbar1 (TGR5) inhibits degradation of type II collagen and aggrecan in human chondrocytes. Eur J Pharmacol 2019;856:172387.a
 control group,
control group,  INT777
INT777