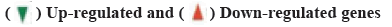- *Corresponding Author:
- Chunyun Wang
Department of Gastrointestinal Surgery, The Second Affiliated Hospital, Hengyang Medical School, University of South China, Hengyang, Hunan 421001
E-mail: 892357467@qq.com
| This article was originally published in a special issue, “Advanced Targeted Therapies in Biomedical and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2023:85(1) Spl Issue “205-213” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
To explore the genes correlated with the prognosis in colorectal cancer is the main objective of the study. Identification of differential genes through series of GSE20970, GSE37182, GSE44861 and GSE64392 from gene expression Omnibus database and then the differential genes were analyzed by gene ontology and Kyoto encyclopedia of genes and genomes enrichment. Through protein-protein interaction networks, screening of key genes was done and then we analyzed the prognosis of colorectal cancer from the cancer genome Atlas database. We obtained the differential genes through limma screening in series data GSE20970, GSE37182, GSE44861 and GSE64392 from gene expression Omnibus database. Venn diagram was used to screen 206 intersecting differentially expressed genes. Gene ontology enrichment analysis terms used were cytosol, transcription, deoxyribonucleic acid-templated, nucleic acid-templated transcription, ribonucleic acid biosynthetic process, nucleobase-containing compound biosynthetic process, heterocycle biosynthetic process, aromatic compound biosynthetic process, regulation of nucleobase-containing compound metabolic process, regulation of ribonucleic acid metabolic process, organic cyclic compound biosynthetic process. Kyoto encyclopedia of genes and genomes pathway terms were interleukin-17 signaling pathway, bladder cancer, phospholipase D signaling pathway, human cytomegalovirus infection, aldosterone-regulated sodium reabsorption, Kaposi’s sarcoma-associated herpesvirus infection, forkhead box O signaling pathway, nuclear factor kappa B signaling pathway, hepatitis B. Protein-protein interaction network has shown 206 key genes. According to the top 50 of betweenness value, we analyzed the prognosis of colorectal cancer from the cancer genome Atlas database. We found Ki-ras2 Kirsten rat sarcoma viral oncogene homolog, C-X-C motif chemokine ligand 8, partner and localizer of breast cancer gene 2, peroxiredoxin 4 and translocase of outer mitochondrial membrane 20 are the key genes which impact prognosis in colorectal cancer. We predict that these genes can promote the survival time of colorectal cancer in future.
Keywords
Colorectal cancer, gene expression Omnibus database, the cancer genome Atlas database, differential genes, gene ontology, Kyoto encyclopedia of genes and genomes
Colorectal Cancer (CRC) is the most common cancer in gastrointestinal tract[1]. It is the second and the third common cancer in woman and men, respectively[2]. In global cancer statistics 2020, more than 1.9 million new CRC (including anus) cases and 935 000 deaths were estimated to occur in 2020[1]. The 5 y and 10 y survival rates are 65 % and 58 % respectively[3], and incidence and mortality rates are 25 % higher in men than in women[4]. 20 % cases show metastatic and 40 % cases recurred with localized CRC[5]. However, the prognosis of metastatic and recurrence CRC is poor, 5 y survival rate is less than 20 %[6]. The incidence rates are approximately 4-fold in transitioned countries than transitioning countries[1]. In recent years, the incidence rate of people >50 y is lower than people with <50 y[3]. Colonoscopy screening reduces and improves the incidence and prognosis of CRC[7]. The prevalence of colon cancer is closely related to living habits, smoking and drinking, and so on[8,9]. In addition, there are 10 %-20 % of patients with CRC who possess a positive family history and ~5 % of cases of CRC are linked to a known hereditary CRC syndrome detectable by germline testing[4].
The development and prognosis of CRC are closely related to genetic factors besides tumor stage[10]. More and more gene targeted drugs have been developed or they are being developed for Vascular Endothelial Growth Factor (VEGF) and Ki-ras2 Kirsten Rat Sarcoma Viral Oncogene Homolog (KRAS)[11,4]. We obtained genes related to the prognosis of CRC to improve survival rate. Gene Expression Omnibus (GEO) and The Cancer Genome Atlas (TCGA) database are the most commonly used tumor database in the global, which includes gene sequencing.
In this study, we used GEO database for the analysis of the Differentially Expressed Genes (DEGs) with tumor tissue and normal tissue in CRC and then validate DEGs expression and analyze the prognosis of CRC from TCGA database. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis was used to analyze DEGs. This study was performed in order to study the prognosis of key DEGs in CRC.
Materials and Methods
Data collection and workflow illustration:
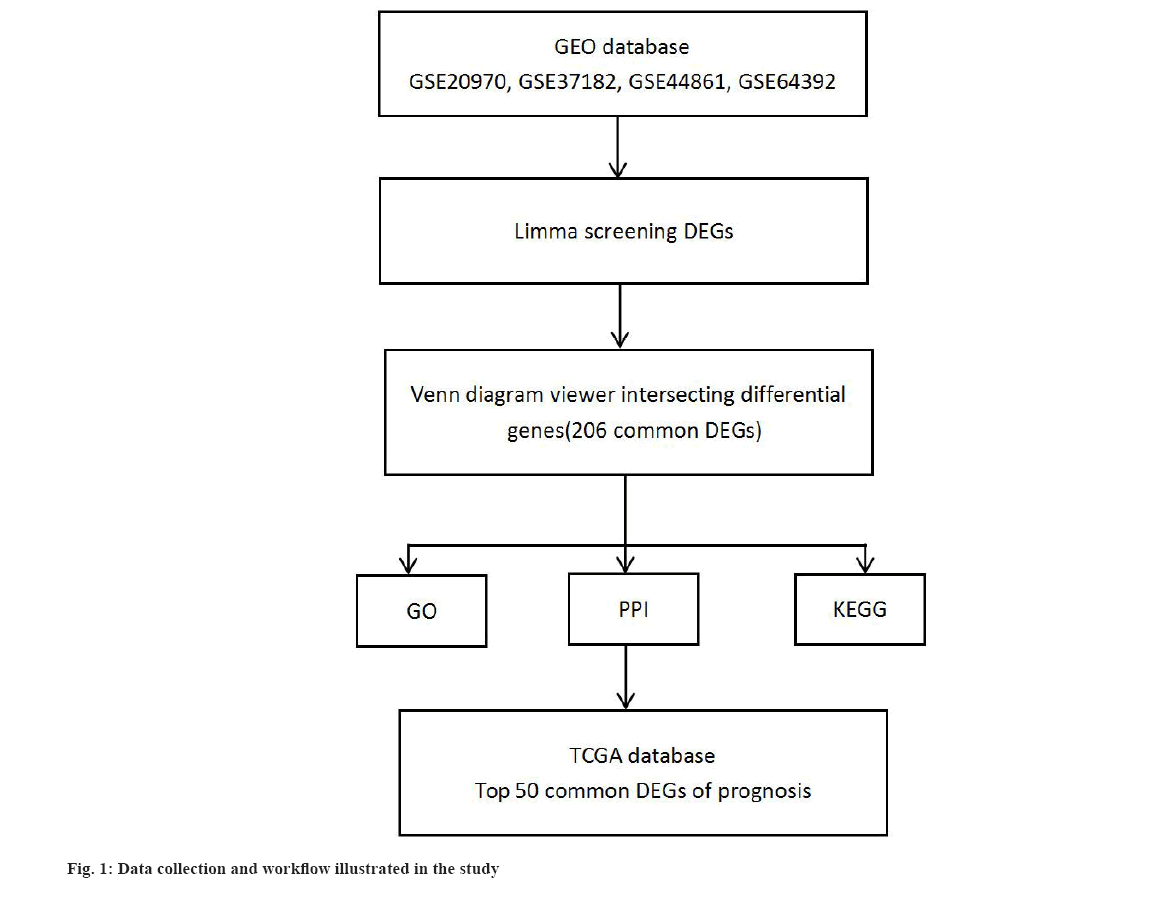
We obtained messenger Ribonucleic Acid (mRNA) transcriptome data in series of GSE20970, GSE37182, GSE44861 and GSE64392 from GEO database (https://www.ncbi.nlm.nih.gov/geo/) and clinical data from TCGA database of CRC, respectively. The workflow is illustrated in fig. 1.
Limma screening of DEGs:
We obtained genes profile through clusterProfiler package in R version 4.0.2 (http://www.r-project.org/) from series of GSE20970, GSE37182, GSE44861 and GSE64392. Limma R package (version 3.40.6) was used to analyze the DEGs. The threshold for identifying significant DEGs was False Discovery Rate (FDR)<0.01 and |log2 (fold change)|≥2. Venn diagram was drawn to show the relationship between DEGs and obtained intersecting differential key genes from four series data.
Key DEGs expression and prognosis in CRC from TCGA database:
From TCGA database, we obtained 455 tumor samples and 45 normal tissues. The key DEGs expression was through clusterProfiler package in R version 4.0.2. The key DEGs of prognosis were obtained from online website, University of Alabama at Birmingham Cancer Data Analysis (UALCAN) (http://ualcan.path.uab.edu/).
GO and KEGG enrichment analysis:
GO analysis is performed and functional enrichment analysis includes Biological Process (BP), Molecular Function (MF) and Cellular Component (CC). KEGG is the advanced function and mechanism involved in the biological system at the molecular level. We analyze the key DEGs function and mechanisms pathway though Database for Annotation, Visualization and Integrated Discovery (DAVID) platform (http://david.ncifcrf.gov/), p value<0.05 and FDR<0.25 were considered statistically significant.
Protein-Protein Interaction (PPI) network and key genes:
The GeneMANIA (http://genemania.org/) prediction server was designed to assess the PPI network[12]. The network was analyzed and visualized by Cytoscape 3.6.1 (http://www.cytoscape.org/). In the network, a high degree value indicated a more essential role for that gene. The degree value of each gene was calculated by the network analyzer tool that was built in the Cytoscape software. The genes whose degree value, closeness centrality and betweenness centrality were greater than the median value were identified as key genes.
Statistical analysis:
The Student’s t-test (R function t-test) was used to determine whether there were significant differences between the two groups and p-value<0.05 was considered to be statistically significant. The ggplot package was used for plotting.
Results and Discussion
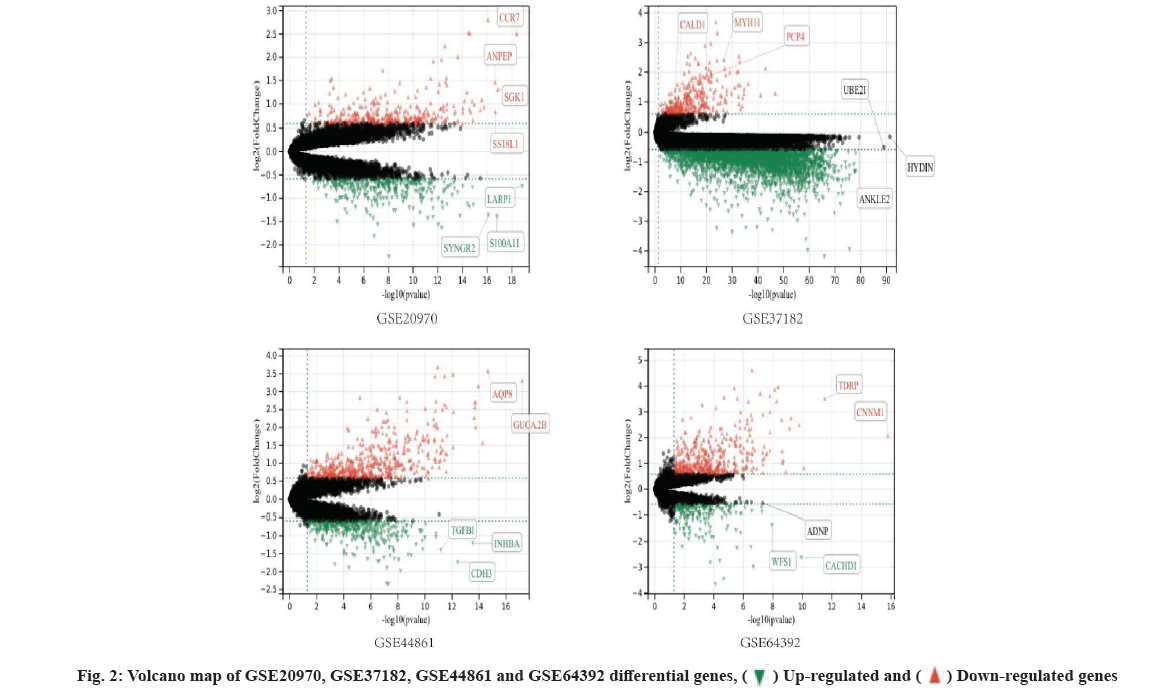
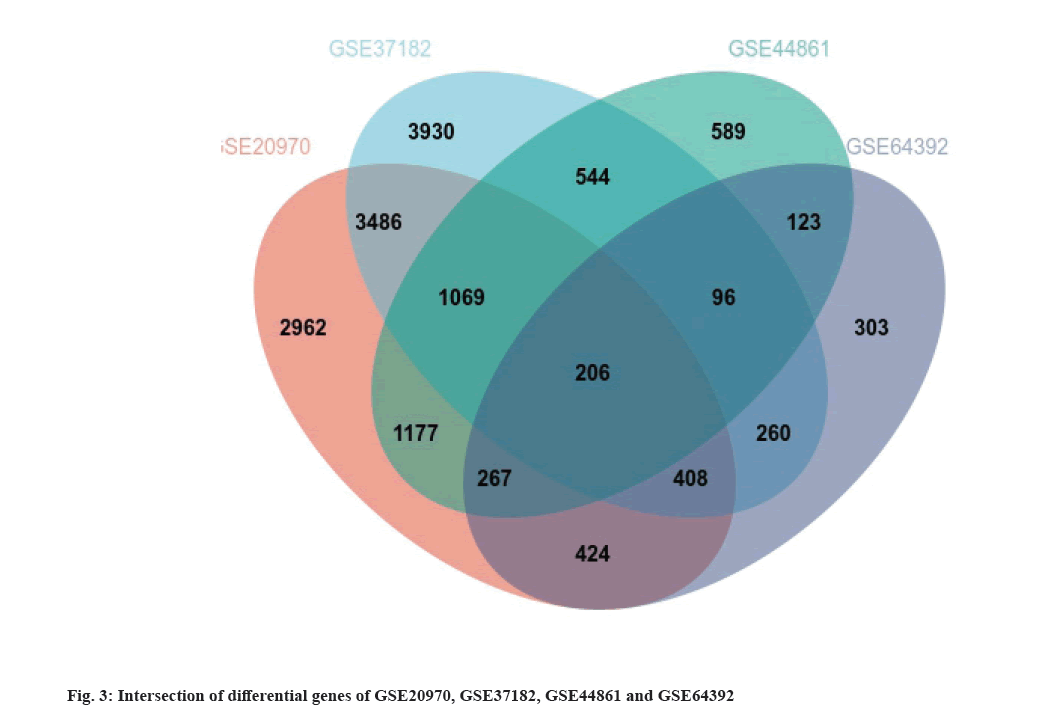
Identification of DEGs was shown here. We used limma screening of DEGs in series data GSE20970, GSE37182, GSE44861 and GSE64392 from GEO database (fig. 2). And then, we use DEGs to draw Venn diagram. There are 206 intersecting key genes (fig. 3).
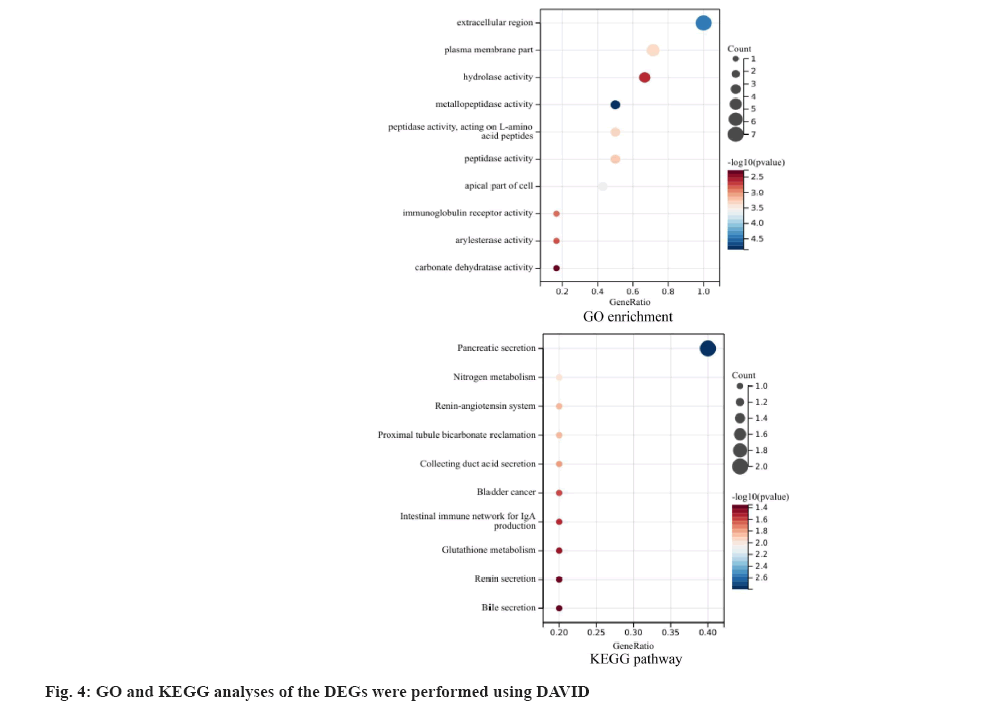
GO and KEGG analyses of the DEGs were performed using DAVID software. GO and KEGG enrichment analysis include GO enrichment terms like cytosol, transcription, DNA-templated, nucleic acid- templated transcription, RNA biosynthetic process, nucleobase-containing compound biosynthetic process, heterocycle biosynthetic process, aromatic compound biosynthetic process, regulation of nucleobase-containing compound metabolic process, regulation of RNA metabolic process, organic cyclic compound biosynthetic process. KEGG pathway include Interleukin-17 (IL-17) signaling pathway, bladder cancer, phospholipase D signaling pathway, human cytomegalovirus infection, aldosterone- regulated sodium reabsorption, Kaposi’s sarcoma- associated herpesvirus infection, Forkhead Box O (FOXO) signaling pathway, Nuclear Factor kappa B (NF-κB) signaling pathway, hepatitis B (fig. 4).
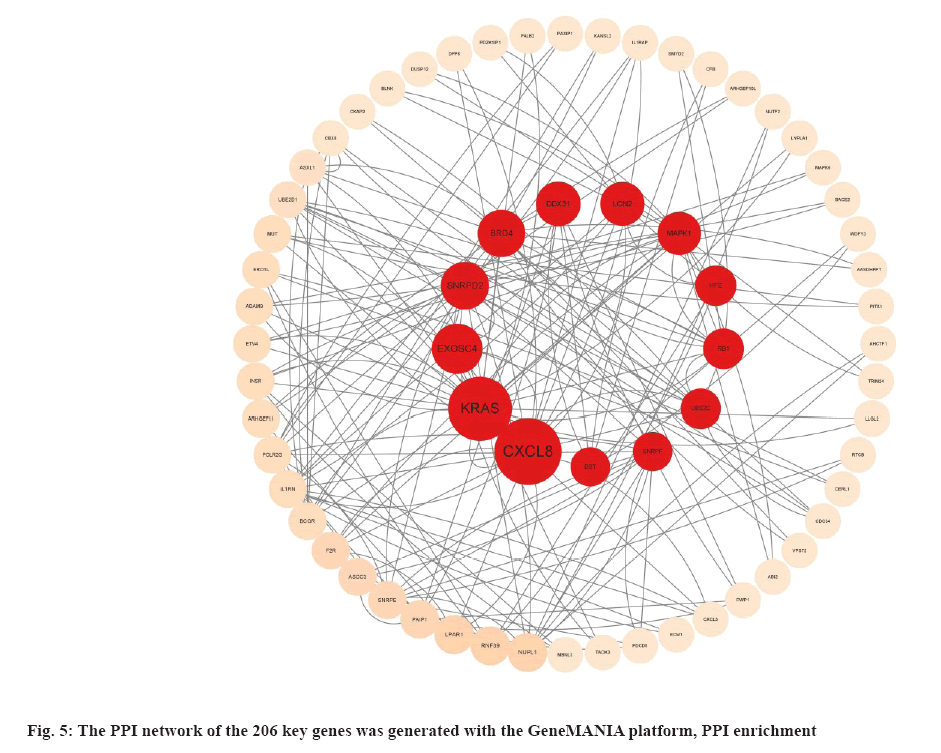
The PPI network of the 206 key genes was generated with the GeneMANIA platform and the PPI enrichment p-value=3.62-05. From the removal of marginal genes, we obtained 52 genes. The PPI network analyzed betweenness value using Cytoscape software (fig. 5).
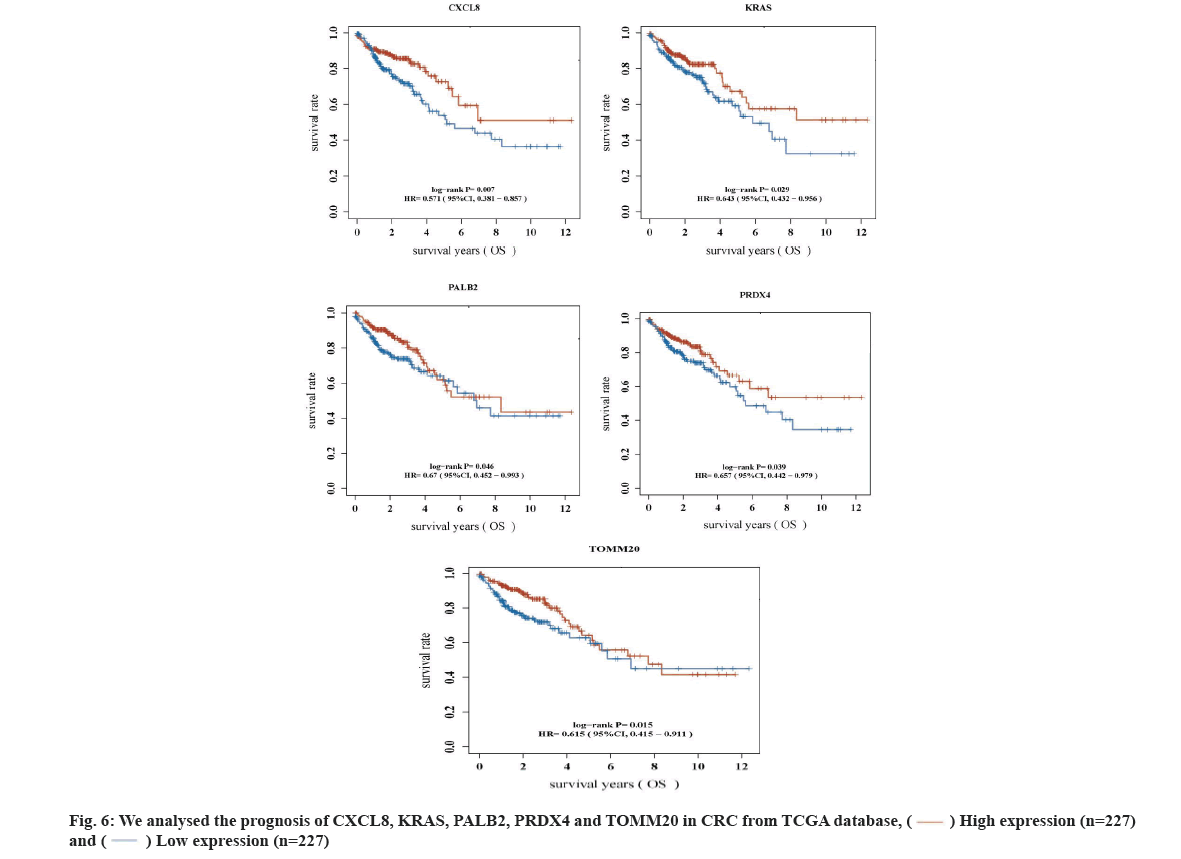
Key DEGs related to prognosis of CRC was shown here. According to Betweenness value of top 50 genes (Table 1), we analyzed their relationship with the prognosis of CRC in TCGA database. Finally, we found that 5 key DEGs were significantly different from the prognosis of CRC (p<0.05) (fig. 6). They are KRAS, C-X-C Motif Chemokine Ligand 8 (CXCL8), Partner and Localizer of Breast Cancer Gene 2 (PALB2), Peroxiredoxin-4 (PRDX4), Translocase of Outer Mitochondrial Membrane 20 (TOMM20), respectively. They are significantly different from the prognosis of CRC i.e. CXCL8, p=0.007, Hazard Ratio (HR)=0.571 (95 % Confidence Interval (CI): 0.381-0.857); KRAS, p=0.029, HR=0.643 (95 % CI: 0.432-0.956); PALB2, p=0.046, HR=0.67 (95 % CI: 0.482-0.993); PRDX4, p=0.039, HR=0.657 (95 % CI: 0.442-0.979) and TOMM20, p=0.015, HR=0.615 (95 % CI: 0.415-0.911), respectively.
| Genes | Betweenness | Closeness |
|---|---|---|
| CXCL8 | 5541.41 | 0.0535009 |
| KRAS | 3970.5308 | 0.053539347 |
| MAPK1 | 2948.6423 | 0.052743364 |
| LCN2 | 2819.2688 | 0.05170021 |
| EXOSC5 | 2787.6616 | 0.052650176 |
| HFE | 2685.3203 | 0.050050385 |
| DDX10 | 2152.4856 | 0.05179006 |
| CCND1 | 2144.006 | 0.053443328 |
| AURKA | 2112.3137 | 0.0520979 |
| NUP153 | 2017.262 | 0.051772065 |
| UBE2C | 1961.2054 | 0.05268741 |
| BRD4 | 1925.4847 | 0.052317414 |
| DST | 1546 | 0.049732976 |
| PNPLA3 | 1524 | 0.048111074 |
| MLXIPL | 1288 | 0.046287667 |
| MAPK8 | 1214.8868 | 0.052706048 |
| ARHGEF11 | 1179.0071 | 0.049932975 |
| LPAR1 | 1134.8159 | 0.051592797 |
| RB1 | 1040.2368 | 0.052575864 |
| ADRM1 | 1030.7721 | 0.049932975 |
| SNRPF | 1007.0974 | 0.051628552 |
| POLR2G | 825.45984 | 0.05018525 |
| PRDX4 | 804.98456 | 0.050783914 |
| GTF2IRD1 | 780 | 0.044544097 |
| CDK4 | 727.26843 | 0.052706048 |
| SNRPE | 700.8439 | 0.051592797 |
| DDX31 | 661.7624 | 0.050818555 |
| ERO1L | 647.8914 | 0.049370445 |
| MMP1 | 621.92694 | 0.052299052 |
| KATNA1 | 588.569 | 0.05049136 |
| TOMM20 | 526 | 0.049866132 |
| GTF2I | 526 | 0.04290239 |
| MUT | 526 | 0.05137931 |
| WDFY3 | 526 | 0.047695264 |
| ABI2 | 526 | 0.05045716 |
| TERF1 | 524 | 0.04798712 |
| TEAD4 | 524 | 0.047879178 |
| SNRPD2 | 519.55206 | 0.051574938 |
| STX6 | 417.98175 | 0.050525602 |
| EXOSC4 | 410.3557 | 0.052116126 |
| RPN2 | 359.62906 | 0.04864512 |
| ASXL1 | 345.27673 | 0.051736113 |
| TRIM5 | 333.8962 | 0.05 |
| PALB2 | 310.83032 | 0.05141477 |
| IL1RN | 290.33334 | 0.051450275 |
| TRIM24 | 283.84546 | 0.05045716 |
| F2R | 278.9583 | 0.051988836 |
| BCOR | 277.04736 | 0.051397033 |
| ESRP2 | 274.84512 | 0.049093906 |
| RNF39 | 272.8513 | 0.048629243 |
Table 1: PPI Network Analyzed Betweenness Value of Top 50 Genes
In our study, we analyzed DEGs through series GSE20970, GSE37182, GSE44861 and GSE64392 from GEO database. We found 206 DEGs have significant role in four series from GEO database. Subsequently, we analyzed 206 DEGs by GO enrichment and KEGG pathway, through PPI network analysis. Finally, we found KRAS, CXCL8, PALB2, PRDX4, TOMM20 genes are key DEGs in the prognosis of CRC. Then we used 206 DEGs analysis and prognosis of CRC in TCGA database. We found that 17 DEGs were significantly different in the prognosis of Liver Hepatocellular Carcinoma (LIHC).
KRAS is a proto oncogene and belongs to Rat Sarcoma Virus (RAS) family. It involve the intracellular signal transmission and relate to tumor generation, proliferation, migration, diffusion and angiogenesis[12]. Although KRAS protein is evenly expressed by most tissues but overexpressed only by a few[13]. However, it most frequently mutate in human cancers. More than 80 % of pancreatic cancers and more than 30 % of CRC, cholangiocarcinoma and lung adenocarcinomas harbor activating mutations of KRAS gene as one of the founder carcinogenic mutation in the genome[14]. Mutation of KRAS is the main pathogenic factor of CRC, about 40 %[14,15]. KRAS mutation has poor prognosis than wild-type mutation in CRC[14-16]. In addition, KRAS mutations, especially G12D are predictive of an inferior response to chemotherapy and a high risk of recurrence[17]. At present, KRAS is used as a routine test in clinic of CRC. More and more drugs research and development for KRAS mutation, such as ARS-853[18], ARS-1620[19], AMG 510[20] and MRTX859[21,22] was carried out.
IL-8, CXCL8 belongs to the neutrophil-specific CXC family of chemokines[23]. It is produced by macrophages and is primarily responsible for neutrophil chemotaxis during the inflammatory process. Recently, many researches has found that CXCL8 play a crucial role in tumor immune escape[24]. Overexpression of CXCL8 promotes the proliferation, migration and invasion of CRC cells[25]. Some studies show that CXCL8 is closely related with metastasis, poor prognosis and poor disease- free survival[26]. In addition, up-regulation of CXCL8 expression is associated with a poor prognosis and enhances tumor cell malignant behaviors in liver cancer[27].
PALB2 as a linker between Breast Cancer gene (BRCA) 1 and BRCA2 bridging protein, it encodes a protein that may function in tumor suppression[28]. In breast cancer, PALB2 has been confirmed as an important carcinogenic factors[29]. In prostate cancer, Woko?orczyk et al. found the PALB2 mutation is associated with high-grade prostate cancer. They suggest PALB2 mutations predispose specifically to aggressive prostate cancers[30].
PRDX4 belongs to the typical 2-Cys PRDXs group and involved cell protection against oxidative injury, regulation of cell proliferation, modulation of intracellular signaling and the pathogenesis of tumors[31]. The PRDX4 show overexpression in many tumors. In breast cancer, the tumor tissues have overexperssion than normal tissues. More than this, the overexpression of PRDX4 have better survival time in breast cancer which show impact and prognosis[32]. In prostate cancer, overexperssion of PRDX4 promoted tumor cell proliferation and invasion[33]. At this time, the overexperssion of PRDX4 have a poor prognosis in prostate cancer[34].
TOMM20 is a receptor that target mitochondria functional protein[35]. Overexperssion of TOMM20 is seen in various cancers such as liver cancer, CRC, breast cancer, lung cancer[36]. In CRC, TOMM20 inhibited cell proliferation, migration and invasion[37]. In Park et al.[38] research in CRC, they suggest TOMM20 as a potential therapeutic target of CRC. Yang et al.[39] found overexperssion of TOMM20 which impacted liver cancer prognosis.
Through our study, the results show KRAS, CXCL8, PALB2, PRDX4, TOMM20 are the key genes and impact prognosis in CRC. We predict that these genes of target drug can promote the survival time of CRC in future.
Conflict of interests:
The authors declared no conflict of interest.
References
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2021;71(3):209-49.
[Crossref] [Google scholar] [PubMed]
- Miller KD, Nogueira L, Mariotto AB, Rowland JH, Yabroff KR, Alfano CM, et al. Cancer treatment and survivorship statistics, 2019. CA Cancer J Clin 2019;69(5):363-85.
[Crossref] [Google scholar] [PubMed]
- Siegel RL, Miller KD, Sauer AG, Fedewa SA, Butterly LF, Anderson JC, et al. Colorectal cancer statistics, 2020. CA Cancer J Clin 2020;70(3):145-64.
[Crossref] [Google scholar] [PubMed]
- Dekker E, Tanis PJ, Vleugels JL, Kasi PM, Wallace M. Colorectal cancer.Lancet 2019;394(10207):1467-80.
[Crossref] [Google scholar] [PubMed]
- Kahi CJ, Boland CR, Dominitz JA, Giardiello FM, Johnson DA, Kaltenbach T, et al. Colonoscopy surveillance after colorectal cancer resection: Recommendations of the US Multi-Society Task Force on Colorectal Cancer. Gastroenterology 2016;150(3):758-68.
[Crossref] [Google scholar] [PubMed]
- Zampino MG, Magni E, Ravenda PS, Cella CA, Bonomo G, Vigna PD, et al. Treatments for colorectal liver metastases: A new focus on a familiar concept. Crit Rev Oncol Hematol 2016;108:154-63.
[Crossref] [Google scholar] [PubMed]
- Wolf AM, Fontham ET, Church TR, Flowers CR, Guerra CE, LaMonte SJ, et al. Colorectal cancer screening for average?risk adults: 2018 guideline update from the American Cancer Society. CA Cancer J Clin 2018;68(4):250-81.
[Crossref] [Google scholar] [PubMed]
- Doubeni CA, Major JM, Laiyemo AO, Schootman M, Zauber AG, Hollenbeck AR, et al. Contribution of behavioral risk factors and obesity to socioeconomic differences in colorectal cancer incidence. J Natl Cancer Inst 2012;104(18):1353-62.
[Crossref] [Google scholar] [PubMed]
- Brenner H, Kloor M, Pox CP. Colorectal cancer. Lancet 2014;383(9927):1490-502.
[Crossref] [Google scholar] [PubMed]
- Li J, Ma X, Chakravarti D, Shalapour S, dePinho RA. Genetic and biological hallmarks of colorectal cancer. Genes Dev 2021;35(11-12):787-820.
[Crossref] [Google scholar] [PubMed]
- Dienstmann R, Mason MJ, Sinicrope FA, Phipps AI, Tejpar S, Nesbakken A, et al. Prediction of overall survival in stage II and III colon cancer beyond TNM system: A retrospective, pooled biomarker study. Ann Oncol 2017;28(5):1023-31.
[Crossref] [Google scholar] [PubMed]
- Liu P, Wang Y, Li X. Targeting the untargetable KRAS in cancer therapy. Acta Pharm Sin B 2019;9(5):871-9.
[Crossref] [Google scholar] [PubMed]
- Zhang Y, Ma JA, Zhang HX, Jiang YN, Luo WH. Cancer vaccines: Targeting KRAS-driven cancers. Expert Rev Vaccines 2020;19(2):163-73.
[Crossref] [Google scholar] [PubMed]
- Dienstmann R, Connor K, Byrne AT, Fridman WH, Lambrechts D, Sadanandam A, et al. Precision therapy in RAS mutant colorectal cancer. Gastroenterology 2020;158(4):806-11.
[Crossref] [Google scholar] [PubMed]
- Mármol I, Sánchez-de-Diego C, Dieste AP, Cerrada E, Yoldi MJR. Colorectal carcinoma: A general overview and future perspectives in colorectal cancer. Int J Mol Sci 2017;18(1):1-39.
[Crossref] [Google scholar] [PubMed]
- Teo MY, Fong JY, Lim WM, In LL. Current advances and trends in KRAS targeted therapies for colorectal cancer. Mol Cancer Res 2022;20(1):30-44.
[Crossref] [Google scholar] [PubMed]
- Biller LH, Schrag D. Diagnosis and treatment of metastatic colorectal cancer: A review. JAMA 2021;325(7):669-85.
[Crossref] [Google scholar] [PubMed]
- Patricelli MP, Janes MR, Li LS, Hansen R, Peters U, Kessler LV, et al. Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer Discov 2016;6(3):316-29.
[Crossref] [Google scholar] [PubMed]
- Janes MR, Zhang J, Li LS, Hansen R, Peters U, Guo X, et al. Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell 2018;172(3):578-89.
[Crossref] [Google scholar] [PubMed]
- Canon J, Rex K, Saiki AY, Mohr C, Cooke K, Bagal D, et al. The clinical KRAS (G12C) inhibitor AMG 510 drives anti-tumour immunity. Nature 2019;575(7781):217-23.
[Crossref] [Google scholar] [PubMed]
- Hallin J, Engstrom LD, Hargis L, Calinisan A, Aranda R, Briere DM, et al. The KRASG12C inhibitor MRTX849 provides insight toward therapeutic susceptibility of KRAS-mutant cancers in mouse models and patients. Cancer Discov 2020;10(1):54-71.
[Crossref] [Google scholar] [PubMed]
- Christensen JG, Olson P, Briere T, Wiel C, Bergo MO. Targeting KRASG12C?mutant cancer with a mutation?specific inhibitor. J Intern Med 2020;288(2):183-91.
[Crossref] [Google scholar] [PubMed]
- Baggiolini M. CXCL8-the first chemokine. Front Immunol 2015;6:1-3.
[Crossref] [Google scholar] [PubMed]
- Russo RC, Garcia CC, Teixeira MM, Amaral FA. The CXCL8/IL-8 chemokine family and its receptors in inflammatory diseases. Expert Rev Clin Immunol 2014;10(5):593-619.
[Crossref] [Google scholar] [PubMed]
- Bie Y, Ge W, Yang Z, Cheng X, Zhao Z, Li S, et al. The crucial role of CXCL8 and its receptors in colorectal liver metastasis. Dis Markers 2019;2019:1-12.
[Crossref] [Google scholar] [PubMed]
- Lee YS, Choi I, Ning Y, Kim NY, Khatchadourian V, Yang D, et al. Interleukin-8 and its receptor CXCR2 in the tumour microenvironment promote colon cancer growth, progression and metastasis. Br J Cancer 2012;106(11):1833-41.
[Crossref] [Google scholar] [PubMed]
- Sun F, Wang J, Sun Q, Li F, Gao H, Xu L, et al. Interleukin-8 promotes integrin β3 upregulation and cell invasion through PI3K/Akt pathway in hepatocellular carcinoma. J Exp Clin Cancer Res 2019;38(1):1-10.
[Crossref] [Google scholar] [PubMed]
- Xia B, Sheng Q, Nakanishi K, Ohashi A, Wu J, Christ N, et al. Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell 2006;22(6):719-29.
[Crossref] [Google scholar] [PubMed]
- Antoniou AC, Casadei S, Heikkinen T, Barrowdale D, Pylkäs K, Roberts J, et al. Breast-cancer risk in families with mutations in PALB2. N Engl J Med 2014;371(6):497-506.
[Crossref] [Google scholar] [PubMed]
- Woko?orczyk D, Klu?niak W, Stempa K, Rusak B, Huzarski T, Gronwald J, et al. PALB2 mutations and prostate cancer risk and survival. Br J Cancer 2021;125(4):569-75.
[Crossref] [Google scholar] [PubMed]
- Park SY, Lee YJ, Park J, Kim TH, Hong SC, Jung EJ, et al. PRDX4 overexpression is associated with poor prognosis in gastric cancer. Oncol Lett 2020;19(5):3522-30.
[Crossref] [Google scholar] [PubMed]
- Tiedemann K, Sadvakassova G, Mikolajewicz N, Juhas M, Sabirova Z, Tabariès S, et al. Exosomal release of L-plastin by breast cancer cells facilitates metastatic bone osteolysis. Transl Oncol 2019;12(3):462-74.
[Crossref] [Google scholar] [PubMed]
- Jia W, Chen P, Cheng Y. PRDX4 and its roles in various cancers. Technol Cancer Res Treat 2019;18:1-10.
[Crossref] [Google scholar] [PubMed]
- Basu A, Banerjee H, Rojas H, Martinez SR, Roy S, Jia Z, et al. Differential expression of peroxiredoxins in prostate cancer: Consistent upregulation of PRDX3 and PRDX4. Prostate 2011;71(7):755-65.
[Crossref] [Google scholar] [PubMed]
- Zhao Z, Han F, He Y, Yang S, Hua L, Wu J, et al. Stromal-epithelial metabolic coupling in gastric cancer: Stromal MCT4 and mitochondrial TOMM20 as poor prognostic factors. Eur J Surg Oncol 2014;40(10):1361-8.
[Crossref] [Google scholar] [PubMed]
- Zhou B, Zhang JY, Liu XS, Chen HZ, Ai YL, Cheng K, et al. Tom20 senses iron-activated ROS signaling to promote melanoma cell pyroptosis. Cell Res 2018;28(12):1171-85.
[Crossref] [Google scholar] [PubMed]
- Park SH, Lee AR, Choi K, Joung S, Yoon JB, Kim S. TOMM20 as a potential therapeutic target of colorectal cancer. BMB Rep 2019;52(12):712-717.
[Crossref] [Google scholar] [PubMed]
- Park SH, Oh SO, Park SS, Roh SJ, Han KS, Kim B, et al. Characteristics of minute T1 colorectal cancer in relevance to pathology and treatment. Ann Surg Treat Res 2020;98(4):199-205.
[Crossref] [Google scholar] [PubMed]
- Yang X, Song D, Zhang J, Feng H, Guo J. PRR34-AS1 sponges miR-498 to facilitate TOMM20 and ITGA6 mediated tumor progression in HCC. Exp Mol Pathol 2021;120:1-8.
[Crossref] [Google scholar] [PubMed]