- *Corresponding Author:
- Xiaohui Xu
School of Life Sciences, Shanghai University, Shanghai 200444, China
E-mail: xhxu@wnmc.edu.cn
| This article was originally published in a special issue,“New Research Outcomes in Drug and Health Sciences” |
| Indian J Pharm Sci 2023:85(6) Spl Issue “102-114” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
This study investigated the feasibility of polypyrimidine tract binding protein 1 knockout mouse spinal cord astrocytes in the treatment of spinal cord injury. We cultured primary astrocytes from the spinal cord of newborn mice, then integrated sh polypyrimidine tract binding protein 1 into the genome by lentiviral transduction and verified their knockout efficiency. Immunofluorescence staining and transcriptome sequencing were performed to test whether the cells differentiated into neurons. The results showed that polypyrimidine tract binding protein 1 knockout could transform spinal cord astrocytes into functional neurons in one step. After whole transcriptome sequencing analysis, gene ontology annotation and Kyoto encyclopedia of genes and genomes pathway enriched differentially expressed genes revealed that mitogen-activated protein kinase, erythroblastic oncogene B and Ras-associated protein-1 signaling pathways may be involved in this transdifferentiating process. Predictive analysis of microRNA target genes revealed microRNAs that may be involved in neural differentiation, as well as long noncoding RNA and circular RNA that indirectly regulate transdifferentiating through these microRNAs. Analysis of the circular RNA-microRNA-messenger RNA network revealed three circular RNA that may play a key role in transdifferentiating, as well as possible regulatory mechanisms in this process. We further constructed a mouse spinal cord injury model and found that 9 w after surgery, the basso mouse scale score of the right hind limb of the sh polypyrimidine tract binding protein 1 lentivirus injection group was significantly higher than that of the scrambled lentivirus injection group. Our results suggest that polypyrimidine tract binding protein 1 knockdown of reactively proliferating spinal cord astrocytes in spinal cord injury promotes neuron generation.
Keywords
Spinal cord injury, astrocytes, transdifferentiation, polypyrimidine tract binding protein 1, whole-transcriptome sequencing
Spinal Cord Injury (SCI) results in the destruction of the anatomical structure of the spinal cord, resulting in a series of pathological reactions such as axonal rupture, neuronal degeneration and necrosis, inflammatory reaction, demyelination and astrogliosis, which eventually leads to severe neurological dysfunction.
The fundamental structural and operational components of the Central Nervous System (CNS) are neurons. Adult neurons are basically irreversible after injury and death. When SCI occurs, astrocytes in the injured area will be activated, resulting in reactive cell hyperplasia and hypertrophy, and finally form a dense glial scar. In the chronic secondary stage of SCI, glial scar formation (gliosis) is a reactive cellular mechanism supported by astrocytes. The body's natural repair mechanism after SCI protects and starts with the scarring of astrocytes. This thick astrocyte scar acts as a barrier that prevents inflammatory cells from spreading from the non-neuropathic core to the remaining nerve cells around it[1].
On the other hand, the presence of glial scars is also an important reason why injured axons fail to regenerate. Glial scars not only form physical barriers, but also secrete axonal growth inhibitors, which affect SCI regeneration and repair. Activated astrocytes secrete a large group of proteoglycans such as tenascin, an endothelial specific growth factor ephrin-B2, and Heparan Sulphate Proteoglycans (HSPG), Dermatan Sulphate Proteoglycan (DSPG), Keratan Sulphate Proteoglycan (KSPG) and Chondroitin Sulphate Proteoglycan (CSPG), and other proteoglycans. These proteoglycans inhibit nerve regeneration after injury, thus forming a chemical barrier to nerve regeneration. Although different neuronal populations in the spinal cord have variable capacities for expanding into proteoglycanrich lesions, near the center of the glial scar lesion, all growing fibers develop dystrophic characteristics and eventually stop developing[2]. As a result, it has been suggested that controlling some of the basic traits of activated astrocytes is a key technique for treating CNS damage.
SCI leads to the irreversible loss of a large number of neurons and the destruction of local neural circuits, which are the main causes of severe neurological dysfunction. Therefore, exploring how to supply these lost neurons and reconstruct local neural circuits is the biggest challenge for effective SCI repair. At present, transplantation of neural precursor cells or neurons derived from stem cells is an suggest ways to treatment of SCI, but it still faces many challenges, such as immune activation, ethical barriers and the risk of tumor formation.
If these over-activated astrocytes can be converted into neurons through cellular reprogramming of the injured spinal cord, it will not only replenish the neurons lost during SCI, but also reduce the formation of glial scar, thereby promoting SCI repair.
In 2020, Xiangdong Fu laboratory found that knockdown of Ribonuclic Acid (RNA)-binding protein Polypyrimidine Tract-Binding (PTB) in astrocytes could directly transdifferentiate them into functional neurons. This one-step transdifferentiating technique can induce the generation of new functional dopamine neurons, reconstruct the damaged neural circuits, restore the dopamine level in the striatum in a mouse model of Parkinson's disease, and effectively treat the movement disorders associated with Parkinson's syndrome. More importantly, use of inhibiting Polypyrimidine Tract-Binding Protein 1 (PTBP1) Antisense Oligonucleotide (ASO) also can achieve similar therapeutic effect[3]. Although the detailed mechanism by which PTBP1 knockdown alone can effectively induce neuronal transdifferentiating is not clear, this simple approach of knockdown of a single gene is more likely to be applied to clinical translation, because similar drugs by ASO and small interfering RNA already have mature Food and Drug Administration (FDA)-approved drug applications. For example, Roche received an implied approval application for a clinical trial of its new drug for Huntington's Disease (HD), RO7234292 injection. This ASO drug, developed by Roche and ion is pharmaceuticals, targets the underlying cause of HD by reducing mutant Huntington (HTT) Protein, is the first of its kind to enter a pivotal trial and has been designated as an orphan drug for HD patients by the US FDA and European Medicines Agency (EMA), Europe. In addition, ONPATTRO® (patisiran), the first therapeutic small interfering RNA (siRNA) approved by FDA in 2019, is used to treat hereditary Transthyroxine Mediated Amyloidosis (hATTR).
Due to the regional heterogeneity of astrocytes, whether astrocytes in the spinal cord can be reprogrammed into spinal-specific neurons by knocking down PTBP1 and reconstructing spinal cord neural circuits and the specific mechanism are still unclear. In this study, we induced neuronal transformation of primary mouse spinal cord astrocytes by lentiviral transduction of small hairpin RNA targeting PTBP1 (shPTBP1), and then explored the related genes and mechanisms of transdifferentiating by whole transcriptome sequencing. Our work may provide new clues for the clinical treatment of SCI in the future.
Materials and Methods
Animals:
All methods for handling and using animals complied with National Institutes of Health (NIH) regulations and were approved by Shanghai University's Animal Experimentation Ethics Committee. Shanghai Jiesijie Laboratory Animal Co., Ltd. sold the KM mice that were used in this investigation. All experimental animals were female mice in order to exclude out sex differences.
Real Time-quantitative Polymerase Chain Reaction (RT-qPCR):
Total RNA was extracted with Trizol (Vitrogen). RNA samples were reverse transcribed into complimentary Deoxyribonucleic Acid (cDNA) using the GoldenstarTMRT6 cDNA Synthesis Kit (TSINGKE, #TSK302S) and used as a template for qPCR. RT–qPCR was performed using the 2× Fast qPCR Mix (SYBR Green I) kit (TSINGKE, #TSE202) on a CFX96 PCR instrument (BIORAD). The primers used are listed as follows; primer sequence (from 5’ to 3’); PTBP1-forward GTACAAAGCGGGGATCTGAC and reverse CGGCTGTCACCTTTGAACTT; Glyceraldehyde 3-Phosphate Dehydrogenase (GAPDH)-forward AGGCCGGTGCTGAGTATGTC and reverse TGCCTGCTTCACCACCTTCT.
Immunofluorescence:
Cells on dishes were fixed with 4 % paraformaldehyde for 20 min at room temperature, followed by three separate washings with 0.01 M Phosphate Buffer Solution (PBS) for 5 min each. After that, cells were exposed to 0.3 % Triton X-100 with 3.0 % Bovine Serum Albumin (BSA) for 30 min at room temperature. After that, cells were washed three times with 0.01 M PBS for 5 min each time, followed by a 12 h–16 h incubation with primary antibodies at 4°. The cells were then given fluorescence-conjugated secondary antibodies, and they were incubated at 37° for 40 min. Hoechst 33258 nuclear stain was applied and let to sit at room temperature for 10 min before two more 5 min PBS rinses were performed. The primary antibodies included anti-Beta (β)-tubulin III (1:2000, Promega, G7121), anti-5-HT (1:5000, ImmunoStar, 20079). The secondary antibodies were AF594 antibodies to mouse Immunoglobulin G (IgG) (1:200) and AF647 antibodies to goat IgG (1:200).
Vectors and virus production:
To construct a lentiviral vector expressing shPTB in mouse astrocytes, the target sequence 5'-GGGTGAAGATCCTGTTCAATA-3 was shuttled to pLKO. 1 Puro vector (Addgene, #8453). Viral particles were packaged in 293FT cells cotransfected with two packaging plasmids: pMD2.G (Addgene, #12259) and psPAX2 (Addgene, #12260).
In vitro cell culture and transdifferentiation:
Mouse astrocytes were isolated from spinal cord of postnatal (P0–P1) pups and plated onto dishes coated with poly-D-lysine (sigma) after incubating with trypsin. Isolated astrocytes were cultured in Dulbecco's Modified Eagle Medium (DMEM) (GIBCO) plus 10 % Fetal Bovine Serum (FBS) and penicillin/streptomycin (GIBCO). After reaching ~90 % confluence, the flask was placed in a shaker at 37° at 200 rpm overnight. Since astrocytes are more adherent, the lowest layer of cells left after an overnight stay were astrocytes. Then cells were passaged with trypsin.
The mouse PTB-targeting lentivirus was used to infect mouse astrocytes, which were then plated on Poly-D-Lysine (PDL)-coated dishes to induce transdifferentiation in vitro. Cells were chosen with puromycin (1.2 g/ml, Thermo Fisher) in new media for 72 h after 24 h. After that, the medium was changed to N3/basal media, which contained 0.4 % B27, 2 % FBS, and a trio of small molecules (1 M ChIR99021, 10 M SB431542, and 1 mM DbcAMP) in addition to 25 g/ml of insulin, 50 g/ml of transferring, and 30 nm of sodium selenite. Every other day, only half of the medium was changed.
RNA-seq and data analysis:
Total RNA of the samples was extracted using TRIzol (Invitrogen) technique and genomic DNA was eliminated using DNase I (TaKara). The quality and quantity of RNA samples were determined using 2100 Bioanalyzer (Agilent) and ND-2000 (nanodroptechnologies) methods to ensure that only qualified samples (Optical Density (OD) 260/280=1.82.2, OD 260/2302.0, RIN6.5, 28S:18S1.0, >1 g) were used for transcriptome sequencing.
The TruSeqTM RNA sample preparation kit from Illumina, San Diego, California, was used to create RNA libraries. First, 1 g of total RNA was utilized to enrich messenger RNA (mRNA) with poly-A tail using magnetic beads and Oligo (dT). The mRNA was then randomly split into fragments of 300 bp in a fragmentation buffer. Then, using reversetranscribed mRNA as a template, a strand of cDNA was created using an Invitrogen (California) SuperScript double-stranded cDNA synthesis kit and six-base random primers (Illumina). After that, a stable double-strand structure is created using two-strand synthesis. When End Repair Mix was applied, the sticky end of the doublestranded cDNA was patched into a flat end, and an A base was added to the 3' end to link the Y-shaped connection. DNA clean beads were employed to screen bands of 200–300 bp after cDNA was enriched by PCR (sample preparation Kit, Illumina, San Diego, California) on the sample. Following TBS380 (Picogreen) quantification, the libraries were sequenced utilizing the highthroughput Illumina HiSeq xten/NovaSeq 6000 sequencing technology with PE150 read length. Website (https://cloud.majorbio.com/) used to perform differential expression gene analysis and target gene prediction analysis.
SCI surgical procedures:
Mice were placed on a continual heating pad to maintain temperature at 37° throughout the SCI process after anesthesia with 0.3 % sodium pentobarbital solution (0.2 ml/10 g, i.p). At the level of T10–T12, a laminectomy was done to expose the dorsal cord surface without damaging the dura. The right half of the spinal cord was then sliced from top to bottom using microsurgical shears. Then, using 4-0 silk and a needle, layers of muscle and skin were sutured. After SCI, the bladder had to be manually emptied twice a day until the ability to urinate was restored. All of the animals in the sham group had laminectomy alone.
7 d later, the skin of mice in the injured group was cut again to expose the spinal cord. Three mice in each group were injected with shPTBPP1 lentivirus and scramble empty vector lentivirus.
Basso Mouse Scale (BMS) scoring analysis:
To find changes in mice's hind limb locomotive function after SCI, BMS scoring analysis was used. All animals were rated from 0 to 9 points using the same scoring criteria (posterior ankle joint mobility, coordination, paw posture, trunk stability, and tail posture) by the same one or two observers who were blind to the experimental condition.
Statistical analysis:
All data were statistically analyzed using a singlefactor Analysis of Variance (ANOVA) GraphPad Prism 5 and a Student t-test. All results are shown as the average Standard Error of the Mean (SEM) of at least three experiments (significant differences were considered when p<0.05).
Results and Discussion
To verify that lentiviruses can effectively knockdown PTBP1 in mouse spinal cord astrocytes, we transduced mouse spinal cord astrocytes with lentiviruses expressing shPTBP1.
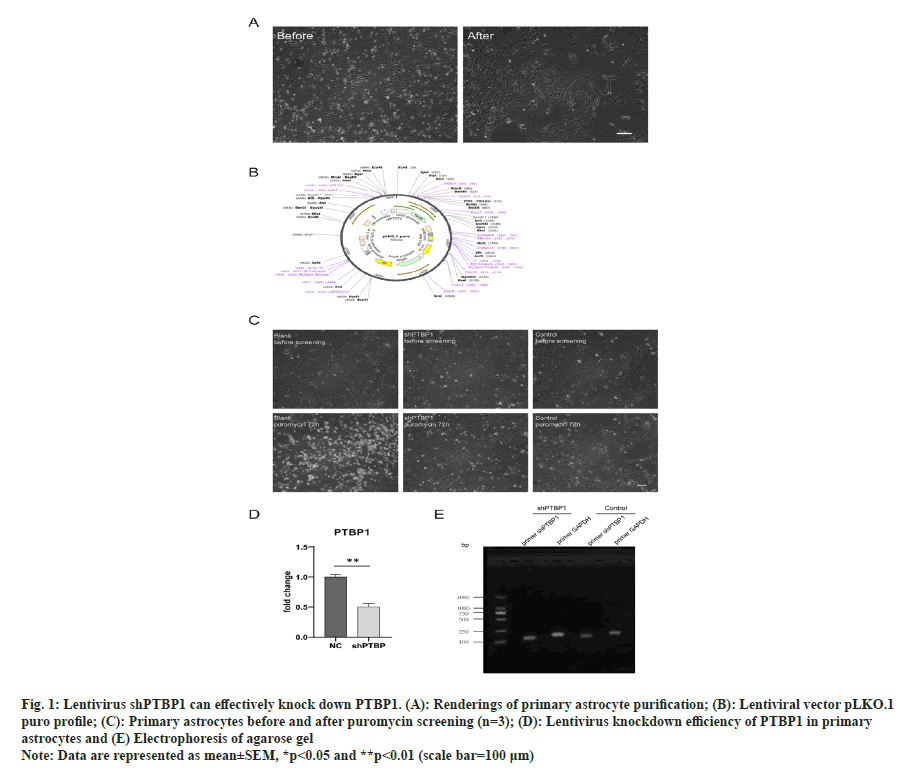
To prepare mouse spinal cord astrocytes, primary mixed cells obtained from the spinal cord of 0 d newborn mice were cultured in PDL-coated flask for about 7 d until the cells almost covered the bottom of the flask and placed in a shaker at 37° at 200 rpm overnight. Because astrocytes were more adherent, astrocytes were the lowest layer of adherent cells left after overnight shaking (fig. 1A).
The lentivirus with a final concentration of 8 μg/ ml was added to improve the efficiency of viral infection. Since the lentivirus is puromycin resistant, we treated cells with puromycin to screen successfully infected cells (fig. 1B). After pre-experiments, it was found that 1.2 μg/ ml of puromycin could kill almost all wild-type astrocytes with a density of >90 % after 72 h (data not show), so we used 1.2 μg/ml of puromycin to screen successfully infected cells, and withdrew the drug when all the cells in the blank group died to end the screening. The transfection efficiency was found to reach >80 % (fig. 1C).
Fig. 1: Lentivirus shPTBP1 can effectively knock down PTBP1. (A): Renderings of primary astrocyte purification; (B): Lentiviral vector pLKO.1
puro profile; (C): Primary astrocytes before and after puromycin screening (n=3); (D): Lentivirus knockdown efficiency of PTBP1 in primary
astrocytes and (E) Electrophoresis of agarose gel
Note: Data are represented as mean±SEM, *p<0.05 and **p<0.01 (scale bar=100 μm)
After successfully infected cells were screened, total RNA was extracted from the cells, followed by reverse transcription and RT-qPCR to verify the knockout efficiency of PTBP1 in the cells. The results showed that the PTBP1 level was significantly reduced (fig. 1D). In order to ensure the specific amplification of the target gene in qPCR, agarose gel electrophoresis was performed with the qPCR product, and the electrophoresis results showed only the band of the target gene (fig. 1E).
Therefore, lentivirus expressing shPTBP1 can effectively knock down PTBP1 in mouse spinal cord astrocytes.
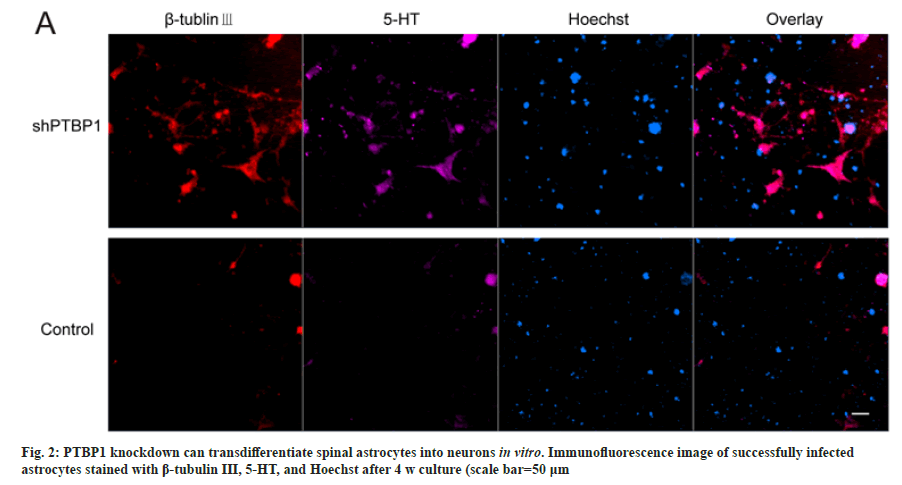
To verify that knocking down PTBP1 transdifferentiated mouse spinal astrocytes into neurons, we repeated the lentiviral transfection experiment described above. And after puromycin screening, the successfully infected cells were cultured for 4 w. referring to the medium of Fu Laboratory[3], we added 25 μg/ml insulin, 50 μg/ ml transferring, 30 nm sodium selenite, 0.4 % B27, 2 % FBS, and three small molecules (1 μm ChIR99021, 10 μm SB431542, and 1 mm dibutyryl Cyclic Adenosine Monophosphate (dBcAMP)) with DMEM/F12 and neurobasal 1:1. The medium was half-changed every the other day. After 4 w, the cells were fixed and immunofluorescent stained with β-tubulin III and 5-HT antibodies. shPTBP1 transduced cells were β-tubulin III positive and most of them also expressed serotonergic neuronal markers. However, astrocytes transduced with control empty vector lentivirus were not positive (fig. 2). These results suggest that PTBP1 knockdown can transform spinal astrocytes into functional neurons in one step.
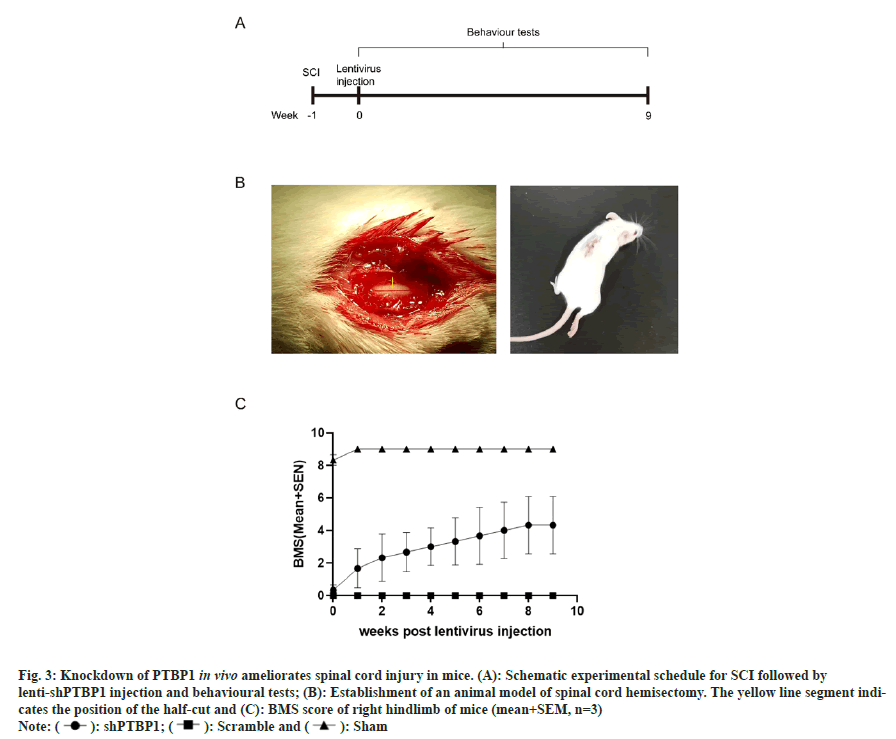
We further constructed a mouse model of SCI (spinal cord hemisectomy) (fig. 3A and fig. 3B), and injected concentrated shPTBP1 lentivirus at the injury site on postoperative d 7. Scramble group was injected with empty vector lentivirus, and sham group was only opened lamina without spinal cord hemisectomy or virus injection. After 9 w, it was found that the mice injected with shPTBP1 lentivirus showed a trend of motor improvement, and the BMS score of the right hindlimb was significantly higher than that of the injected scramble lentivirus group (fig. 3C). Our results suggest that after PTBP1 knockdown in reactive proliferative spinal astrocytes at SCI, the reprogrammed neurons may reconstruct the neural circuitry at the site of SCI and ultimately promote the recovery of spinal nerve function.
Fig. 3: Knockdown of PTBP1 in vivo ameliorates spinal cord injury in mice. (A): Schematic experimental schedule for SCI followed by
lenti-shPTBP1 injection and behavioural tests; (B): Establishment of an animal model of spinal cord hemisectomy. The yellow line segment indicates
the position of the half-cut and (C): BMS score of right hindlimb of mice (mean+SEM, n=3)
Note: ( ): shPTBP1; (
): shPTBP1; ( ): Scramble and (
): Scramble and ( ): Sham
): Sham
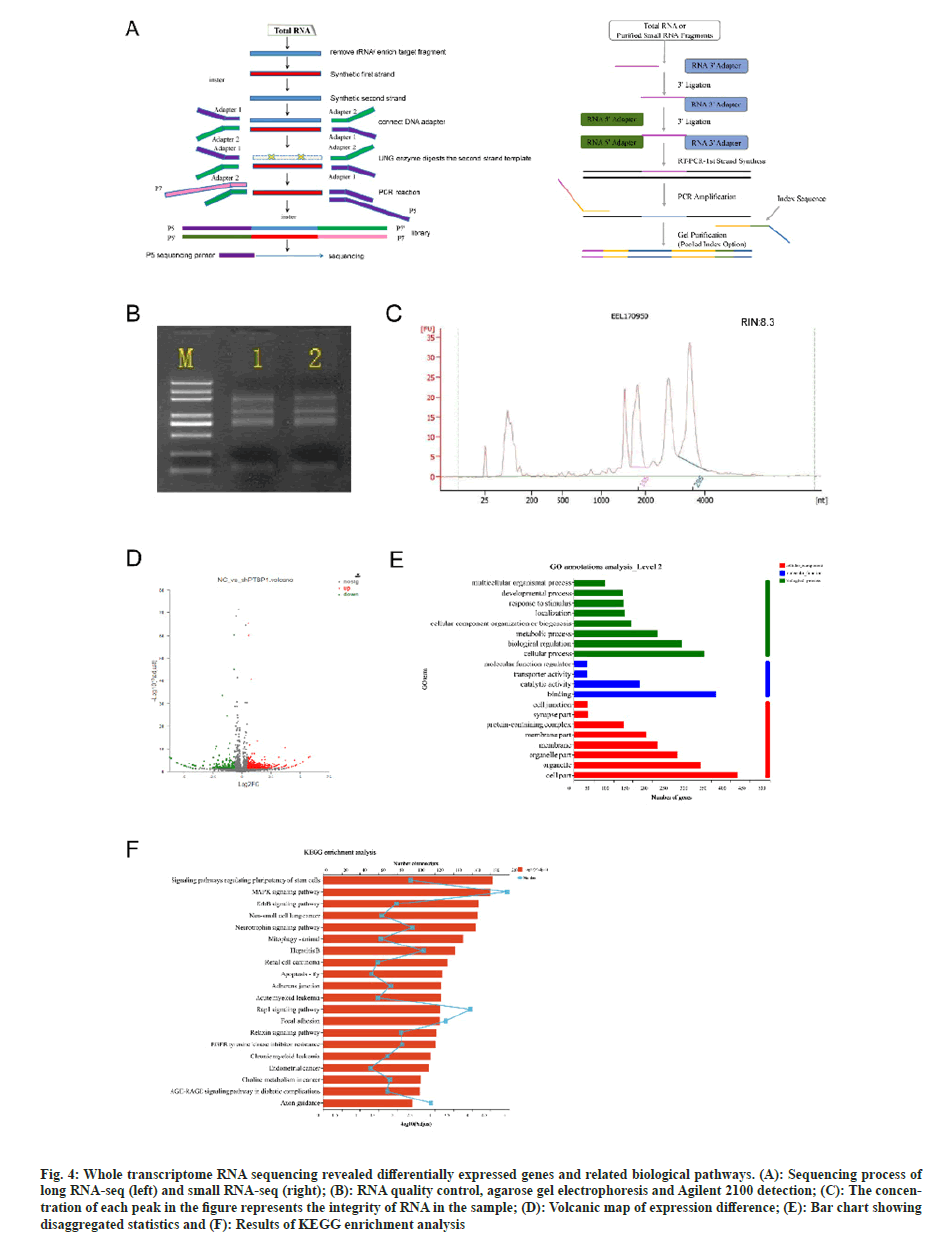
The whole transcriptome is the collection of all transcription products (mRNA, long noncoding RNA (lncRNA), circular RNA (circRNA) and microRNA) of a specific cell or tissue in a certain functional state. With the help of high-throughput sequencing technology, the information of transcription products in samples can be comprehensively obtained, joint analysis can be carried out, and the transcriptional level regulation network can be deeply mined. In this sequencing experiment, strand specific libraries were used to construct libraries and sequenced on Illumina platform, and two libraries were established for long RNA-seq and small RNA-seq (fig. 4A).
The sequencing samples needed to be checked before loading, with NanoDrop 2000 to detect RNA concentration and purity, agarose gel electrophoresis to detect RNA integrity, and Agilent 2100 Nano to detect RNA Integrity Number (RIN) values for samples. RIN values indicate the integrity of RNA samples, ranging from 1 to 10, with higher values indicating better RNA integrity. The quality inspection results show that the RNA bands are clear, without pigment, protein, sugar and other impurities contamination. The brightness of 28/23S is greater than 18/16S, and RIN value is 8.3, OD260/280≥1.8, and OD260/230≥1.0, indicating that the quality meets the requirements of standard library construction (fig. 4B and fig. 4C).
Fig. 4: Whole transcriptome RNA sequencing revealed differentially expressed genes and related biological pathways. (A): Sequencing process of long RNA-seq (left) and small RNA-seq (right); (B): RNA quality control, agarose gel electrophoresis and Agilent 2100 detection; (C): The concentration of each peak in the figure represents the integrity of RNA in the sample; (D): Volcanic map of expression difference; (E): Bar chart showing disaggregated statistics and (F): Results of KEGG enrichment analysis
The gene expression profiles of lentiviral-treated astrocytes that had PTBP1 knocked down were evaluated using RNA sequence analysis. 634 genes with differential expression in all were found. Their 465 genes are up-regulated and their 169 genes are down-regulated (fig. 4D).
Gene Ontology (GO) association created the GO database, which can categorize and annotate the genes in the chosen gene set and explain the biological activities of genes from three perspectives; cell component, molecular function, and biological process. Through the GO annotation analysis of the differentially expressed genes in this study, a total of 47 GO entries were generated, of which the top 20 GO entries were shown in fig. 4E. The main GO functions of these differentially expressed genes are as follows; metabolic processes, biological regulation, cellular processes, catalytic activity, binding, organelle and cell part, etc.
Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway is one of the many databases that make up the KEGG database, which carefully examines the metabolic pathways of gene products in cells. Understanding the significantly altered metabolic pathways under experimental conditions requires pathway analysis of differentially expressed genes. We used KEGG functional enrichment analysis to identify the pathways in which differentially expressed gene transcripts primarily participated. These pathways included those that control the pluripotency of stem cells, the Mitogen-Activated Protein Kinase (MAPK) signaling pathway, the Erythroblastic oncogene B (ErbB) signaling pathway, the non-small cell lung cancer pathway, the neurotrophin signaling pathway, the Ras-associated protein-1 (Rap1) signaling pathway, and the axonal guidance (fig. 4F). Previous studies have reported that MAPK signaling contributes to the acceleration of neuronal differentiation, and ErbB is associated with neuronal axon growth and differentiation phenotype, and Rap1 signaling is important for migration, differentiation, axon growth and neuronal polarity[4-11]. Therefore, PTBP1 may be involved in regulating neuronal differentiation through the above pathways.
Genes are influenced by many regulations in the process of changing from DNA to proteins. It mainly includes five levels of regulation; gene level, transcription level, post-transcriptional level, translation level and post-translational level. Post-transcriptional regulation is the process of converting primary transcriptional RNA into mature RNA in eukaryotic cells, including conventional regulation of alternative splicing of RNA, m6A methylation, 5' capping and 3' capping of mRNA, microRNA regulation and so on. The process of post-transcriptional regulation mainly depends on RNA-Binding Proteins (RBP).
A well-researched RNA-binding protein called PTBP1 is involved in a number of posttranscriptional processes that control how genes are expressed. Although originally classified as a pre-mRNA splicing regulator, PTBP1 has been widely recognized as a multifunctional protein shuttling between the nucleus and cytoplasm. As a result, certain RNA targets, structural components, and proteins can interact with PTBP1[12].
The ability of PTBP1 to transform fibroblasts into neuron-like cells engaged in the functioning of the microRNA circuits it regulates was demonstrated by Xue et al.[13]. The reduction of PTBP1-mediated inhibition of the action of microRNAs on various REST complex members, which uninhibited a huge number of neuronal genes, including miR-124 and numerous transcription factors specific to neurons in non-neuronal cells, is a crucial event during the induction of neurons. This causes the cell to undergo reprogramming into a neuronal lineage by changing the negative feedback loop into a positive feedback loop. Therefore, we speculate that in the process of knocking down PTBP1 to transdifferentiate astrocytes into neurons, there are also related microRNAs involved in regulation.
Differential analysis of microRNA expression showed that a total of 46 microRNAs had significant changes in expression, including known miRNAs and new miRNAs. Among them, 17 miRNAs were up-regulated 29 were downregulated.
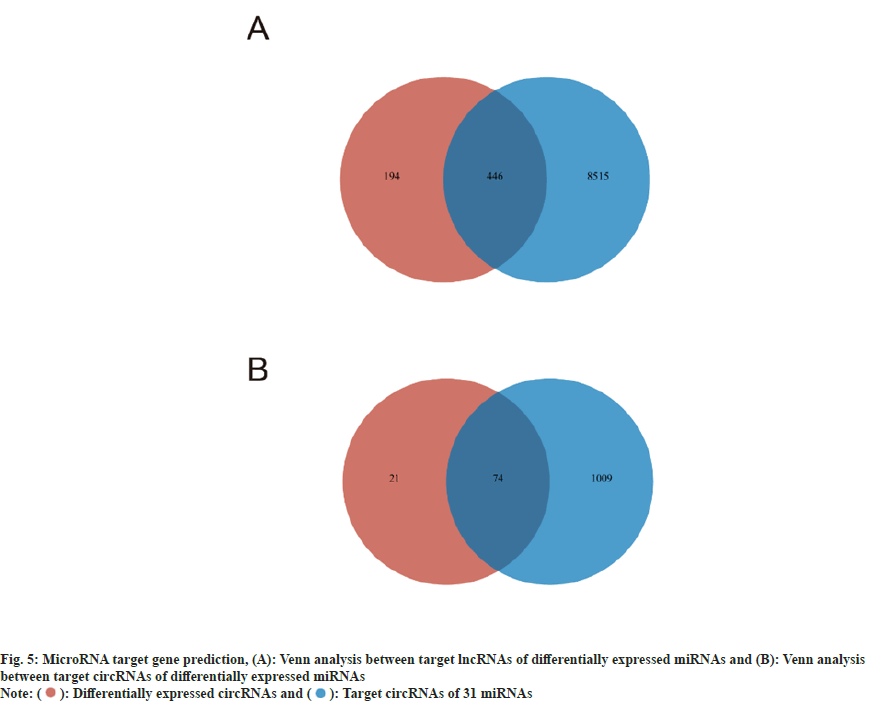
Post-transcriptional control of their target genes is how miRNAs work. One miRNA may control several target genes, and numerous miRNAs of various kinds may control the same target gene. Creating a transcript set of 142 mRNAs of known neural differentiation-specific factors[14], and running target gene prediction with the above 46 differentially expressed miRNAs yielded 31 miRNAs, which regulate 138 target mRNAs involved in neural differentiation. Because knockdown of PTBP1 will also cause differential expression of ncRNA, we want to further understand whether the differentially expressed ncRNA can regulate the neuronal transdifferentiating of astrocytes by sponge these 31 miRNAs.
In order to explore which lncRNAs these 31 miRNAs may interact with, we created a transcript set of all lncRNAs in the sample and ran target gene prediction with these 31 miRNAs and obtained 8961 lncRNAs. Venn analysis was conducted between them and 640 differentially expressed lncRNAs, and 446 lncRNAs were obtained (fig. 5A). These differentially expressed lncRNAs may indirectly regulate neuronal differentiation by interacting with the 31 miRNAs described above.
Similarly, in order to explore which circRNAs these 31 miRNAs may interact with, we created a transcript set of all circRNAs in the sample and ran target gene prediction with these 31 miRNAs. 1083 circRNAs were obtained. Venn analysis was performed between them and 95 differentially expressed circRNAs, and 74 circRNAs were obtained (fig. 5B). These differentially expressed circRNAs may indirectly regulate neuronal differentiation by interacting with the above 31 miRNAs.
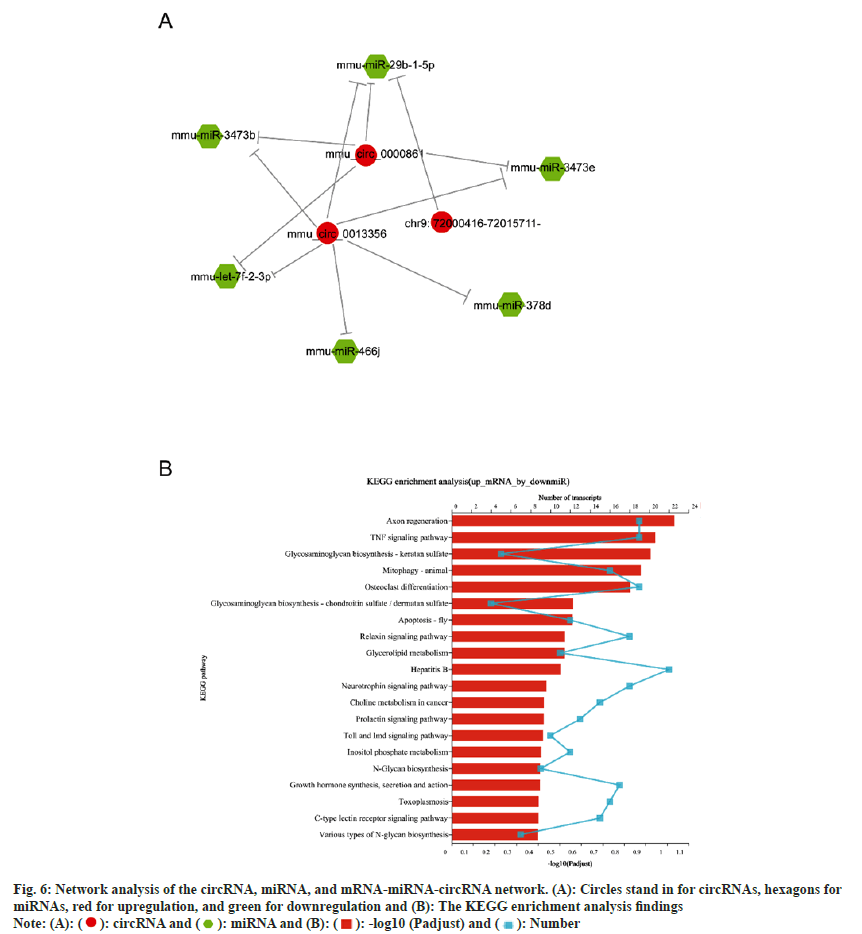
According to studies, circRNAs operate as a "miRNA sponge" to absorb miRNA and so accurately regulate miRNA levels. To learn more about circRNA functions and determine which circRNAs act as miRNA sponges in neuronal development, we performed predictions of interactions of top 3 significantly up-regulated circRNAs with differentially expressed miRNAs using popular miRNA target gene prediction software. These 3 circRNAs are 18_35610871_35613502 (mmu_ circ_0000861), 6_38440455_38491931 (mmu_ circ_0013356), and 9_72000416_72015711 (chr9: 72000416-72015711-). Proprietary software based on Miranda and RNA hybrid was used to reversely predict miRNAs, as well as target mRNAs of significantly down-regulated miRNAs. The results showed that 6 miRNAs were significantly down-regulated by the 3 circRNAs (fig. 6A). KEGG functional enrichment analysis of their up-regulated target mRNAs found that they played important roles in axon regeneration, TNF signaling pathway, and neurotrophin signaling pathway (fig. 6B).
Fig. 6: Network analysis of the circRNA, miRNA, and mRNA-miRNA-circRNA network. (A): Circles stand in for circRNAs, hexagons for
miRNAs, red for upregulation, and green for downregulation and (B): The KEGG enrichment analysis findings
Note: (A): ( ): circRNA and (
): circRNA and ( ): miRNA and (B): (
): miRNA and (B): ( ): -log10 (Padjust) and (
): -log10 (Padjust) and ( ): Number
): Number
In conclusion, the 3 significantly up-regulated circRNAs mentioned above may indirectly increase the expression of some mRNAs related to neural differentiation and axon regeneration by acting on 6 miRNAs to down-regulate them, and eventually lead to the transdifferentiation of astrocytes to neurons.
A significant development in regenerative medicine has just been made thanks to in vivo neural reprogramming. Due to the adult CNS limited capacity for neurogenesis, this groundbreaking technology may be utilized to heal damage to the CNS by turning endogenous glial cells into functioning neurons. This strategy totally solves the crucial issue of immunological rejection that cell transplantation techniques encounter. Additionally, other teams have successfully used various disease models to demonstrate in vivo neural reprogramming[15].
In vivo neural reprogramming has shown effects on stroke-induced brain damage[16], chemically induced PD[17,18] and genetic mouse models of blindness[19]. If these developments are validated, they may open the door to previously unheardof regeneration therapy for numerous additional nerve lesions and neurological conditions. The function of newly reprogrammed neurons in the process of functional recovery is uncertain because glial cell reprogramming not only creates new neurons but also alters the microenvironment. Reactive gliosis regulates tissue damage and nerve regeneration following injury or degeneration[20-23]. However, the functional recovery following nerve injury is inhibited by reactive glial cell scarring and Chondroitin Sulfate Proteoglycan (CSPG) secretion. Post-traumatic regeneration may be enhanced by reducing reactive gliosis or CSPG activity[24,25]. Determining how new neurons and environmental factors affect functional recovery following injury or degeneration is therefore critical.
On the other hand, despite recent significant advances in neural reprogramming in vivo, there are still critical issues that need to be considered to advance the field. For example, whether glial transformed neurons have any deleterious effects remains to be determined, as these new neurons may lead to the disruption of preexisting neural circuits or the formation of aberrant circuits/PTBP1 itself is an RNA-binding protein, and changes in its expression affect mRNA and noncoding RNA splicing in vivo.
In this study, we transduced mouse spinal cord primary astrocytes with lentivirus expressing shPTBP1 to knockdown PTBP1 and explore the feasibility of transdifferentiating into neurons for the treatment of SCI[26,27].
The medium used was referred to the transdifferentiating medium of Xiangdong Fu laboratory[3]. The results of qPCR showed that the level of PTBP1 was significantly knocked down by about 50 %. We also performed agarose gel electrophoresis of the qPCR products to ensure that the target gene was amplified specifically in qPCR. Immunofluorescence showed that shPTBP1 virus-transduced cells were positive for the neuronal marker β-tubulin III, and most of them also expressed the serotonergic neuronal marker, while astrocytes transduced with the control empty vector lentivirus were not positive. We further constructed a mouse model of SCI (cord hemisectomy). It was found that the score of right hind limb of mice in shPTBP lentivirus injection group was significantly higher than that in scramble lentivirus injection group at 9 w after operation. We found that mice injected with shPTBP lentivirus began to show improvement in motor function 1 w after injection, which was speculated to be due to changes in the characteristics of activated astrocytes and decreased secretion of inhibitory factors after PTBP1 knockdown. Therefore, our results strongly support that knockdown of PTBP1 in vivo promotes the transdifferentiation of astrocytes into neurons and promotes the repair of SCI.
Following analyzing the entire transcriptome sequencing results for differential expression, GO annotation, and KEGG enrichment, we discovered that 465 genes were significantly up-regulated and 169 genes were significantly down-regulated following PTBP1 knockdown. The primary GO functions of these differentially expressed genes were metabolism, biological control, cellular activities, catalysis, binding, organelle and cell parts, etc. Its transcripts mostly participate in the following signaling pathways; pathways regulating the pluripotency of stem cells; pathways involving MAPK; pathways involving ErbB; pathways involving non-small cell lung cancer; pathways involving neurotrophin; pathways involving Rap1 and pathways involving axonal guidance. Therefore, PTBP1 may be involved in regulating neuronal differentiation through these pathways.
PTBP1 is an RNA-binding protein that was originally identified as a precursor mRNA splicing regulator and has been shown to have the ability to reprogram fibroblasts into neuron-like cells by regulating the activity of specific microRNA circuits[13]. Therefore, we speculate that there are also related microRNAs involved in the process of PTBP1 knockdown to transdifferentiate astrocytes into neurons. Analysis of the difference in microRNA expression in the sequencing data showed that a total of 26 miRNAs had significant changes, of which 9 were up-regulated and 17 were down-regulated. We ran target gene prediction with 142 mRNAs of known neural differentiation-specific factors with these 26 differentially expressed miRNAs to obtain 10 miRNAs that regulate 39 target genes involved in neural differentiation. By predicting target genes for these 10 miRNAs, we also get differentially expressed circRNAs and lncRNAs that may interact with them, which may indirectly regulate neuronal differentiation.
In summary, lentivirus expressing shPTBP1 can effectively knock down PTBP1 in mouse spinal cell astrocytes and convert it into functional neurons in one step, and in vivo knockdown PTBP1 can promote motor function recovery in spinal hemi section mice. We also explored the genes, related pathways and biological pathways that may be involved in the transdifferentiating process. Analysis of circRNA-miRNA-mRNA network reveals 3 circRNAs that may play a key role in the transdifferentiation, as well as possible regulatory mechanisms in the process. These results provide new research ideas for the treatment of SCI.
Ethical approval:
All animals in this study were from the joint venture Sipper BK laboratory animals (Shanghai, China). The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Ethics Committee of Shanghai University (ECSHU 2020-001, 31st March 2020).
Authors’ contribution:
Xiaohui Xu and Zhaohuan Zhang designed experiments; Yunyi Zhu and Zhaohuan Zhang carried out the astrocytes isolation, culture and transdifferentiation experiments. Changling Yue conducted immunofluorescence experiments. Qianwen Yang did the RT-PCR assay. Yunyi Zhu performed the mouse SCI modeling and bioinformatics analysis. Changling Yue and Qianwen Yang contributed to the performance of behaviour tests. Xiaohui Xu analyzed the data. Yunyi Zhu and Xiaohui Xu wrote the manuscript. Changling Yue and Qianwen Yang have contributed equally to this work. All authors have read and agreed to the published version of the manuscript.
Funding:
This work was supported by National Natural Science Foundation of China (No. 81671263), the Anhui Provincial Natural Science Foundation (2208085MH221), and the Major projects of Education Department of Anhui Province (KJ2021ZD0094).
Conflict of interests:
The authors declared no conflict of interests.
References
- Mekhail M, Almazan G, Tabrizian M. Oligodendrocyte-protection and remyelination post-spinal cord injuries: A review. Progress Neurobiol 2012;96(3):322-39.
[Crossref] [Google Scholar] [PubMed]
- Silver J, Miller JH. Regeneration beyond the glial scar. Nat Rev Neurosci 2004;5(2):146-56.
[Crossref] [Google Scholar] [PubMed]
- Qian H, Kang X, Hu J, Zhang D, Liang Z, Meng F, et al. Reversing a model of Parkinson?s disease with in situ converted nigral neurons. Nature 2020;582(7813):550-6.
[Crossref] [Google Scholar] [PubMed]
- Yang H, Xia Y, Lu SQ, Soong TW, Feng ZW. Basic fibroblast growth factor-induced neuronal differentiation of mouse bone marrow stromal cells requires FGFR-1, MAPK/ERK, and transcription factor AP-1. J Biol Chem 2008;283(9):5287-95.
[Crossref] [Google Scholar] [PubMed]
- Utreras E, Henriquez D, Contreras-Vallejos E, Olmos C, di Genova A, Maass A, et al. Cdk5 regulates Rap1 activity. Neurochem Int 2013;62(6):848-53.
[Crossref] [Google Scholar] [PubMed]
- Pinkas-Kramarski R, Eilam R, Alroy I, Levkowitz G, Lonai P, Yarden Y. Differential expression of NDF/neuregulin receptors ErbB-3 and ErbB-4 and involvement in inhibition of neuronal differentiation. Oncogene 1997;15(23):2803-15.
[Crossref] [Google Scholar] [PubMed]
- Rueda D, Navarro B, Mart??nez-Serrano A, Guzma?n M, Galve-Roperh I. The endocannabinoid anandamide inhibits neuronal progenitor cell differentiation through attenuation of the Rap1/B-Raf/ERK pathway. J Biol Chem 2002;277(48):46645-50.
[Crossref] [Google Scholar] [PubMed]
- Garc?a L, Castillo C, Carballo J, Rodr?guez Y, Forsyth P, Medina R, et al. ErbB receptors and PKC regulate PC12 neuronal-like differentiation and sodium current elicitation. Neuroscience 2013;236:88-98.
[Crossref] [Google Scholar] [PubMed]
- Ledonne A, Mercuri NB. On the modulatory roles of neuregulins/ErbB signaling on synaptic plasticity. Int J Mol Sci 2019;21(1):275.
[Crossref] [Google Scholar] [PubMed]
- Lin C, Chen PY, Chan HC, Huang YP, Chang NW. Peroxisome proliferator-activated receptor alpha accelerates neuronal differentiation and this might involve the mitogen-activated protein kinase pathway. Int J Devel Neurosci 2018;71:46-51.
[Crossref] [Google Scholar] [PubMed]
- Yang HJ, Ma SP, Ju F, Zhang YP, Li ZC, Zhang BB, et al. Thrombospondin-4 promotes neuronal differentiation of NG2 cells via the ERK/MAPK pathway. J Mol Neurosci 2016;60(4):517-24.
[Crossref] [Google Scholar] [PubMed]
- Romanelli MG, Diani E, Lievens PM. New insights into functional roles of the polypyrimidine tract-binding protein. Int J Mol Sci 2013;14(11):22906-32.
[Crossref] [Google Scholar] [PubMed]
- Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell 2013;152(1):82-96.
[Crossref] [Google Scholar] [PubMed]
- Vieira MS, Santos AK, Vasconcellos R, Goulart VA, Parreira RC, Kihara AH, et al. Neural stem cell differentiation into mature neurons: Mechanisms of regulation and biotechnological applications. Biotechnol Adv 2018;36(7):1946-70.
[Crossref] [Google Scholar] [PubMed]
- Tai W, Xu XM, Zhang CL. Regeneration through in vivo cell fate reprogramming for neural repair. Front Cell Neurosci 2020;14:107.
[Crossref] [Google Scholar] [PubMed]
- Chen YC, Ma NX, Pei ZF, Wu Z, Do-Monte FH, Keefe S, et al. A NeuroD1 AAV-based gene therapy for functional brain repair after ischemic injury through in vivo astrocyte-to-neuron conversion. Mol Ther 2020;28(1):217-34.
- Rivetti Di Val Cervo P, Romanov RA, Spigolon G, Masini D, Mart?n-Monta?ez E, Toledo EM, et al. Induction of functional dopamine neurons from human astrocytes in vitro and mouse astrocytes in a Parkinson's disease model. Nat Biotechnol 2017;35(5):444-52.
[Crossref] [Google Scholar] [PubMed]
- Yoo J, Lee E, Kim HY, Youn DH, Jung J, Kim H, et al. Electromagnetized gold nanoparticles mediate direct lineage reprogramming into induced dopamine neurons in vivo for Parkinson's disease therapy. Nat Nanotechnol 2017;12(10):1006-14.
[Crossref] [Google Scholar] [PubMed]
- Yao K, Qiu S, Wang YV, Park SJ, Mohns EJ, Mehta B, et al. Restoration of vision after de novo genesis of rod photoreceptors in mammalian retinas. Nature 2018;560(7719):484-8.
[Crossref] [Google Scholar] [PubMed]
- Karimi-Abdolrezaee S, Billakanti R. Reactive astrogliosis after spinal cord injury?beneficial and detrimental effects. Mol Neurobiol 2012;46(2):251-64.
[Crossref] [Google Scholar] [PubMed]
- Robel S, Berninger B, G?tz M. The stem cell potential of glia: Lessons from reactive gliosis. Nat Rev Neurosci 2011;12(2):88-104.
[Crossref] [Google Scholar] [PubMed]
- Okada S, Nakamura M, Katoh H, Miyao T, Shimazaki T, Ishii K, et al. Conditional ablation of Stat3 or Socs3 discloses a dual role for reactive astrocytes after spinal cord injury. Nat Med 2006;12(7):829-34.
[Crossref] [Google Scholar] [PubMed]
- Sofroniew MV. Molecular dissection of reactive astrogliosis and glial scar formation. Trends Neurosci 2009;32(12):638-47.
[Crossref] [Google Scholar] [PubMed]
- Lang BT, Cregg JM, dePaul MA, Tran AP, Xu K, Dyck SM, et al. Modulation of the proteoglycan receptor PTP? promotes recovery after spinal cord injury. Nature 2015;518(7539):404-8.
[Crossref] [Google Scholar] [PubMed]
- Wilhelmsson U, Li L, Pekna M, Berthold CH, Blom S, Eliasson C, et al. Absence of glial fibrillary acidic protein and vimentin prevents hypertrophy of astrocytic processes and improves post-traumatic regeneration. J Neurosci 2004;24(21):5016-21.
[Crossref] [Google Scholar] [PubMed]
- Zhou K, Zheng Z, Li Y, Han W, Zhang J, Mao Y, et al. TFE3, a potential therapeutic target for spinal cord injury via augmenting autophagy flux and alleviating ER stress. Theranostics 2020;10(20):9280-302.
[Crossref] [Google Scholar] [PubMed]
- Basso DM, Fisher LC, Anderson AJ, Jakeman LB, Mctigue DM, Popovich PG. Basso Mouse Scale for locomotion detects differences in recovery after spinal cord injury in five common mouse strains. J Neurotrauma. 2006;23(5):635-59.
[Crossref] [Google Scholar] [PubMed]
 ): Differentially expressed circRNAs and (
): Differentially expressed circRNAs and ( ): Target circRNAs of 31 miRNAs
): Target circRNAs of 31 miRNAs