- *Corresponding Author:
- F. Wang
School of Management Science and Engineering
Anhui University of Technology
Maanshan 243032, China
E-mail: ahutwangfuyu@yeah.net
| This article was originally published in a special issue, “Trending Topics in Biomedical Research and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2022:84(1) Spl Issue “270-276” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
To investigate the expression of ubiquitin fusion degradation 1 like protein in gastric cancer and its relationship with the clinicopathological characteristics. Analyzed the data of ubiquitin fusion degradation 1 like protein in gastric cancer and normal tissues in the Cancer Genome Atlas database and the University of Alabama Cancer database. The expression of ubiquitin fusion degradation 1 like protein from fresh samples was detected by real-time quantitative polymerase chain reaction and Western blot, the expression of ubiquitin fusion degradation 1 like protein from paraffin samples was detected by immunohistochemistry. Observed the relationship between the expression level of ubiquitin fusion degradation 1 like protein and clinicopathological characteristics of patients. Data from the Cancer Genome Atlas and the University of Alabama Cancer Databases suggested that ubiquitin fusion degradation 1 like protein messenger ribonucleic acid expression in gastric cancer tissues was significantly higher than that in normal gastric tissues (p<0.05); Western blot and real-time fluorescence quantitative polymerase chain reaction confirmed that ubiquitin fusion degradation 1 like protein and messenger ribonucleic acid expression in fresh gastric cancer tissues were significantly higher in adjacent tissues (p<0.05). Immunohistochemistry results showed that 88 cases of gastric cancer had positive protein expression in 71 cases and negative expression in 17 cases. The expression difference of ubiquitin fusion degradation 1 like protein in gastric cancer and non-tumor tissues adjacent to the cancer was statistically significant (p<0.05) and it was significantly correlated with tumor, nodes and metastases stage and differentiation degree (p<0.05). The expression level of ubiquitin fusion degradation 1 like protein in gastric cancer tissues is significantly increasing and associated with disease progression, suggesting ubiquitin fusion degradation 1 like protein may be involved in the development and progression of gastric cancer.
Keywords
Ubiquitin fusion degradation 1 like protein, gastric cancer, chemotherapy, antibody
Gastric cancer is one of the common malignant tumors of digestive system[1]. At present, surgical resection is the main treatment for gastric cancer and the only method to cure gastric cancer[2]. In China, the incidence rate of gastric cancer is relatively high, but most cases are relatively poor because of late discovery. Therefore, it is of great significance to explore ideal molecular markers for the diagnosis, treatment and prognosis of gastric cancer[3]. Ubiquitin Fusion Degradation 1 Like Protein (UFD1L) participates in the ubiquitin proteasome pathway, which is closely related to the physiological function and pathological state of cells[4]. The gene is also involved in the regulation of polyadenylation of messenger Ribonucleic Acid (mRNA). After binding with p97, the UFD1L/Nuclear Protein Localization-4 (NPL4) binary complex is ubiquitin dependent and participates in the mitotic process[5]. At present, there is no study on the relationship between UFD1L and gastric cancer. This study mainly discusses the expression of UFD1L protein in clinical samples of gastric cancer from protein and mRNA levels, and analyzes the relationship between UFD1L protein and clinicopathological characteristics of gastric cancer. It provides a new idea for further finding the mechanism of gastric cancer.
Materials and Methods
General information:
From March 2016 to April 2019, 25 cases of gastric cancer and 20 cases of normal gastric tissue adjacent to cancer were collected from Ma’anshan people’s hospital. The paraffin embedded tissues of 88 cases of gastric cancer and 60 cases of normal gastric cancer adjacent to cancer were collected. All patients did not receive chemotherapy, radiotherapy or biotherapy before operation. Fresh specimens were immediately placed in -80° refrigerator for cryopreservation.
Methods:
The Cancer Genome Atlas (TCGA) database: TCGA (https://cancergenome.nih.gov/) compared with normal tissues, many differentially expressed genes in tumor samples were identified by gene expression profile analysis. Extracting TCGA database data: Mining high expression genes in stad in TCGA database. Set filter criteria: Search The Cancer Genome Atlas Stomach Adenocarcinoma (TCGA-STAD) in projects project. After screening UFD1L gene from TCGA database, the search criteria were set as project ID is TCGA-STAD and gene is UFD1L in the exploration project.
The University of Alabama Cancer (UALCAN) database: According to the data of UALCAN (http://ualcan.path.uab.edu/) RNA sequencing (RNA-Seq) data, the expression of UFD1L mRNA in normal gastric tissues and gastric cancer tissues was analyzed. The median of UFD1L transcriptome data was used as the cut-off value to distinguish the high and low expression of UFD1L.
Protein collection and Western blot detection: The fresh samples of gastric cancer tissues were collected. After grinding and crushing, the total protein was extracted by lysis fluid. The 10 % Sodium Dodecyl Sulphate–Polyacrylamide Gel Electrophoresis (SDSPAGE) was used to transfer to the Polyvinylidene Difluoride (PVDF) membrane under constant current of 300 mA and 90 min, then 5 %. The milk was sealed for 1 h and then the primary antibody was incubated overnight at 4°. The next day, the membrane was washed with Phosphate Buffered Saline with Tween® 20 (PBST) solution and the second antibody was washed with PBST solution again. Finally, the developer was added for exposure and image acquisition. The results were analyzed by gray scale and the data were statistically analyzed.
The expression of UFD1L mRNA was detected by Real-Time quantitative Polymerase Chain Reaction (RT-qPCR): The total RNA was extracted from gastric cancer tissues and adjacent tissues according to the instructions of the kit and the RNA was reverse transcribed into complementary Deoxyribonucleic Acid (cDNA) according to the reverse transcription kit. The forward primer sequence of UFD1L was 5’-TAGGAACTTTGCCTGTCTG-3’ and the reverse primer sequence was 5’-TCGGCTTCACCTTCTGTC -3’. The reaction system of RT-qPCR was 20 UL. RT-qPCR reaction conditions: 95° 2 min, 95° 30 s, 59° 30 s, 72° 30 s, a total of 30 cycles. After the reaction, the data were collected to obtain the Cycle Threshold (CT) value of the sample and the relative expression of UFD1L mRNA was calculated by relative quantitative method using 2−ΔΔΔ.
The expression of UFD1L protein was detected by immunohistochemistry: The resected gastric cancer tissues and adjacent tissues were fixed with 10 % neutral formaldehyde, dehydrated with gradient ethanol, embedded in paraffin and sectioned continuously (thickness of 4 sex a). According to the instructions of immunohistochemistry kit, the samples were repaired with citrate buffer for 15 min, blocked with hydrogen peroxide (3 %) for 15 min and goat serum was sealed for 20 min at room temperature. The Rabbit anti human polyclonal antibody with dilution ratio of 1:200 UFD1L (Wuhan Sanying company) was added and incubated at room temperature for 1 h. After washing with Phosphate Buffered Saline (PBS), Diaminobenzidine (DAB) color developing solution was added, 20 % hematoxylin was dyed, neutral resin was sealed and observed under microscope. 5-10 visual fields were randomly selected for observation under 200x microscope and evaluated according to the degree of staining and the percentage of stained cells. Among them, colorless was 0, light yellow was 1, yellow was 2 and brown was 3; the percentage of stained cells in total count cells was less than 5 %, 5 %-25 % was 1 point, 26 %-50 % was 2 points and >51 % was 3 points. The total score of the two groups ≤3 points was negative and >3 points was positive.
Statistical treatment:
Statistical Package for the Social Sciences (SPSS) 22.0 statistical software was used to analyze the data and draw graphs. The data of measurement data in accordance with normal distribution were measured with mean±standard deviation (univariate analysis of variance was used for comparison between groups and t test was used for comparison between two groups). The count data were represented by use case (%) and the chi square test was used.
Results and Discussion
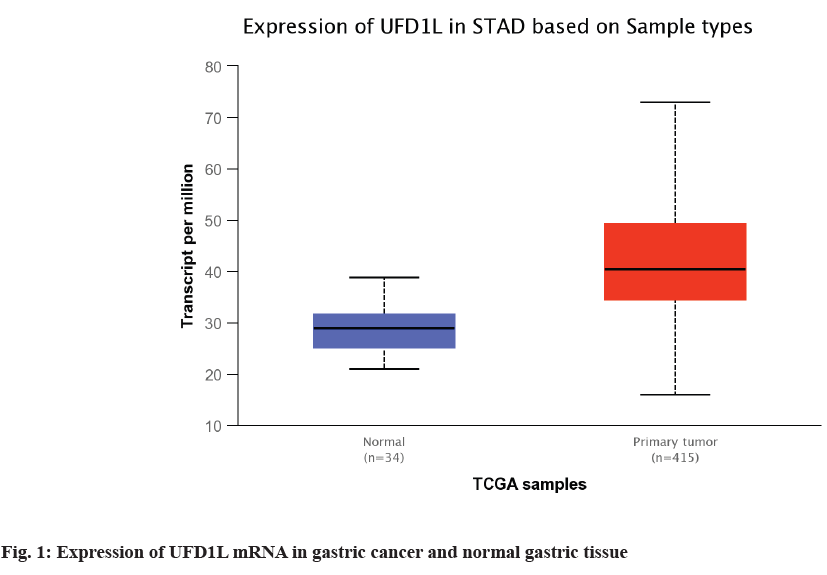
The expression of UFD1L in 34 normal gastric tissues and 415 gastric cancer tissues in TCGA database was compared. Combined with the results of UFD1L mRNA expression in normal gastric tissues and gastric cancer tissues in UALCAN database, the expression of UFD1L mRNA in gastric cancer tissues was significantly higher than that in normal gastric tissues (p<0.01, fig.1).
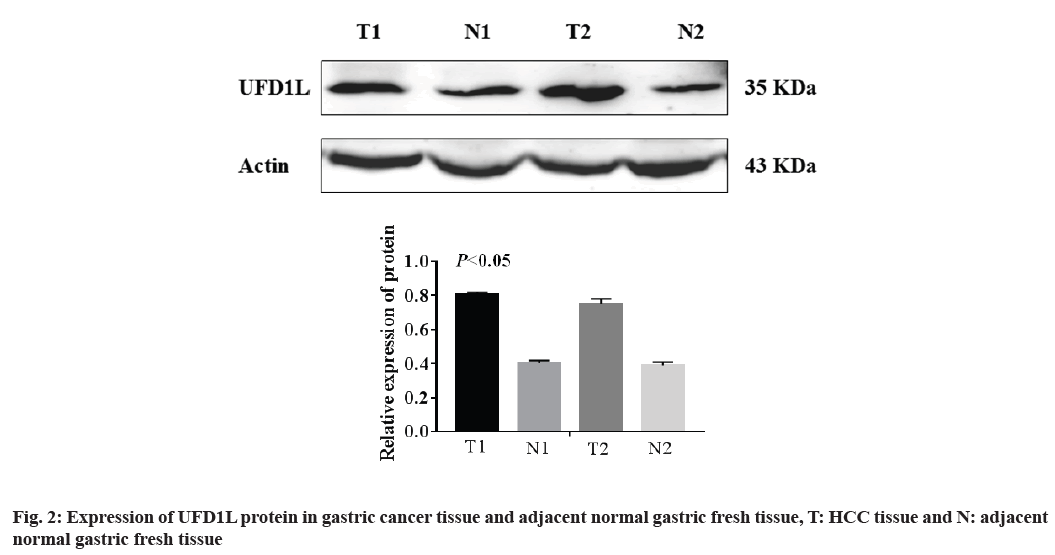
Western blot was used to detect the expression of UFD1L protein in 25 cases of gastric cancer and 20 cases of adjacent normal gastric tissue. The results showed that the expression of UFD1L was up-regulated in gastric cancer tissues (p<0.01, fig. 2).
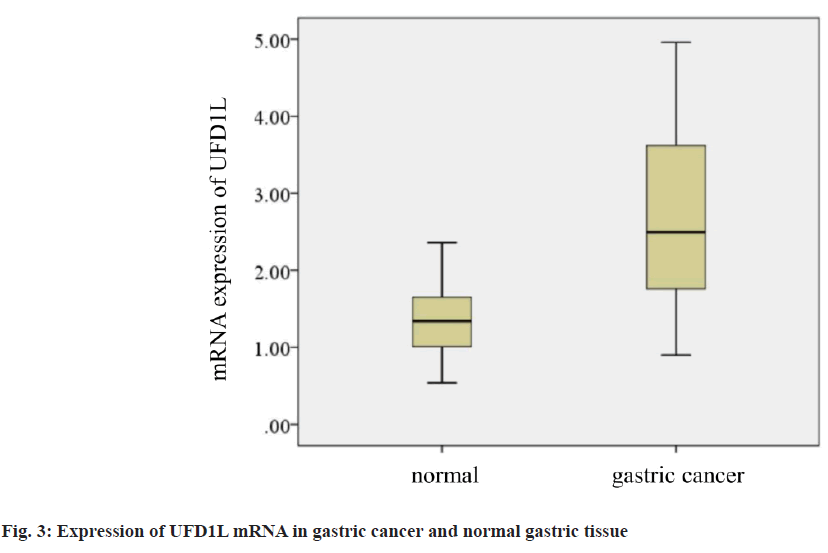
Secondly, the mRNA expression of UFD1L in gastric cancer and adjacent normal gastric tissue was detected by fluorescence quantitative PCR. The results showed that the mRNA expression level of UFD1L in gastric cancer tissue was significantly higher than that in normal adjacent gastric tissue (p<0.05; fig. 3).
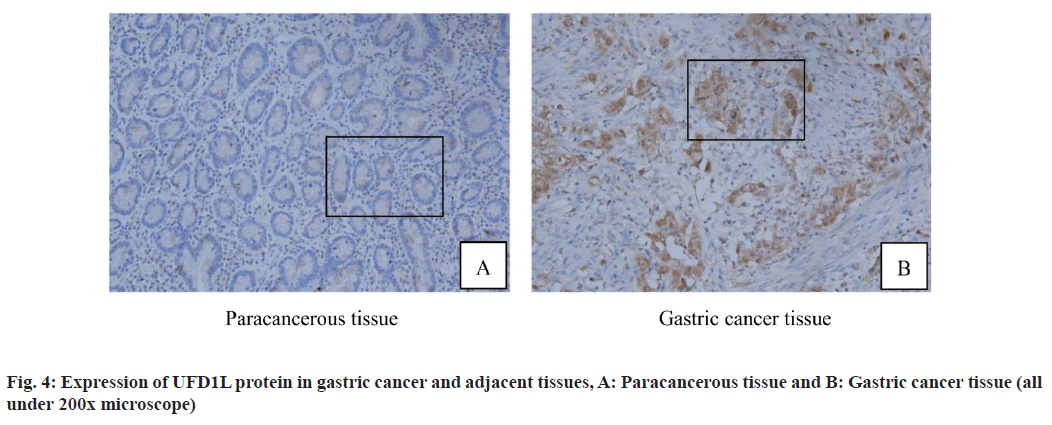
In addition, immunohistochemistry was used to detect the expression of UFD1L protein in 88 cases of gastric cancer and 60 cases of adjacent normal gastric cancer tissues. The results showed that UFD1L protein particles were mainly located in the cytoplasm and the appearance of diffuse homogeneous light yellow to brown particles was regarded as positive. Five high power visual fields were randomly selected from each section and judged by semi quantitative scoring standard. Immunohistochemical results showed that UFD1L protein was positive in gastric cancer tissues (fig. 4A and fig. 4B). The positive expression rate of UFD1L protein in gastric cancer group was 86.44 % and that in adjacent normal gastric tissue was 40.00 %, the difference was statistically significant (p<0.05; fig. 4, Table 1).
| Group | UFD1L/case | Positive rate/% | |
|---|---|---|---|
| Negative | Positive | ||
| Paracancerous group | 39 | 21 | 35.00 |
| Gastric cancer group | 17 | 71 | 80.68 |
| p | - | - | <0.01 |
Note: UFD1L: Ubiquitin Fusion Degradation 1 Like Protein
Table 1: Expression of UFD1L in Gastric Cancer and Adjacent Tissues (n=148)
By analyzing the clinicopathological parameters of 88 cases of gastric cancer and 60 cases of adjacent normal gastric tissue, we found that UFD1L protein expression was significantly correlated with Tumor, Nodes and Metastases (TNM) stage and differentiation degree of gastric cancer (p<0.05), but not with age, gender, lymph node metastasis, vascular invasion and tumor size (all p>0.05, Table 2).
| Clinicopathological parameters | Number of cases | UFD1L positive | χ2 value | p value |
|---|---|---|---|---|
| (n) | n (%) | |||
| Gender | 0.459 | 0.646 | ||
| Male | 48 | 27 (56.25 %) | ||
| Female | 40 | 19 (47.50 %) | ||
| Age (years) | 0.228 | 0.819 | ||
| ≤55 | 61 | 31 (50.82 %) | ||
| <55 | 27 | 15 (55.56 %) |
||
| Differentiation Degree | — | 0.047 | ||
| High | 37 | 10 (27.03 %) | ||
| Medium | 27 | 16 (59.26 %) | ||
| Low | 24 | 20 (83.33 %) | ||
| Lymphatic metastasis | 0.254 | 0.779 | ||
| Yes | 54 | 28 (51.85 %) | ||
| No | 34 | 16 (47.06 %) | ||
| Vascular invasion | 0.063 | 0.949 | ||
| Yes | 39 | 21 (53.85 %) | ||
| No | 49 | 27 (55.10 %) | ||
| TNM staging | 2.012 | 0.004 | ||
| I+II | 29 | 9 (31.03 %) | ||
| III+IV | 59 | 43 (72.88 %) | ||
| Tumor size | 0.482 | 0.63 | ||
| <4cm | 57 | 29 (50.88 %) | ||
| ≥4cm | 31 | 13 (41.94 %) |
Table 2: Relationship between UFD1L Expression and Clinicopathological Features of Gastric Cancer
Gastric cancer originated from epithelial malignant tumor of stomach. Due to the occult onset, there are often no typical clinical symptoms and signs and most patients have advanced gastric cancer[3]. Despite the increasing popularity of gastroscopy, the proportion of early gastric cancer is gradually increasing, but how to improve the surgical treatment and prognosis of patients with advanced gastric cancer is still a hot spot in clinical research[6]. At present, surgery is still the main treatment for gastric cancer, while molecular targeted therapy is still in its infancy[2]. Some studies have shown that the abnormal expression of Phosphatase and Tensin Homolog (PTEN) protein in gastric cancer patients is significantly related to tumor differentiation, staging and chemotherapy resistance. Phosphatidylinositol 3-Kinase (PI3K) signaling pathway inhibitors can inhibit PI3K/Protein Kinase B (Akt)/mammalian Target of Rapamycin (mTOR) signal pathway is used to reduce the expression of PTEN protein, so as to improve the efficacy of chemotherapy drugs and effectively prolong the survival period of patients. So far, molecular targeted therapy has gradually become the top priority in the research[7]. In addition, it also provides a new idea for the treatment of gastric cancer. At present, targeted drugs targeting known targets are still in or will enter clinical trials. Searching for molecular markers related to the prognosis of gastric cancer not only helps to predict the outcome of clinical prognosis, but also can be used as a new molecular target to provide better treatment for gastric cancer patients.
UFD1L, located on human chromosome 22q11.2, is a human homologous gene encoding yeast ubiquitin fusion degradation 1 protein. It is also an important part of ubiquitin dependent protein degradation pathway and also participates in the mRNA processing process[4,5]. Ubiquitin proteasome pathway is one of the proteolysis pathways. UFD1L forms heterodimer by binding with NPL4 to maintain the integrity of p97 protein complex and participate in the degradation of endoplasmic reticulum related protein and nucleoprotein. I kappa B alpha (IκBα) is a transcription factor that regulates inflammatory response. After being transferred to the nucleus, IκBα activates a series of pro-inflammatory genes, which can be degraded by 26S proteasome. P97- UFD1L-NPL4 protein complex can mediate cytokine regulated IκBα protein degradation by binding with 26S proteasome. UFD1L gene is the cofactor of p97 and Ubiquitination Binding Domain (UBD) plays an important role. Under physiological conditions, UFD1L is mainly expressed in neural crest derived cells of heart, bronchial arch, pharyngeal sac, craniofacial bone, thymus and parathyroid gland, and may be related to the development of hippocampus[8]. At the same time, during embryogenesis, UFD1L is highly expressed in the cardiac outflow tract. Relevant studies have shown that double valve aortic valve is related to the downregulation of UFD1L gene expression[9]. In addition, the deletion of UFD1L gene may lead to 22q11.2 deletion syndrome, which further confirms that UFD1L gene is closely related to human genetic pathology[10].
This study was the first time to detect the expression of UFD1L protein in gastric cancer. The results of Western blot and real-time fluorescence quantitative PCR showed that UFD1L protein was highly expressed in gastric cancer tissues, suggesting that UFD1L may be involved in the occurrence and development of gastric cancer. At present, studies have found that UFD1L is highly expressed in subependymoma, but not in mucinous papillary ependymoma. Moreover, the expression level of UFD1L has no significant relationship with tumor grade and prognosis[11]. At the same time, the expression level of UFD1L protein in Oral Squamous Cell Carcinoma (OSCC) was significantly higher than that in adjacent tissues. However, there is no clear evidence that UFD1L is related to the prognosis of OSCC. Studies suggest that UFD1L may be an independent tumor promoting factor and may interact with other genes to participate in the process of cancer[12]. In order to further explore the relationship between UFD1L expression and clinicopathological features of gastric cancer, immunohistochemistry was used to detect the relationship between UFD1L protein expression and clinicopathological characteristics of gastric cancer patients. The results showed that UFD1L expression was positively correlated with TNM stage and differentiation degree of gastric cancer, suggesting that UFD1L may be involved in the progression of gastric cancer. This is consistent with the result that UFD1L expression is significantly up-regulated in adrenocortical adenocarcinoma[13]. These results suggest that UFD1L may be one of the potential oncogenes. However, the role of this gene in the regulation of mRNA polyadenylation is still unclear, but its upregulation indicates that the endoplasmic reticulum is in acute stress state, which may be significantly related to the occurrence, development and mechanism of cancer[5,14]. UFD1L may be a new index to evaluate the malignant degree of gastric cancer.
In conclusion, the expression level of UFD1L in gastric cancer was significantly increased and it was closely related to the differentiation degree and TNM stage of gastric cancer. Since then, we speculated that UFD1L may be involved in the occurrence and development of gastric cancer and it is expected to become a new molecular target for the evaluation of malignant degree and treatment of gastric cancer. However, there are still some deficiencies in this study and the molecular mechanism of UFD1L in gastric cancer is not clear, which needs to be further explored in the follow-up experiments.
Acknowledgement:
This work was supported by the National Natural Science Foundation of China (No.71872002).
Conflicts of interest:
The authors declared no conflicts of interest.
References
- Waldum HL, Fossmark R. Types of gastric carcinomas. Int J Mol Sci 2018;19(12):4102-9.
[Crossref] [Google Scholar] [PubMed]
- National Health Commission. Diagnostic criteria for gastric cancer (2018 Edition). Electron J Comprehensive Treatment Cancer 2019;5(1):55-82.
- Huang S, Dai Y, Gao JJ. Molecular epidemiology of gastric cancer. Chin Oncol Clin 2019;46(1):16-21.
- Li JM, Wu H, Zhang W, Blackburn MR, Jin J. The p97-UFD1L-NPL4 protein complex mediates cytokine-induced IκBα proteolysis. Mol Cell Biol 2014;34(3):335-47.
[Crossref] [Google Scholar] [PubMed]
- Botta A, Tandoi C, Fini G, Calabrese G, Dallapiccola B, Novelli G. Cloning and characterization of the gene encoding human NPL4, a protein interacting with the ubiquitin fusion-degradation protein (UFD1L). Gene 2001;275(1):39-46.
[Crossref] [Google Scholar] [PubMed]
- Huang Q, Fang C, Shi J, Sun Q, Wu H, Gold JS, et al. Differences in clinicopathology of early gastric carcinoma between proximal and distal location in 438 Chinese patients. Sci Rep 2015;5(1):1-2.
- Li JH, Xiong JP. Advances in molecular targeted therapy of gastric cancer. Chin Oncol Clin 2015;42(23):1118-23.
- Xie L, Ye L, Ju G, Xu Q, Zhang X, Liu S, et al. A family‐ and population‐based study of the UFD1L gene for schizophrenia. Am J Med Genet B Neuropsychiatr Genet 2008;147(7):1076-9.
[Crossref] [Google Scholar] [PubMed]
- Mohamed SA, Hanke T, Schlueter C, Bullerdiek J, Sievers HH. Ubiquitin fusion degradation 1–like gene dysregulation in bicuspid aortic valve. J Thorac Cardiovasc Surg 2005;130(6):1531-6.
[Crossref] [Google Scholar] [PubMed]
- Novelli G, Mari A, Amati F, Colosimo A, Sangiuolo F, Bengala M, et al. Structure and expression of the human ubiquitin fusion–degradation gene (UFD1L). Biochim Biophys Acta Gene Regul Mech 1998;1396(2):158-62.
[Crossref] [Google Scholar] [PubMed]
- Zangen IL, Kneitz S, Monoranu CM, Rutkowski S, Hinkes B, Vince GH, et al. Ependymoma gene expression profiles associated with histological subtype, proliferation and patient survival. Acta Neuropathol 2007;113(3):325-37.
[Crossref] [Google Scholar] [PubMed]
- Hsu CW, Yu JS, Peng PH, Liu SC, Chang YS, Chang KP, et al. Secretome profiling of primary cells reveals that THBS2 is a salivary biomarker of oral cavity squamous cell carcinoma. J Proteome Res 2014;13(11):4796-807.
[Crossref] [Google Scholar] [PubMed]
- Velázquez-Fernández D, Laurell C, Geli J, Höög A, Odeberg J, Kjellman M, et al. Expression profiling of adrenocortical neoplasms suggests a molecular signature of malignancy. Surgery 2005;138(6):1087-94.
[Crossref] [Google Scholar] [PubMed]
- Santoro ML, Gadelha A, Ota VK, Cunha GR, Asevedo E, Noto CS, et al. Gene expression analysis in blood of ultra-high risk subjects compared to first-episode of psychosis patients and controls. World J Biol Psychiatry 2015;16(6):441-6.
[Crossref] [Google Scholar] [PubMed]