- *Corresponding Author:
- S. Harakeh
Department of Biological Sciences, Faculty of Science, King Abdulaziz University, Jeddah, Saudi Arabia
E-mail: sharakeh@gmail.com
| This article was originally published in a special issue, “Current Trends in Pharmaceutical and Biomedical Sciences” |
| Indian J Pharm Sci 2022:84(5) Spl Issue “49-58” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
In silico 16S ribosomal RNA gene-based restriction enzyme approach is a computer-simulated polymerase chain reaction-restriction fragment length polymorphism analysis method, namely customizable in silico sequence evaluation for restriction sites. The purpose of this study was to identify lactic acid bacteria using restriction enzyme analysis of 16S gene (16S ribosomal RNA polymerase chain reaction-restriction fragment length polymorphism). An in silico investigation was conducted to detect species-specific molecular markers in different lactic acid bacteria strains. We selected six out of 20 16S ribosomal RNA gene sequences from probiotic-potential lactic acid bacteria strains. Initially, the corresponding 16S ribosomal RNA gene sequences were selected from the database aligned using the Clustal Omega program and their evolutionary relationships were determined. The results indicated that the studied lactic acid bacteria strains belonged to 6 distinct clades except for the genus of Lactobacillus. Furthermore, the in silico polymerase chain reactionrestriction fragment length polymorphism was conducted for further taxonomic classification. Different restriction enzymes were used to recognize sites present on the lactic acid bacteria 16S ribosomal RNA genes. Customizable in silico sequence evaluation for restriction sites output comprised a predicted agarose gel as virtual polymerase chain reaction-restriction fragment length polymorphism patterns, for proper selection of the appropriate enzyme (s) to be used for polymerase chain reaction-restriction fragment length polymorphism marker generation. Our study demonstrated that ApyPI, CchIII and NheI produced distinct restriction patterns as those of Ku324937_C species. The most abundant species-specific markers were seen for Ku324928_C (10 markers), followed by Ku324908_C (8 markers) of order Hemiptera, then Ku324909_C (5 markers), Ku324898_C (4 markers) and finally, Ku324937_C (3 markers). The results demonstrated that customizable in silico sequence evaluation for restriction sites is a powerful tool to detect species-specific markers to provide better understanding of organisms found in raw and/or fermented milk.
Keywords
Lactic acid bacteria, 16S ribosomal RNA, polymerase chain reaction-restriction fragment length polymorphism, probiotics, in silico, restriction enzyme
With the advent of new technologies, interest in studying microbial communities that are specifically related to human health and food has boomed over the last decade. Recently, in silico approaches have emerged in parallel with the newly introduced microbiological tools to identify microorganisms. Such approaches, based on 16S ribosomal Deoxyribonucleic Acid (rDNA) sequences, have been used for the identification of Lactic Acid Bacteria (LAB), including probiotic ones. There are nine “hypervariable regions” (V1-V9) contained within bacterial 16S ribosomal Ribonucleic Acid (rRNA) genes having major sequence diversity among various bacterial species. Among the nine regions, V2 and V3 are the most specific in distinguishing species of bacteria to the genus level[1].
LAB has been used in food fermentation. Historically, a wide variety of fermented dairy products (also known as cultured dairy foods) are still a very important part of the daily food in the world[2]. Fermented milk is the main source of lactic acid fermentations in which LAB predominate. LAB are naturally associated with fermentation and present in many foods including cereal, vegetables, meat and plants[3]. LAB has been used in food preservation (improving texture, flavor) as well as in the modification of the organoleptic characteristics of foods[4,5]. Recent evidence based on in vitro studies, showed that LAB have the potential to enhance and improve immune responses[3]. LAB are ecofriendly and have bio-conservative potential[6].
More recently, candidates of probiotic strains of Lactobacillus and Bifidobacterium are available as dietary supplements and in dairy products that are beneficial to health and help in the treatment of diarrheal diseases[7]. LAB strains, especially the genera Lactobacillus, Bifidobacterium, Enterococcus, Streptococcus, Lactococcus, Leuconostoc and Pediococcus, are commonly isolated from fermented foods[8]. LAB starter cultures are commonly used in milk fermentations[5]. In recent years, LAB strains from Lactobacillus genera have been utilized in food preservation due to their antimicrobial potential[9]. Accurate identification of Lactobacillus by conventional phenotypic methods is rather tedious and is not always reliable[3,5]. For the past 100 y, the classification of LAB relied on old techniques which were recently replaced by modern molecular based techniques using the 16S rDNA sequencing[4,10,11]. These techniques not only are simple and rapid but also successfully overcame the limitations of the traditional methods[12]. 16S rRNA sequencing in combination with high-scored Basic Local Alignment Search Tool (BLAST) searches facilitates the identification of sequence homology down to the species level for some types of bacteria. However, using 16S rRNA gene sequences as standard markers for the differentiation of Lactobacillus species offers a very limited scope because several Lactobacillus species share similar 16S rRNA gene sequences. Precise identification of these bacteria to the species level is not an easy task, for example, when two different bacterial species share almost the same 16S rRNA sequence, this technique would not be useful in differentiating among the closely related bacterial strains. However, rRNA sequencing in combination with high-scored BLAST allows the identification of sequence homology down to the species level for some types of bacteria but using 16S rRNA gene sequences as standard markers will allow the differentiation of Lactobacillus to the species level. In other words, only species-specific Polymerase Chain Reaction (PCR) primer pairs could efficiently differentiate among the different species LAB as compared to the universal primer used with the 16S rRNA gene detection[13].
In the current study, LAB classification was carried out by utilizing a computer-simulated PCR-Restriction Fragment Length Polymorphism (virtual PCR-RFLP) namely Customizable in silico Sequence Evaluation for Restriction Sites (CisSERS) analysis of the 16S rRNA gene which allows to select the most suitable restriction enzymes and subsequent analysis of the size and distribution of the obtained fragments to distinguish among the different closely related species, and to predict the number of enzymes required to ensure differentiation between known and unknown species. The combination of the best ranked enzymes exhibits higher discriminating resolution, even at the species level, in contrast to their solo usage[14]. The 16S rRNA gene sequences were used to detect the species and genus-specific molecular markers. For this purpose, we used 20 LAB strains which were previously isolated from raw milk and in traditionally fermented milk isolated from local animals procured in Jeddah province of Saudi Arabia[15]. Out of those, six LAB strains have been selected based on the highest match of a partial DNA sequence of the 16S rRNA gene deposited in the GenBank database.
Materials and Methods
Reference strains of LAB:
The LAB isolates were isolated in a previous published study from our group[15]. Sequences of the 16S rRNA gene were selected from the National Center for Biotechnology Information (NCBI) database[15]. Initially, 20 LAB species were chosen and their 16S rRNA gene sequences were retrieved from the GenBank and six of those undergone further characterization.
Nucleotide sequence accession numbers:
The accession numbers, 16S rRNA nucleotide sequences of 20 LAB isolates including the source of the isolates submitted to the GenBank database are shown in Table 1.
| Strain No. | Accession number | Identity | Source | Similarity (%) |
|---|---|---|---|---|
| 1 | KU324909 | Lactococcus lactis strain HadRami9 | Stirred yogurt | 99.9 |
| 2 | KU324896 | Lactobacillus casei MSJ1 | Cow cheese | 99.4 |
| 3 | KU324896 | Lactobacillus casei Dwan5 | Cow cheese | 98.5 |
| 4 | KU324901 | Lactobacillus plantarum EyLan2 | Cow cheese | 98.8 |
| 5 | KU324906 | Enterococcus faecium Gail-BawZir8 | Stirred yogurt | 98.3 |
| 6 | KU324914 | Streptococcus thermophilus MaNaL33 | Cream | 98.3 |
| 7 | KU324921 | Streptococcus equinus Omer9 | Camel milk | 98.3 |
| 8 | KU324908 | Streptococcus thermophilus BinSlman8 | Stirred yogurt | 98.3 |
| 9 | KU324898 | Lactobacillus casei BgShn3 | Cow cheese | 98.2 |
| 10 | KU324920 | Enterococcus faecium BagHom4 | Camel milk | 98.1 |
| 11 | KU324928 | Weissella confusa AhMd8 | Cow butter | 98.3 |
| 12 | KU324937 | Lactococcus garvieae ZSJ5 | Cow butter | 98.2 |
| 13 | KU324926 | Streptococcus equinus JmaL3 | Camel frozen milk | 98.2 |
| 14 | KU324927 | Streptococcus equinus Foad7 | Camel frozen milk | 98.2 |
| 15 | KU324901 | Lactobacillus plantarum EyLan2 | Cow cheese | 98.2 |
| 16 | KU324902 | Lactobacillus futsaii strain EMBM2 | Cow cooked cheese | 98.2 |
| 17 | KU324903 | Weissella confusa SaEd-7 | Cow cooked cheese | 98.3 |
| 18 | KU324930 | Weissella confusa NooR1 | Cow butter | 98.2 |
| 19 | KU324931 | Weissella confusa SaYun2 | Cow butter | 98.2 |
| 20 | KU324933 | Weissella confusa SYary1 | Cow butter | 98.2 |
Table 1: Matches of the Available 16S rRNA Gene Sequence in the NCBI Genbank Database, Blast similarity scores ranged between 98 %-100 %[15].
In silico evaluation:
Sequence data preprocessing: Sample sequence contaminants from DNA preparation or host genomic DNA was removed to achieve better resolution of sequence alignment and infer accurate phylogeny. The best 16S rRNA gene sequences of the 6 out of the 20 selected strains were Streptococcus thermophilus BinSlman8-KU324908/Lactobacillus casei BgShn3-KU324898/Lactococcus lactis HadRami9-KU324909/Enterococcus faecium BagHom4-KU324920/Weissella confusa and AhMd8-KU324928/Lactococcus garvieae ZSJ5-KU324937. BLAST (NCBI) website (http://www.ncbi.nlm.nih.gov/BLAST) was used for searching the highest sequence identity score. Samples were assigned to species retrieved from BLAST hits with >98 % similarity score.
In silico sequence alignment of 16s rRNA gene sequences:
The Clustal Omega program was used for alignment of multiple sequences (http://www.ebi.ac.uk/Tools/msa/clustalo/). We aligned both the sample sequence and 20 matched sequences available in the databases in order to obtain the “consensus” fragment. Clustal Omega uses the HHalign algorithm and its default settings as its core alignment engine. Default value is ClustalW with character counts. We defined parameters in mBed-like clustering guide-tree (yes), mBed-like clustering iteration (yes), number of combined iterations (default [0]), max guide tree iterations (default [-1]) and max Hidden Markov Model (HMM) iterations (default [-1]). The order in which the sequences appear in the final alignment was aligned, will read in the file, align the sequences and output the alignment to screen (default) in the default Fast Adaptive Shrinkage Threshold Algorithm (FASTA) format. After performing a sequence alignment with the Clustal Omega program, a consensus sequence is determined from a multiple alignment to narrow down the species from 20 LAB strains to 6 representative strains (Table 2).
| Strain No. | Strain name | Accession number | Reference |
|---|---|---|---|
| 1 | Streptococcus thermophilus BinSlman8 | KU324908 | [15] |
| 2 | Lactobacillus casei BgShn3 | KU324898 | [15] |
| 3 | Lactococcus lactis HadRami9 | KU324909 | [15] |
| 4 | Enterococcus faecium BagHom4 | KU324920 | [15] |
| 5 | Weissella confusa AhMd8 | KU324928 | [15] |
| 6 | Lactococcus garvieae ZSJ5 | KU324937 | [15] |
Table 2: The Selected 6 Isolated Lab based on 16S rRNA Gene Sequences with Accession Numbers submitted to the NCBI Genbank Database.
Consensus sequence:
After performing sequence alignment with the Clustal Omega program, a consensus sequence is determined from a multiple alignment to narrow down the 20 species of LAB strains to 6 as shows in Table 2.
In silico design of PCR-RFLP for 16S rDNA sequences and restriction enzymes:
Simulations of polymorphism analysis of the restriction fragment length of rRNA (PCR-RFLP) were based on the use of CisSERS software[1]. CisSERS was first applied to 6 reference strains representing 20 species to verify whether the method can efficiently differentiate among the different species of the Lactobacillus genus with very high 16S rRNA sequence homology. CisSERS is a computational analysis tool based on a graphical user interface for high-throughput analysis of mature multiple sequences for the identification of restriction enzyme site analysis.
16S rRNA gene sequences of LAB alignment:
16S rRNA gene sequences of LAB were aligned using the Clustal Omega program and the aligned fragments were exported to CisSERS software that can process single or multiple sequences in simple FASTA formatted file containing the reference sequences. Output describes the fragment counts and locations for each restriction site and dynamically created predicted gel images. The main list of enzymes (237 in the present study) is retrieved from the Restriction Enzyme Database (REBASE). A set of formulas used via CisSERS were used to produce fragment size. Further, the motif detection feature of CisSERS was previously validated for genes of Nostoc and ATPC1.
Phylogenetic analysis based on 16S rDNA sequence analysis:
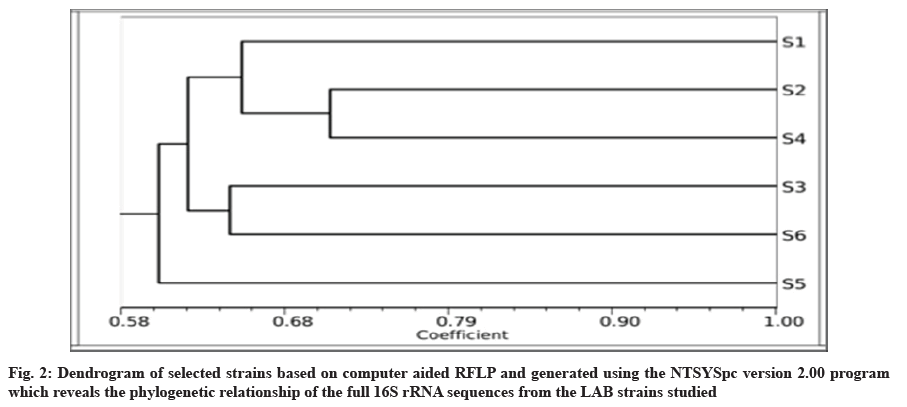
The phylogenetic tree among species was constructed using Numerical Taxonomy System for personal computer (NTSYSpc) based on sequencing of 16S rRNA with TotalLab gel work 1D Advan. Dendrograms that showed genetic similarities between the six LAB species were closely constructed using the similarity coefficients=Simple Matching (SM), which was used to evaluate the results obtained, based on the 16S rRNA markers. The estimated similarity coefficients were calculated with the NTSYSpc 2.01 software and indicated the maximum similarity (0.66) between the S1 and S2 species. However, the minimum genetic similarity (0.71) was observed between S2 and S4.
Results and Discussion
20 LAB isolates, deposited in the NCBI GenBank database, included in this study were characterized based on the following-morphological, physiology, biochemical characteristics as well as on the molecular level using 16S rRNA gene sequences as described in a recent study by Bin Masalam et al.[15]. They were categorized into two main genera Lactobacillus and Streptococcus using the selective media. Almost all isolates were confirmed as Gram-positive (92.47 %) and catalase negative bacteria. Based on the cultural and primary and secondary biochemical tests, there were similarities as well as differences among the 20 tested LAB strains[15]. According to their phenotypes, the isolates were divided into classes at the beginning and then they were divided into groups by using RFLP analysis and based on the sequencing of the 16S rRNA four bacterial groups (A to D) were identified based on biochemical characteristics led to the tests and phenotypic features. The isolates Lactobacillus casei MSJ1, Lactobacillus casei Dwan5, Lactobacillus plantarum EyLan2 and Enterococcus faecium Gail-BawZir8 were most abundant of LAB with the best for their probiotic efficacy. In the previous work, antibiotic resistance testing was also used as a criterion to differentiate among the different strains.
According to the antibody resistance testing, Weissella confusa showed resistance for cefoxitin, oxacillin and nalidixic acid. Five of the tested strains were resistant to penicillin G (Streptococcus thermophilus BinSlman8, Streptococcus thermophilus MaNaL33, Streptococcus equinus Omer9, Streptococcus equinus JmaL3 and Streptococcus equinus Foad7) while 15 strains showed vancomycin resistance. These strains belonged to Lactobacillus casei (4 strains), Lacticaseibacillus paracasei NMBM1, Lactobacillus plantarum EyLan2, Lactobacillus futsaii EMBM2 and Weissella confusa (8 strains)[15]. The previous data also revealed that Streptococcus thermophilus BinSlman8, Streptococcus thermophilus MaNaL33, strains had alpha (α)-hemolytic activity[15]. 20 of the 20 tested strains tolerated acidic conditions at various levels. Most tolerant acidic strains included enterococci such as Enterococcus faecium SMBM3, BagHom4, ZiNb3, Gail-BawZir8, ESJ4, NSJ2, Marwh2, SSJ3, Etimad1 and other LAB genera, such as Weissella confusa SaEd-7, AhMd8, Tarim4, NooR1, SaYun2, SYary1, Lactobacillus casei MSJ1, BgShn3, Dwan5, Lactobacillus futsaii EMBM2 and Lactococcus lactis HadRami9. These results indicated that Lactobacillus casei MSJ1, Lactobacillus casei Dwan5, Lactobacillus plantarum EyLan2 and Enterococcus faecium Gail-BawZir8 isolates derived from fermented or raw Saudi milk, especially as in the case of camel’s yogurt (Laban) had a very good probiotics activity[15]. On the other hand, the results in Table 3 and fig. 1 indicated that the nucleotide sequences of strains aligned with the 16S rRNA sequences of 6 different species belonging to five genera, namely, Enterococcus, Lactococcus, Lactobacillus, Streptococcus and Weissella. Based on the above, the CisSERS approach offered an alternative to conventional methods, permitting accurate identification and grouping of LAB strains to the species level. In addition, the present method produced results on LAB comparable to those obtained using conventional identification methods. However, individual strains within a species can differ significantly in key genotypic and phenotypic characteristics, such as antibiotic resistance, virulence and growth rate.
| S. No. | Strain enzymes | Ku324908_C | Ku324898_C | Ku324909_C | Ku324920_C | Ku324928_C | Ku324937_C |
|---|---|---|---|---|---|---|---|
| 1 | AccBSI | 1270//230 | - | - | - | - | - |
| 2 | ApaI | - | - | - | - | 1500 | - |
| 3 | ApyPI | - | - | - | - | - | 750//7500 |
| 4 | BaeI | - | - | 800//700 | - | - | - |
| 5 | BamHI | 900//600 | - | - | - | - | - |
| 6 | BbvCI | - | - | - | - | 720//780 | - |
| 7 | BcII | - | 1100//400 | - | - | - | - |
| 8 | BfuI | - | 950//580 | - | - | - | - |
| 9 | BInI | 1250//280 | - | - | - | - | - |
| 10 | Bso31I | - | - | - | - | 900//600 | - |
| 11 | Bst107I | - | - | - | - | 1250//250 | - |
| 12 | BstAPI | - | - | - | - | 1300//200 | - |
| 13 | CchIII | - | - | - | - | - | 950//550 |
| 14 | DarIII | 80//700 | - | - | - | - | |
| 15 | Eco47III | - | - | 1450//50 | - | - | - |
| 16 | GasI | - | 1100//400 | - | - | - | - |
| 17 | HincII | - | 1500 | - | - | - | - |
| 18 | KfII | - | - | - | - | 1000//500 | - |
| 19 | NcoI | - | - | 1200//400 | - | - | - |
| 20 | NheI | - | - | - | - | - | 750//7500 |
| 21 | NemAIII | - | 1500 | - | - | - | - |
| 22 | NruI | - | - | 1300//200 | - | - | - |
| 23 | PasI | - | - | - | - | 1000//500 | - |
| 24 | PciI | 1400//100 | - | - | - | - | - |
| 25 | PciSI | 1450//50 | - | - | - | - | - |
| 26 | PfoI | 800//700 | - | - | - | - | - |
| 27 | PpiI | - | 1450//50 | - | - | - | - |
| 28 | PspPRI | - | - | - | - | 900//600 | - |
| 29 | PstI | - | - | - | 850//650 | - | - |
| 30 | PstNI | 1300//200 | - | - | - | - | - |
| 31 | RdeGBII | - | - | 1000/500 | - | 700//800 | - |
| 32 | RpaB5I | - | - | - | - | 1500 | - |
| 33 | SdeAI | - | - | - | - | - | - |
| 34 | SfiI | - | - | 900//600 | - | - | |
| 35 | TstI | 1500 | - | - | - | - | - |
Note: //: Integer Division
Table 3: List of Restriction Enzymes with Recognition Sites within 16S rRNA Gene along with Lab Strains.
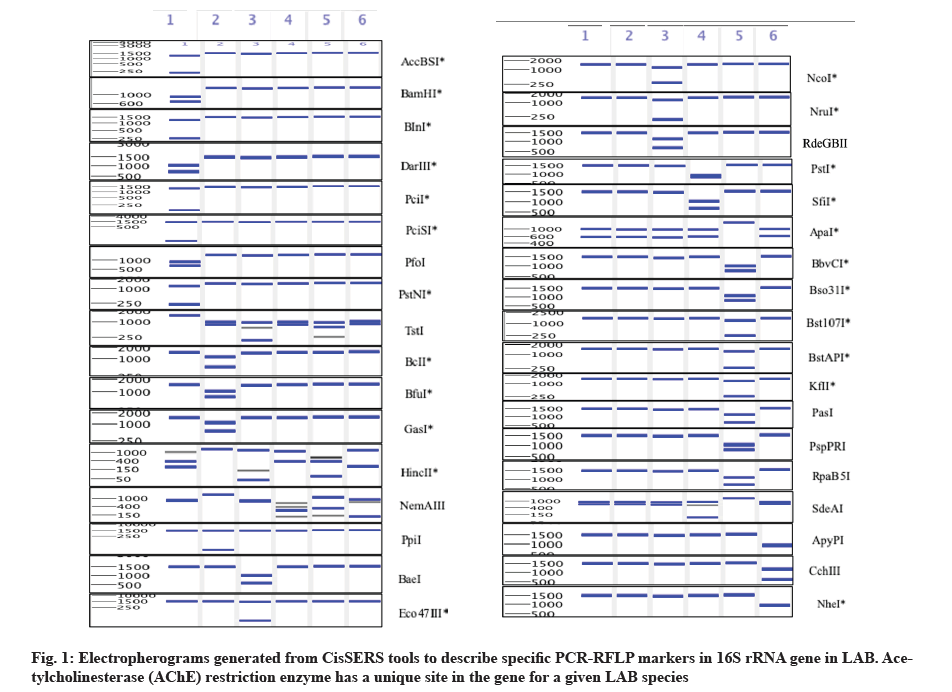
In the present study, we utilized virtual PCR-RFLP analysis of the 16S rRNAs gene, used as a fast and effective approach to differentiate and classify the LAB species present in large numbers. In silico 16S rRNA PCR-RFLP, uses certain identified restriction sites located on a fragment of the target gene (e.g., 16S rRNA) which will serve as markers for the quick identification and the LAB species classification and related genera, which reveals more information about their phylogenetic relationships. PCR-RFLP is a relatively simple and inexpensive method for genotyping of Single Nucleotide Polymorphisms (SNPs), which is based on the number and sizes of the in silico cleaved fragment of any given species. RFLP analysis was performed to narrow down the identification of LAB strains to the species level. Cumulative results from a limited number of studies suggested that 16S rRNA gene sequencing provides identification at the genus level (90 %) but less so concerning species level (65 to 83 %), with 1 %-14 % of the isolates remaining unidentified after testing[12].
In the current study, we selected 6 out of 20 species based on the similarity of the sequences which are retrieved from the GenBank database. The similarity scores were between 98 % and 100 %. High 16S rRNA sequence similarity (98 %-99 %) identified Lactobacillus species (Table 2). The Clustal Omega alignment program was used to find the best-matching sequences to get the “consensus” fragment. Consensus sequences used was extremely relevant in deciphering the evolutionary relationships among different bacteria. These sequences were exposed to CisSERS in order to produce the virtual PCR-RFLP patterns and the specific restriction enzymes recognition sites that can either individually cut DNA of a single species or may lead to the production of a specific DNA fragment which is specific to an individual single LAB species. The digested sequences were visualized by the virtual gel output and the resulting patterns were used to differentiate among them based on the digested DNA size with respect to each of the specific restriction enzymes used (fig. 1). Bacterial identification results are shown in Table 1 based on similarity scores and were then assigned at the species or genus level based on the similarity score of ≥99 % and from 90 % to 99 %, respectively[16,17].
BLAST does not enable the identification of bacteria to the species level. This is because of the presence of sequence homologies among some species. BLAST is based on matching similar regions among sequences and the identification of related genes will give related evolutionary and functional information concerning the relationships of the cloned genes or even within whole genomes, which is a prerequisite for most phylogenetic analyses[18]. These tests, therefore, showed that BLAST is an accurate, fast and sensitive tool used for the analysis of the sequence alignments. However, it did not differentiate well in the identification of closely related species. In this study, the resulted consensus sequences showed significant similarities of 98 %-99 % among the six samples tested. The six isolated bacteria belonged to the genera of Lactobacillus, Weissella, Enterococcus, Lactococcus and Streptococcus as shown in both Table 1 and Table 2. In a published study, the species of the Streptococcus mitis, including Streptococcus pneumoniae, were almost indistinguishable from one another based on their BLAST analysis on 16S rRNA genes, with sequence similarities ranging from 99 %-100 %[16].
The 16S rRNA partial gene sequence (~1500 bp) generates adequate phylogenetic information, however, better and more precise identification of bacteria will be achieved upon using the full 16S rRNA gene sequence[19]. Published data indicated that the 16S rRNA gene sequences of LAB strains showed high sequence similarities[20]. To differentiate among the closely related LAB, PCR-RFLP analysis was used to achieve a more precise discrimination among the large number of isolates[21]. In the current study, using computer-simulation analyses, we were able to identify a group of restriction enzymes that could produce unique LAB 16S rRNA band patterns, different from that generated by other bacteria. These findings indicated that CisSERS was precise in the identification and discrimination among the different bacterial species. No single approach used can provide sufficient information for intra and inter species differentiation, hence, the current approach to be followed is multiphasic in order to identify and characterize the different LAB strains in a precise fashion[22].
In silico analyses of the locations and sequences of the restriction sites to be used for the differentiation among the various LAB was explained here. The virtual PCR-RFLP patterns obtained for the 6 reference strains with 36 restriction enzymes approach are shown in fig. 1 and summarized in Table 3.
16S rRNA gene sequences contain hypervariable regions that could provide species-specific signature sequences useful for identification of a certain species. The largest number of species-specific markers are shown for KU324928_C (10 markers) of Streptococcus thermophilus BinSlman8, followed by KU324908_C (8 markers) of Lactobacillus casei BgShn3, then KU324909_C (5 markers) of Lactococcus lactis HadRami9, KU324898_C (4 markers) of Enterococcus faecium BagHom4 and finally, KU324937_C (3 markers) of Lactococcus garvieae ZSJ5as, shown in Table 4.
| S. No. | Species | Enzymes | Species-specific marker (base pair (bp)) 5’…………….3’ |
|---|---|---|---|
| 1 | Ku324908_C | AccBSI | 1280//220 |
| BamHI | 900/600 | ||
| BInI | 280/1220 | ||
| DarIII | 800/700 | ||
| PciI | 1400/100 | ||
| PciSI | 1450/50 | ||
| PfoI | 800/700 | ||
| PstNI | 1300/200 | ||
| TstI | 1500 | ||
| 2 | Ku324898_C | BcII | 1100/400 |
| BfuI | 920/580 | ||
| GasI | 1100/400 | ||
| HincII | 1500 | ||
| NemAIII | 1500 | ||
| PpiI | 1450/50 | ||
| 3 | Ku324909_C | BaeI | 800/700 |
| Eco47III | 1450/50 | ||
| NcoI | 1200/300 | ||
| NruI | 1300/200 | ||
| RdeGBII | 1000/500 | ||
| 4 | Ku324920_C | PstI | 850/650 |
| SfiI | 900/600 | ||
| 5 | Ku324928_C | ApaI | 1500 |
| BbvCI | 780/220 | ||
| Bso31I | 900/600 | ||
| Bst107I | 1250/250 | ||
| BstAPI | 1300/200 | ||
| KfII | 1000/500 | ||
| PasI | 1000/500 | ||
| PspPRI | 900/600 | ||
| RpaB5I | 800/700 | ||
| SdeAI | 1500 | ||
| 6 | Ku324937_C | ApyPI | 750/750 |
| CchIII | 950/550 | ||
| NheI | 750/750 |
Table 4: List of Lab Strains along with Restriction Enzymes with Recognition Sites within 16S rRNA Gene.
Based on the results of the in silico studies, a group of 10 enzymes were identified (PstI, SfiI, BbvCI, Bso31I, Bst107I, BstAPI, KfII, PasI, PspPRI and RpaB5I) that produced fragments, unique to the examined KU324928_C strains (Table 4). The combination of 6 out of the 10 discriminatory enzymes resulted on average in 99.5 % differentiation among the different sequences. Hence, a minimum of six enzymes were used to achieve an acceptable differentiation among the different LAB species. Digestion of the 16S rRNA gene with the ApyPI, CchIII and NheI produced clear, distinctive patterns for KU324937_C species of the 6 species. However, the other species had a similar RFLP pattern. Subsequent digestion of the 16S rRNA gene from KU324909_C species with the BaeI, Eco47III, NcoI, NruI and/or RdeGBII generated species-specific RFLP patterns. Furthermore, the PCR-RFLP patterns within species were correctly predicted by the in silico investigation, hence enabling their unequivocal identification.
The phylogenetic tree was constructed after sequencing of 16S rRNA to determine the relationship among LAB strains. The program NTSYSpc was used to reconstruct phylogenetic relationships of most of the LAB. The phylogenetic tree is known to reveal a high degree of consistency of the relationships among organisms[23]. In this study, the constructed phylogeny revealed that all the strains had a common phylogenetic coherent cluster. Notably, the 2s and 4s strains appeared, related to species with an average similarity of 71 %, as well as 3s strain were very close to 6s that showed clustered with an average similarity of 65 % (range being 70 %-100 % in all the phylogenetic trees). 5s level strain were grouped into separate clusters and performed close clade with remaining. The 6 strains belonged to different species (fig. 2).
In the current study, a computer-based approach was employed to evaluate the efficacy of 237 different restriction enzymes, when used individually or in combination, to differentiate among the LAB species based on the resulted restriction patterns. The final aim was to propose a screening approach for the selection of the enzymes and to predict the number of enzymes required to ensure differentiation of known and unknown species. CisSERS is a time-saving and user-friendly tool to use in phylogenic studies. Firstly, it narrows down the number of restriction enzymes to select those that show the highest possible discriminatory power in terms of studying any microbial population bacterial phylogeny and taxonomy. Secondly, it allows for the identification of the microbial composition of biological samples at unprecedented resolution. Thirdly, it supports the usage of the wet-lab PCR-RFLP as a fast fragment analysis method that requires less equipment, relatively affordable and easy to use. PCR-RFLP is a useful approach for characterizing the dominant taxa within complex microbial populations. Fourthly, it gives species-specific markers used as a first level of species detection. Such an approach can be adopted for other types of universal markers in prokaryotic as well as eukaryotic organisms. Concretely, in silico RFLP-PCR is a powerful tool to detect species-specific markers that can be further utilized to provide better understanding of microbial communities found in raw and fermented milk. Future studies should include more samples sized on more dairy based foods.
In this preliminary study, we have randomly selected 20 strains and included them in this study. Based on the matches of the available 16S rRNA gene sequence in the NCBI GenBank database, BLAST similarity scores ranged between 98 % to 100 %. For this reason, we opted to focus on six of those which are considered by some as a small sample size. However, in future studies, more samples will be evaluated to get more precise information and better conclusions. Our recommendations are that more studies will be conducted in the future with more samples and various consumable dairy-based foods.
Acknowledgements:
This project was funded by the Deanship of Scientific Research (DSR) at King Abdulaziz University, Jeddah, under grant no: G-451-130-38. The authors therefore, acknowledge with thanks DSR for technical and financial support.
Conflict of interests:
The authors declare that they have no conflicts of interest.
References
- Sharpe RM, Koepke T, Harper A, Grimes J, Galli M, Satoh-Cruz M, et al. Cis SERS: Customizable in silico sequence evaluation for restriction sites. PLoS One 2016;11(4):e0152404.
- Juodeikiene G, Bartkiene E, Viskelis P, Urbonaviciene D, Eidukonyte D, Bobinas C. Fermentation processes using lactic acid bacteria producing bacteriocins for preservation and improving functional properties of food products. Adv Appl Biotechnol 2012;2012:63-100.
- Klaenhammer TR, Barrangou R, Buck BL, Azcarate-Peril MA, Altermann E. Genomic features of lactic acid bacteria effecting bioprocessing and health. FEMS Microbiol Rev 2005;29(3):393-409.
- Adams M, Moss MO, McClure P. Food Microbiology. 4th ed. London: Royal Society of Chemistry; 2016.
- Leroy F, de Vuyst L. Lactic acid bacteria as functional starter cultures for the food fermentation industry. Trends Food Sci Technol 2004;15(2):67-78.
- Sánchez ÓJ, Barragán PJ, Serna L. Review of Lactobacillus in the food industry and their culture media. Rev Colomb Biotecnol 2019;21(2):63-76.
- Khalid K. An overview of lactic acid bacteria. Int J Biosci 2011;1(3):1-3.
- Huang CH, Li SW, Huang L, Watanabe K. Identification and classification for the Lactobacillus casei group. Front Microbiol 2018;9:1974.
- Mokoena MP. Lactic acid bacteria and their bacteriocins: Classification, biosynthesis and applications against uropathogens: A mini-review. Molecules 2017;22(8):1255.
[Crossref] [Google scholar] [PubMed]
- Desai AR. Strain identification, viability and probiotics properties of Lactobacillus casei. Victoria University; 2019.
- Winand R, Bogaerts B, Hoffman S, Lefevre L, Delvoye M, van Braekel J, et al. Targeting the 16s rRNA gene for bacterial identification in complex mixed samples: Comparative evaluation of second (illumina) and third (oxford nanopore technologies) generation sequencing technologies. Int J Mol Sci 2019;21(1):298.
[Crossref] [Google scholar] [PubMed]
- Janda JM, Abbott SL. 16S rRNA gene sequencing for bacterial identification in the diagnostic laboratory: Pluses, perils and pitfalls. J Clin Microbiol 2007;45(9):2761-4.
[Crossref] [Google scholar] [PubMed]
- Moreira JL, Mota RM, Horta MF, Teixeira SM, Neumann E, Nicoli JR, et al. Identification to the species level of Lactobacillus isolated in probiotic prospecting studies of human, animal or food origin by 16S-23S rRNA restriction profiling. BMC Microbiol 2005;5(1):1-9.
[Crossref] [Google scholar] [PubMed]
- Neyra M, Khbaya B, de Lajudie P, Dreyfus B, Normand P. Computer-assisted selection of restriction enzymes for rrs genes PCR-RFLP discrimination of rhizobial species. Genet Sel Evol 1998;30:S297-309.
- Bin Masalam MS, Bahieldin A, Alharbi MG, Al-Masaudi S, Al-Jaouni SK, Harakeh SM, et al. Isolation, molecular characterization and probiotic potential of lactic acid bacteria in Saudi raw and fermented milk. Evid Based Complement Alternat Med 2018;2018:1-12.
[Crossref] [Google scholar] [PubMed]
- Peker N, Garcia-Croes S, Dijkhuizen B, Wiersma HH, van Zanten E, Wisselink G, et al. A comparison of three different bioinformatics analyses of the 16S-23S rRNA encoding region for bacterial identification. Front Microbiol 2019;10:620.
[Crossref] [Google scholar] [PubMed]
- Clarridge III JE. Impact of 16S rRNA gene sequence analysis for identification of bacteria on clinical microbiology and infectious diseases. Clin Microbiol Rev 2004;17(4):840-62.
[Crossref] [Google scholar] [PubMed]
- McGinnis S, Madden TL. BLAST: At the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res 2004;32(2):20-5.
[Crossref] [Google scholar] [PubMed]
- Lee CM, Sieo CC, Wong CM, Abdullah N, Ho YW. Sequence analysis of 16S rRNA gene and 16S–23S rRNA gene intergenic spacer region for differentiation of probiotics Lactobacillus strains isolated from the gastrointestinal tract of chicken. Ann Microbiol 2008;58(1):133-40.
- Cihan AC, Tekin N, Ozcan B, Cokmus C. The genetic diversity of genus Bacillus and the related genera revealed by 16S rRNA gene sequences and ardra analyses isolated from geothermal regions of turkey. Braz J Microbiol 2012;43:309-24.
- Scheidegger EM, Fracalanzza SA, Teixeira LM, Cardarelli-Leite P. RFLP analysis of a PCR-amplified fragment of the 16S rRNA gene as a tool to identify Enterococcus strains. Mem Inst Oswaldo Cruz 2009;104:1003-8.
[Crossref] [Google scholar] [PubMed]
- Sharma A, Lee S, Park YS. Molecular typing tools for identifying and characterizing lactic acid bacteria: A review. Food Sci Biotechnol 2020;29(10):1301-18.
- Adhikari A, Nandi S, Bhattacharya I, de Roy M, Mandal T, Dutta S. Phylogenetic analysis based evolutionary study of 16S rRNA in known Pseudomonas sp. Bioinformation 2015;11(10):474-80.
[Google scholar] [PubMed]