- *Corresponding Author:
- Urmila J. Joshi
Department of Pharmaceutical Chemistry, Prin. K. M. Kundnani College of Pharmacy, Cuffe Parade, Mumbai - 400 005, India
E-mail: urmilajjoshi@hotmail.com
| Date of Submission | 11 April 2005 |
| Date of Revision | 17 September 2007 |
| Date of Acceptance | 6 December 2007 |
| Indian J. Pharm. Sci., 2007, 69 (6): 800-804 |
Abstract
Of the various structurally diverse compounds known to bind 5-HT 1A receptor sites, arylpiperazine derivatives represent one of the most important classes. This article deals with the development of a QSAR equation relating the ligand binding activity of various literature reported arylpiperazines acting as agonists at the 5-HT 1A receptor to their 2D descriptors. Significant equation was generated using MOE 2004.03 and validated subsequently using leave one out and test set prediction methods. The equation revealed the importance of combination of electronic and lipophilic parameters in explaining the observed variance.
Keywords
5-HT1A receptor, arylpiperazines, agonists, QSAR
Serotonin (5-hydroxytryptamine, 5-HT), a neurotransmitter in brain has drawn considerable amount of attention in past few years because of detection of multiple, central 5-HT receptors [1,2]. 5-HT1A receptor is the best characterized receptor among these due to the availability of a specific partial agonist, 8-hydroxy-2-(di-n-propylamino)tetraline [3] (8-OH DPAT). The 5-HT1A receptor is a G-protein coupled receptor [4] and is reported to be associated with inducing sleep, cognition, sensory perception, motor activity, temperature regulation, nociception, appetite, sexual behavior and hormone secretion [5]. This receptor is also involved in the psychiatric disorders like depression and anxiety. Therefore modulation of 5- HT1A receptor activity will be an important therapeutic approach in the treatment of these disorders in future. No clinically used specific agonist for 5-HT1A receptor is available currently.
The design of a drug involves a multidisciplinary approach and requires assimilation of information generated by diverse techniques. SAR of arylpiperazines as 5-HT1A agonists [6] and the pharmacophore elements for 5-HT1A receptor agonist [7-10] have been reported in the literature. However a complete X-ray crystallographic data indicating the structure of the 5-HT1A receptor and the receptor bound conformation of arylpiperazine ligands is not available.
In the absence of availability of receptor bound conformation, the ligand based methods of analysis like QSAR form one of the major approaches for understanding the available biological data and developing predictive correlations. This can help to keep the number of synthesized and tested compounds to the minimum, apart from assisting in the prediction of the nature of the receptor. 3D QSAR studies of some arylpiperazines are reported [11]. However unambiguous 3D alignments frequently remain a difficult task particularly for flexible molecules such as arylpiperazines containing a 2-3 carbon alkyl chain. Therefore it was decided to initiate the QSAR studies with 2D descriptors and generate QSAR equations which will help in further development of 5-HT1A agonists.
Materials and Methods
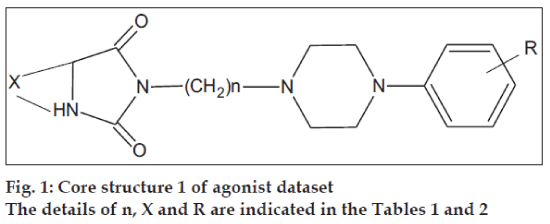
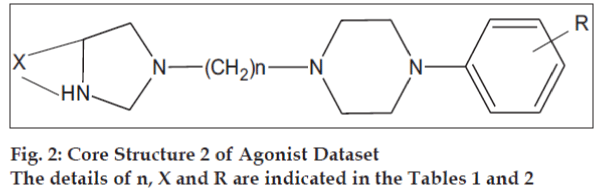
The initial dataset used consisted of 44 literature reported molecules [12,13] of arylpiperazine class, core structures of which are as given in figs. 1 and 2, along with their reported pKi values. This Dataset was then randomly divided into a training set of 37 compounds, structures of which are tabulated in Table 1 and a test set of 7 compounds, structures of which are tabulated in Table 2, taking care that the mean pKi values of the two sets remain comparable. Molecules were built within MOE 2004.0314. The structures were refined using ‘Wash’ module. Molecules were then energy minimized using MMFF94x force field. Due to the unavailability of the receptor bound conformations of these molecules, the 3D molecular descriptors were excluded and only 2D molecular descriptors were calculated for the dataset using ‘QSAR_descriptor’ functionality. Descriptors were pruned using genetic algorithm (GA) and QSAR contingency modules. Table 3 gives the definitions of the descriptors used for deriving QSAR equation. QSAR equation was generated with an assumption that all the compounds in the dataset posses same mechanism of action. The statistical method used for generating QSAR equation was Partial Least Squares (PLS) method. QSAR model was validated using leave one out method, by randomizing the Y-responses [15] and test set predictions. Standard related statistical parameters [16] like correlation coefficient r, standard error s, F-test value, q2 value were calculated for the equation.
| Compdno. | Corestructure | X | n | R | pKi (obs) | pKi (pred) |
|---|---|---|---|---|---|---|
| 1 | 1 | (CH2)4 | 1 | p-F | 6.35 | 6.13 |
| 2 | 1 | (CH2)3 | 2 | m-CF3 | 6.91 | 7.18 |
| 3 | 1 | (CH2)4 | 2 | o-OCH3 | 7.34 | 7.32 |
| 4 | 1 | (CH2)4 | 2 | m-Cl | 6.89 | 6.84 |
| 5 | 1 | (CH2)4 | 2 | p-F | 7.18 | 7.18 |
| 6 | 1 | (CH2)3 | 3 | H | 7.72 | 7.59 |
| 7 | 1 | (CH2)3 | 3 | m-Cl | 7.25 | 7.59 |
| 8 | 1 | (CH2)4 | 3 | m-Cl | 7.27 | 7.60 |
| 9 | 1 | (CH2)4 | 3 | p-F | 6.22 | 6.74 |
| 10 | 1 | (CH2)3 | 4 | H | 7.60 | 7.87 |
| 11 | 1 | (CH2)3 | 1 | o-OCH3 | 7.46 | 7.46 |
| 12 | 1 | (CH2)3 | 4 | p-F | 7.04 | 7.01 |
| 13 | 1 | (CH2)4 | 4 | p-F | 7.23 | 7.01 |
| 14 | 1 | (CH2)3 | 1 | m-Cl | 7.23 | 6.99 |
| 15 | 1 | (CH2)3 | 1 | m-CF3 | 6.92 | 7.32 |
| 16 | 1 | (CH2)3 | 1 | p-F | 6.30 | 6.13 |
| 17 | 1 | (CH2)4 | 1 | H | 6.99 | 6.99 |
| 18 | 1 | (CH2)4 | 1 | m-CF3 | 7.10 | 7.32 |
| 19 | 2 | (CH2)3 | 3 | m-CF3 | 7.53 | 7.63 |
| 20 | 2 | (CH2)4 | 3 | o-OCH3 | 7.85 | 7.78 |
| 21 | 2 | (CH2)4 | 3 | m-CF3 | 7.61 | 7.63 |
| 22 | 2 | (CH2)3 | 4 | o-OCH3 | 7.76 | 8.05 |
| 23 | 2 | (CH2)3 | 4 | m-Cl | 7.55 | 7.58 |
| 24 | 2 | (CH2)3 | 4 | m-CF3 | 7.91 | 7.91 |
| 25 | 2 | (CH2)4 | 4 | o-OCH3 | 8.29 | 8.05 |
| 26 | 2 | (CH2)4 | 4 | m-CF3 | 7.81 | 7.91 |
| 27 | 1 | (CH2)3 | 3 | m-CF3 | 8.42 | 7.92 |
| 28 | 1 | (CH2)4 | 3 | m-CF3 | 8.24 | 7.93 |
| 29 | 1 | (CH2)3 | 4 | o-OCH3 | 8.26 | 8.35 |
| 30 | 1 | (CH2)3 | 4 | m-Cl | 7.95 | 7.87 |
| 31 | 1 | (CH2)3 | 4 | m-CF3 | 8.62 | 8.20 |
| 32 | 1 | (CH2)4 | 4 | o-OCH3 | 8.06 | 8.35 |
| 33 | 1 | (CH2)4 | 4 | m-Cl | 8.14 | 7.87 |
| 34 | 1 | (CH2)4 | 4 | m-CF3 | 8.00 | 8.20 |
| 35 | 2 | (CH2)3 | 3 | o-OCH3 | 7.91 | 7.78 |
| 36 | 1 | (CH2)3 | 3 | o-OCH3 | 8.35 | 8.07 |
| 37 | 1 | (CH2)3 | 1 | H | 7.07 | 6.99 |
The core structures are indicated in figs. 1 and 2. pKi (obs) indicates the observed pKi for the training set and pKi (pred) indicates the pKi calculated using the equation.
Table 1: Structures of training set
| Compd no. | Core structure | X | n | R | pKi (obs) | pKi (pred) | ||
|---|---|---|---|---|---|---|---|---|
| 1 | 1 | (CH2)3 | 2 | o-OCH3 | 6.63 | 7.32 | ||
| 2 | 1 | (CH2)3 | 2 | m-Cl | 6.38 | 6.84 | ||
| 3 | 1 | (CH2)4 | 3 | H | 6.81 | 7.59 | ||
| 4 | 1 | (CH2)4 | 1 | o-OCH3 | 7.49 | 7.46 | ||
| 5 | 1 | (CH2)4 | 1 | m-Cl | 7.23 | 6.99 | ||
| 6 | 2 | (CH2)4 | 4 | m-Cl | 7.61 | 7.58 | ||
| 7 | 1 | (CH2)4 | 3 | o-OCH3 | 8.38 | 8.07 | ||
pKi (obs) indicates the observed pKi for the test set and pKi (pred) indicates the pKi calculated for the test set using the generated QSAR equation.
Table 2: Structures of test set
| Abbreviation | Definition of the descriptor | ||||||
|---|---|---|---|---|---|---|---|
| b_1rotN: | A bound count descriptor; it indicates number of rotatable single bonds. | ||||||
| Diameter: | A steric parameter; it indicates the largest value in the distance matrix. | ||||||
| PC+: | A partial charge descriptor; it indicates the total positive partial charge | ||||||
| Q_VSA_POL: | A partial charge descriptor; it indicates the total polar van der Waals surface area. | ||||||
| SlogP_VSA7: | A subdivided surface area descriptor; it indicates the contribution to lipophilicity | ||||||
| SMR_VSA7: | A subdivided surface area descriptor; it indicates the contribution to molar refractivity. | ||||||
| I1: | Indicator variable for carbonyl oxygens; its value is one when carbonyl oxygen is present and is zero when the carbonyl oxygen is absent. | ||||||
Descriptor definitions are as indicated in the molecular modeling software MOE
Table 3: Descriptor definitions
Results and Discussion
QSAR Equation
pKi = 34.22 − 2.04 (diameter) + 2.31 (b_1rotN) − 0.056 (PEOE_VSA+1) + 5.62 (PC+) − 0.16 (Q_VSA_ POL) − 0.084 (SlogP_VSA7) − 0.040 (SMR_VSA7), n= 37; r = 0.92; r2 = 0.84; s = 0.23; F = 22.3781; q2 = 0.74.
The correlation matrix (Table 4) indicates a high correlation between PC+ and Q_VSA_POL and diameter and b_1rotN. However for resolving the issue of collinearity, Randic recommends that the descriptors which are different in their information content can be retained even if they are highly correlated [17]. As can be seen from the definition of the descriptors the information conveyed by these descriptors is different. Therefore it was decided to retain these descriptors.
| Diameter | b_1rotN | PEOE_VSA+1 | PC+ | Q_VSA POL | SlogP_VSA7 | SMR_VSA7 | |
|---|---|---|---|---|---|---|---|
| Diameter | 1 | ||||||
| b_1rotN | 0.74 | 1 | |||||
| PEOE_VSA+1 | -0.47 | 0.3 | 1 | ||||
| PC+ | 0.10 | -0.5 | -0.29 | 1 | |||
| Q_VSA_POL | 0.18 | 0.32 | -0.10 | 0.88 | 1 | ||
| SlogP_VSA7 | -0.18 | -0.47 | -0.28 | -0.33 | -0.54 | 1 | |
| SMR_VSA7 | -0.25 | 0.20 | 0.29 | -0.56 | -0.31 | -0.38 | 1 |
Table 4: Correlation matrix of descriptors
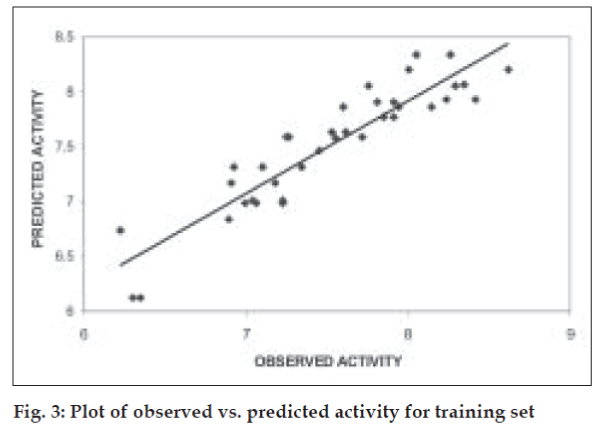
The QSAR equation derived showed a very good r (correlation coefficient) value of 0.92 with 84.37% of the variation in biological activity being explained by the equation. This is associated with a low value of standard error of estimate, s, of 0.23. The equation is found to be highly statistically significant with F-test value of 22.378, critical F-test value at 99.9% confidence limit being 4.82. The model when validated using leave one out method showed good internal predictivity with the q2 value being 0.74 indicating good predictivity of the model. Fig. 3 shows the plot of observed v/s predicted pKi values for training set using the above mentioned equation.
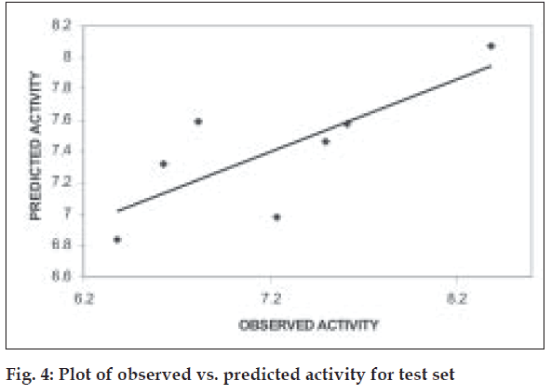
Further validation of the model was carried out by randomization of Y- responses (pKi values) to ensure the absence of chance correlation. Randomization was carried out in triplicates. For each randomization the r value decreased drastically indicating the absence of chance correlation. The mean r2ran was found to be 0.32. When the model was subjected to test set predictions it also exhibited good external predictivity with the r2test value being 0.60. Table 2 shows the predicted pKi values and (fig. 4) shows the plot of observed v/s predicted pKi values for test set.
The descriptors diameter, b_1rotN and PC+ were found to contribute more significantly to the activity as indicated by their higher coefficients in the equation. This suggests that size and shape of the molecule along with its flexibility and the total positive partial charge on the molecule play a significant role in determining the interaction with the receptor. These findings are in agreement with the pharmacophore model reported for 5-HT1A agonist which states that aromatic ring and basic nitrogen are two structural features necessary for ligand recognition7,8. SAR studies reported for arylpiperazines state that presence of alkyl chain improves the activity. This is also proved mathematically by the equation where number of rotatable bonds is a descriptor of greater significance and is positively correlated with biological activity. Other reported pharmacophore models state that the third structural feature of the pharmacophore is an oxygen atom which can act as H-bond acceptor9,10. These pharmacophore models were developed using aminotetralin derivatives without inclusion of the arylpiperazines containing the oxygen atom. A closer look at the structures of the present dataset shows that some of the arylpiperazines of the training set show presence of amide oxygen. This atom may be involved in the H-bonding interaction with the receptor. To model this interaction it was decided to include an indicator variable term I1 which assumed a value of 1 when amide oxygen is present and 0 when amide oxygen is absent. The equation thus generated was as follows: pKi = 42.46 − 2.37 (diameter) + 2.63 (b_1rotN) − 0.065 (PEOE_VSA+1) − 0.12 (SlogP_VSA7) − 0.054 (SMR_VSA7) − 0.18 (Q_VSA_POL) + 6.12 (PC+) + 0.20 (I1), n= 37; r = 0.9194; r2 = 0.81773; s = 0.23361; F = 19.1370.
Although the coefficients of the descriptors have increased slightly in the above equation, there is no significant improvement in the r, s and F values of the equation by inclusion of indicator variable. Thus it may be stated that presence of amide oxygen in this series of arylpiperazines is not an absolute must.
QSAR studies on the arylpiperazines have been reported in the past. These include both 2D QSAR as well as 3D QSAR studies. The 2D QSAR studies reported for the arylpiperazines [18] acting as agonists were done using a subset of the present dataset and the biological activity was carried out in a different way from that for the present dataset. These studies report the importance of o-substituent only. The 3D QSAR studies were done on the arylpiperazines without differentiating them into agonists and antagonist, by assuming a common binding pattern for all. Therefore it is difficult to relate these results with the QSAR of the agonists only.
A different set of descriptors may give different correlation with activity and this should be considered while interpreting the equation. The QSAR equation generated presently for arylpiperazine is in agreement with the literature reports for ligand requirements based on the SAR and pharmacophore generation technique. Further, this equation provides a mathematical tool for designing compounds with better activity.
Acknowledgements
The authors are thankful to AICTE for sanction of grant for the software. Authors are also thankful to Hyderabad (Sindh) National Collegiate Board and Dr. S. R. Naik, Principal of Prin. K. M. Kundnani College of Pharmacy for providing necessary facilities.
References
- Glennon R. Central serotonin receptors as targets for drug research. J Med Chem 1987;30:1-12. .
- Glennon R. Higher-end serotonin receptors: 5-HT 5 , 5-HT 6 , and 5-HT 7. J Med Chem 2003;46:2795-812.
- Arvidsson LE, Johansson AM, Hacksell U, Nilsson LG, Svensson K, Hjorth S, et al. (+)-cis-8-hydroxy-1-methyl-2-(di-n-propylamino) tetralin: A Potent and Highly Stereoselective 5-Hydroxytryptamine Receptor Agonist. J Med Chem 1987; 30:2105-9
- Sanders-Bush E, Mayer S. 5-Hydroxytryptamine (Serotonin): Receptor agonists and antagonists. In ; Hardman JG, Limbird LE, Gilman AG, editors. Goodman and Gilman's The Pharmacological basis of Therapeutics. 10th ed. New York: McGraw-Hill; 2001. p. 269
- Peroutka S. In ; Serotonin receptor subtypes. Basic and clinical aspects. New York: Wiley-Liss; 1991. p. 1.
- Lopez-Rodriguez ML, Ayala D, Benhamu B, Morcillo MJ, Viso A. Arylpiperazine derivatives acting at 5-HT 1A receptors. Curr Med Chem 2002;9:443-69.
- Hilbert MF, McDermott I, Middlemiss DN, Mir AK, Fozard JR. Radioligand binding study of a series of 5-HT 1A receptor agonists and definition of a steric model of this site. Eur J Med Chem 1989;24:31-7
- Mellin C, Vallgarda J, Nelson DL, Bjork YH, Anden NE, Csoregh I, et al. A 3-D Model for 5-HT 1A receptor agonists based on stereoselective methyl- substituted and conformationally restricted analogues of 8-hydroxy-2-(di-n-propylamino) tetralin. J Med Chem 1991;34:497-510
- Chidester CG, Lin CH, Lahti RA, Haadsma-Svensson SR, Smith MW. Comparison of 5-HT 1A and Dopamine Pharmacophores: X-ray crystal structure and affinities of conformationally constrained ligands. J Med Chem 1993;36:1301-5.
- Jain AN, Harris N, Park JY. Quantitative binding site model generation: Compass applied to multiple chemotypestargetting the 5-HT 1A receptor. J Med Chem 1995;38:1295-308.
- Gillard P, Carrupt P, Testa B, Schambel P. Binding of arylpiperazines, aryloxypropanolamines and tetrahydropyridylindoles to the 5-HT 1A receptor: Contribution of the molecular lipophilicity potential to three dimensional quantitative structure-affinity relationship models. J Med Chem 1996;39:126-34
- Lopez-Rodriguez ML, Rosado ML, Benhamu B. Synthesis and Structure-Activity Relationships of a New Model of Arylpiperazines. 1.2-{[4-(o-methoxyphenyl)piperazin-1-yl]methyl}-1,3-dioxoperhydroimidazo[1,5-a]pyridine: A selective 5-HT 1A receptor agonist. J Med Chem 1996;39:4439-50.
- Lopez-Rodriguez ML, Morcillo MJ, Fernandez E. Synthesis and Structure-Activity Relationships of a New Model of Arylpiperazines. 3. 1 2-[-(4-Arylpiperazin-1-yl) alkyl] perhydropyrrolo-[1,2- c ]imidazoles and -perhydroimidazo[1,5- a ]pyridines: Study of the Influence of the Terminal Amide Fragment on 5-HT 1A Affinity/Selectivity. J Med Chem 1997;40:2653-6.
- MOE. 2004. 03 (Molecular Operating Environment), Molecular Modeling System, Chemical Computing Group Inc: 1010 Sherbrooke Street West, Suite 91, Montreal, Quebec, Canada; p. H3A 2R7.
- Campillo N, Goya P, Paez JA. Novel Arylpyrazino[2,3- c ][1,2,6]thiadiazine 2,2-Dioxides as Platelet Aggregation Inhibitors. 2. Optimization by Quantitative Structure-Activity Relationships. J Med Chem 1999;42:3279-88
- Farr CD, Tabet MR, Ball Jr WJ. Three-dimensional quantitative structure-activity relationship analysis of ligand binding to human sequence antidigoxin monoclonal antibodies using comparative molecular field analysis. J Med Chem 2002;45:3257-70.
- Jaiswal M, Khadikar PV, Scozzafava A. Carbonic anhydrase inhibitors: The first QSAR study on inhibition of tumor-associated isoenzyme IX with aromatic and heterocyclic sulfonamides. Bioorg Med ChemLett 2004;14:3283-90
- Lopez-Rodriguez ML, Morcillo MJ, Fernandez E, Rosado ML, Pardo L, Schaper KJ. Synthesis and structure-activity relationships of a new model of arylpiperazines. 6. Study of the 5-HT 1A /x 1 -adrenergic receptor affinity by classical hansch analysis, artificial neural networks and computational simulation of ligand recognition. J Med Chem 2001;44:198-207.