- *Corresponding Author:
- Yang Mooi Lim
M Kandiah Faculty of Medicine and Health Sciences, Universiti Tunku Abdul Rahman, Kampar, Perak 31900, China
E-mail: ymlim@utar.edu.my
| Date of Received | 19 April 2022 |
| Date of Revision | 24 March 2023 |
| Date of Acceptance | 04 October 2023 |
| Indian J Pharm Sci 2023;85(5):1485-1503 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
In Malaysia, nasopharyngeal carcinoma is the 5th (4.0 %) most prevalent malignant tumor and it is the 3rd highest in male for cancer occurrence rate (8.4 %). Therefore, discovery of novel anti-cancer herbal drugs is of importance. In this study, the cytotoxic effect of traditional Chinese herbal prescription Xiao Xian Xiong decoction which made up of Huanglian (Coptis chinensis Franch.), Banxia (Pinellia ternata (Thunb.) Breit.), and Gualou (Trichosanthes kirilowaii Maxim) was tested. The cytotoxic effect Xiao Xian Xiong decoction was studied on 8 nasopharyngeal cancer cell lines (TWO-1, TWO-4, HONE-1, SUNE-1, CNE-2, HK-1, CNE-1 and C666-10). Global gene expression analysis was conducted using nCounter XT gene expression assay. The half-maximal inhibitory concentration values obtained for Xiao Xian Xiong decoction-treated HONE-1 and CNE-2 cell lines were 88.55 and 92.42 μg/ml respectively. Global gene expression showed Xiao Xian Xiong decoction significantly downregulated genes that are associated with cell growth and proliferation, whereas upregulated genes that are associated with induced cell cycle arrest and apoptosis in HONE-1 cells.
Keywords
Huanglian, malignant, nasopharyngeal carcinoma, Xiao Xian Xiong decoction
Nasopharyngeal Carcinoma (NPC) affects 1.5 per 100 000 individuals globally. China made up 59 % (60 558) of the world’s cases, followed by Indonesia (18 %, 17 992 cases), Vietnam (6 %, 6212 cases), India (5 %, 5086 cases), and Philippines (3 %, 2913 cases)[1]. Due to its deep-seated nature, majority of NPC patients do not detect the disease until a mass is found in lymph nodes of the neck. It was previously reported that natural plants can be a useful source of new anti-cancer agent[2]. To date, Chinese herbal medicine is conventionally being used as an adjunct to Western medical intervention[3]. Present study aims to investigate the cytotoxic effect of Xiao Xian Xiong Decoction (XXXD) which is a commonly used Chinese medicinal herb on NPC cancer cell lines.
XXXD is a Chinese medicine prescription originates from “Shang Han Lun”, a renowned Chinese medical masterpiece written by Master Zhang Zhongjing of ancient Chinese Han Dynasty (216 AD). The decoction is made up of three Chinese medicinal herbs derived from natural plants, namely Huanglian (Coptis chinensis Franch.), Banxia (Pinellia ternate (Thunb.) Breit.), and Gualou (Trichosanthes kirilowaii Maxim). Huanglian initially recorded in the Shennong Bencao Jing (Emperor Shennong’s classical medical material) and listed as one of the effective herbs[4]. Banxia, in the perspective of Traditional Chinese Medicine (TCM), is acrid, warm, and toxic in nature. It is mainly used to treat coughing, dyspnea, dizziness, nausea and vomiting, fullness in chest and palpitation caused by phlegm and fluid retention[5]. Gualou is a flowering plant, and the fruit is used to clear body heat and transform phlegm-heat, unbind the chest, reduces abscesses and dissipate nodules[6].
XXXD had been proven to be effective in eliminating heat, resolving phlegm, relieving stuffiness in the chest, and dispersing stagnancy[7]. It had been effectively used in the past centuries to treat indigestion, respiratory and cardiovascular disorders. In recent years, the clinical application of XXXD has been explored to inhibiting tumor growth and promoting body immunity[8]. Modern pharmacological studies had reported XXXD to exhibit distinctive clinical effects, such as anti- inflammation, anti-ulcer, anti-cancer and immunity enhancement. Previous in vitro studies also reported XXXD to show inhibitory effect against Ehrlich Ascites Carcinoma (EAC) and Non-Small Cell Lung Cancer (NSCLC)[9,10]. Nevertheless, the therapeutic value of XXXD on NPC has never been scientifically scrutinized.
Overall, this research comprised the preparation of aqueous extraction of individual and combination extracts of Huanglian, Banxia and Gualou. Subsequently, cytotoxicity of these herbal extracts were screened on a panel of 8 NPC cell lines, namely TWO-1, TWO-4, HONE-1, SUNE-1, CNE-2, HK-1, CNE-1 and C666-1 using 3-(4,5-Dimethylthiazol-2- yl)-2,5-Diphenyltetrazolium Bromide (MTT) assay. Global gene expression on 770 genes representing 13 canonical cancer pathways was carried out in XXXD-treated HONE-1 cell line.
Materials and Methods
Materials:
Roswell Park Memorial Institute 1640 (RPMI-1640) powder purchased from Mediatech, Inc. (Manassas, United States of America (USA)) was prepared at 10.39 g/l in ultrapure water and added with sodium bicarbonates 2 g/l. Fetal Bovine Serum (FBS) was purchased from Lonza Inc. (Allendale, New Jersey, USA). Dulbecco Modified Eagle medium (DMEM) supplemented with 4.5 g glucose/l and 300 mg/l glutamine was purchased from Hyclone Laboratories Inc. (Logan, Utah, United States). MTT reagent and Phosphate Buffer Saline (PBS) were obtained from Merck (Germany). All other chemicals were of analytical grade and commercially available.
Aqueous-extraction of the individual and combination herbs:
Chinese herbs (Huanglian, Banxia and Gualou) were bought from TCM medical hall (Tongrentang (M) Sdn. Bhd.) at Kuala Lumpur. The amount of Huanglian, Banxia and Gualou used for preparing XXXD was based on a ratio of 1:2:5 as stated in Shan Han Lun. Hence, the weight of Huanglian, Banxia, Gualou were 60 g, 120 g, 300 g respectively. All the herbs were soaked in deionized water for 30 min. Then, the herbs were boiled for 30 min in a 3 l customized boiling device. After cooling, the supernatant was collected and filtered with Whatman paper. The filtrates were lyophilized and kept in -20° prior use.
NPC cancer cell lines:
A total of 8 NPC cancer cells were used in this study. TWO-4, HONE-1, SUNE-1, CNE-2, HK-1 and C666-1 cells were cultured in RPMI-1640 medium whereas TWO-1 and CNE-1 were cultured in DMEM. All these media were supplemented with 10 % FBS. Cells were grown as monolayers and were maintained in a humidified Carbon dioxide (CO2) incubator at 37°.
Determination of optimal cell seeding concentration:
MTT assay was used to determine the optimum cell seeding density of each cell line. A total of 2.5×103, 5×103, 1×104, 2×104, 4×104 and 8×104 cells/ml were seeded in a 96-well plate and incubated for 72 h prior to cytotoxicity evaluation by MTT assay. Each well was read at 570 nm for absorbance reading. Absorbance vs. cell density graph was plotted. The optimum cell concentration was determined approximately at the middle point of exponential phase.
MTT assay:
XXXD was tested for their cytotoxicity effect on the NPC cell lines. A total of 100 µl of cells at the optimum cell concentration were seeded into each well of 96- well plate. Three wells containing 100 µl medium only were included as a negative control and three more wells containing 100 µl cell solution (untreated with herbs) served as positive control. After 24 h of incubation, each well was added with 10 µl mixture of fresh medium containing XXXD at the concentrations of 100, 50, 25, 12.5, 6.25 and 3.125 µg/ml using MTT assay and then topped up with 90 µl of medium. The cytotoxicity of the herbal extracts was determined at 72 h of incubation. The assays were conducted in triplicate and repeated in three independent experiments. The average absorbance against number of cells/ml graph was plotted and Half-Maximal Inhibitory Concentration (IC50) value was calculated.
Ribonucleic Acid (RNA) extraction:
The total RNA was extracted with Direct-zol™ RNA Miniprep Plus (Zymo Research, California, USA) following the recommendations of the manufacturer. The extracted RNA was stored at <-70°. Purity and concentration of the total RNA were determined by spectrophotometry at the 260/280 nm ratio (Nano Drop 1000, Thermo Scientific, USA) and 1 % agarose gel electrophoresis.
NanoString sample preparation:
The preparation of hybridization was carried out using nCounter XT gene expression assays (Nano string Technologies, Seattle, Washington). Initially, 70 μl of hybridization buffer was added to reporter CodeSet to create a Master Mix. Secondly, the hybridization reaction was set up by adding 5 μl of pre-prepared RNA samples to 8 μl of Master Mix to make up 13 μl of total volume in a tube. Thereafter, 2 μl of capture probe set was added to the tube before making the final volume of 15 μl. The tube was then capped, well mixed and briefly spined. Finally, a total of 12 tubes (triplicate at time point of 0, 4, 8, 12 h) were incubated in thermocycler at 65° for about 16 h. The next steps were purification and immobilization by using the nCounter MAX/FLEX system which was carried out in the nCounter Prep Station to remove excess probes and immobilize target-probe complexes onto nCounter cartridges. The cartridges were scanned on the nCounter Digital Analyzer for 2.5 h. Reporter Code Count (RCC) file was an output file generated by this nCounter instruments. One RCC file was produced for each sample tested; one file contains the barcode counts from each gene and control in the CodeSet. A Reporter Library File (RLF) is a file specific to the CodeSet. It provided nCounter instruments and the nSolver 4.0 software application with valuable information about the CodeSet, such as the assignment of probe to gene.
After importing to nSolver 4.0 software (NanoString Technologies), the RCC data files were stored under the corresponding RLF file code set on the raw data tab. All samples were normalized using the geometric mean of the housekeeping genes RBM45, SLC4A1AP, AGK, FCF1, MRPS5, DDX50, HDAC3, EIF2B4 and NUBP1. Differentially expressed genes between the treated and the untreated control cells were given as fold change expression. Gene expression data were mapped onto Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway graphs by Pathview Function of the Pan Cancer Pathways Advanced Analysis (PCPAA), providing intuitive views of regulation at the pathway level. Regulated KEGG pathways were identified via NanoString Advance Analysis software.
Results and Discussion
After having incubated for 72 h, each cell line was tested for its optimal cell density. The optimum cell seeding density of each cell line is ranging from 2.5×103 to 5×103 cells.
The IC50 values attained for cytotoxic effects of XXXD against 8 NPC cell lines are summarized in Table 1. The herbal sample XXXD only exhibited its cytotoxic effects on HONE-1 and CNE-2 with the IC50 values of 88.55 and 92.95 µg/ml respectively, whereas it did not show any cytotoxic effect upon TWO-1, C666-1, CNE- 1, HK-1, SUNE-1 and TWO-4.
| Cell lines | Concentration of XXXD (μg/ml) | IC50 (μg/ml) | ||||||
|---|---|---|---|---|---|---|---|---|
| 0 | 3.125 | 6.25 | 12.5 | 25 | 50 | 100 | ||
| TWO 1 | 100 % | 97.60 % | 99.28 % | 97.37 % | 95.33 % | 84.51 % | 69.85 % | Not found |
| C6661 | 100 % | 85.87 % | 88.81 % | 85.55 % | 76.74 % | 82.89 % | 82.51 % | Not found |
| CNE 1 | 100 % | 75.73 % | 89.86 % | 91.42 % | 81.70 % | 79.51 % | 63.02 % | Not found |
| CNE 2 | 100 % | 69.91 % | 88.21 % | 96.80 % | 71.86 % | 55.54 % | 49.00 % | 92.95 |
| HONE 1 | 100 % | 124.07 % | 109.82 % | 114.20 % | 107.10 % | 65.43 % | 45.44 % | 88.55 |
| HK1 | 100 % | 90.25 % | 84.34 % | 72.31 % | 70.01 % | 69.87 % | 75.20 % | Not found |
| SUNE 1 | 100 % | 88.02 % | 87.31 % | 89.94 % | 89.41 % | 82.02 % | 64.09 % | Not found |
| TWO 4 | 100 % | 65.88 % | 67.08 % | 73.37 % | 66.64 % | 64.43 % | 65.13 % | Not found |
Table 1: Percentage of Cell Viability at Different Concentrations (µg/ml) and IC50 values (µg/ml) Attained for the Cytotoxic Effects XXXD against 8 NPC Cell Lines
NanoString pan cancer pathways panel gene expression analysis was used to quantify transcript levels of 770 genes representing 13 canonical cancer pathways in HONE-1 cells upon treatment with XXXD. The gene expression was analyzed at 0 vs. 4 and 0 vs. 8 treatments. The total RNA extracted from HONE-1 cells at different time points were hybridized to the code set with approximately one million raw counts tallied for all genes in each sample. Raw data were processed and normalized using nSolver 4.0 following manufacturer’s guidelines.
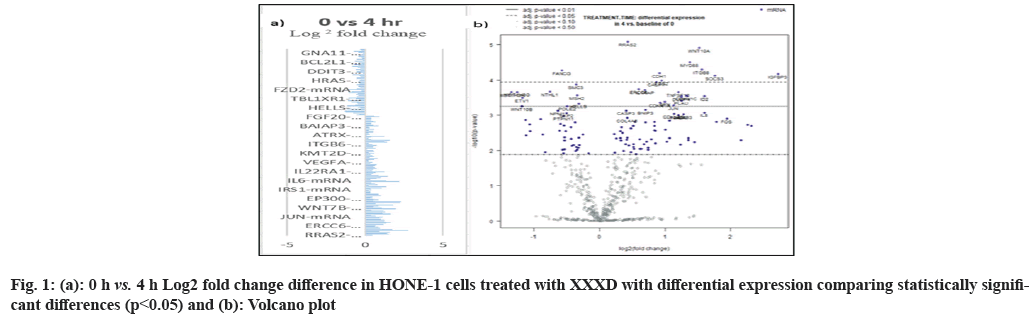
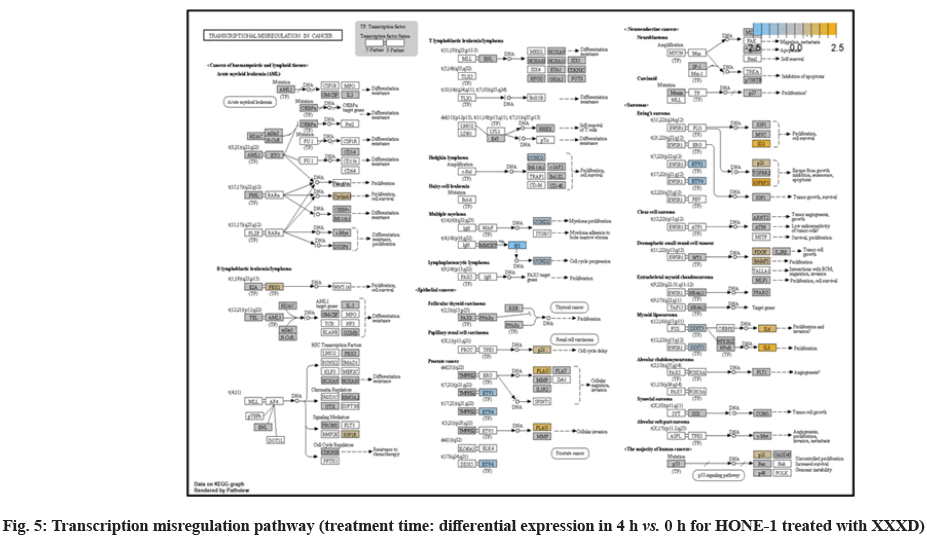
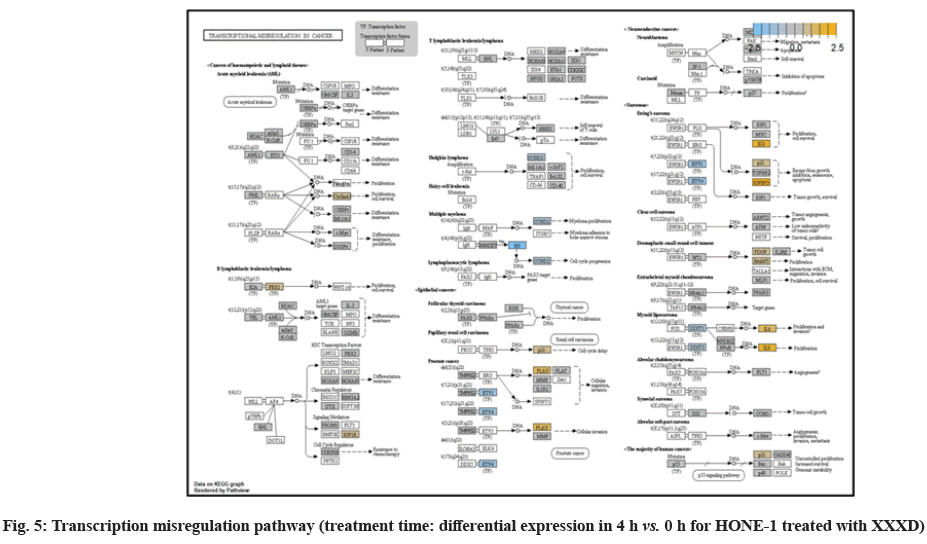
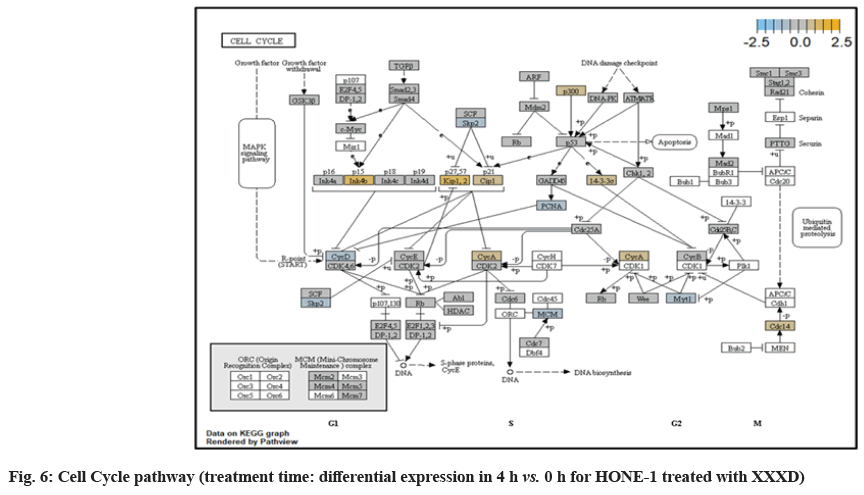
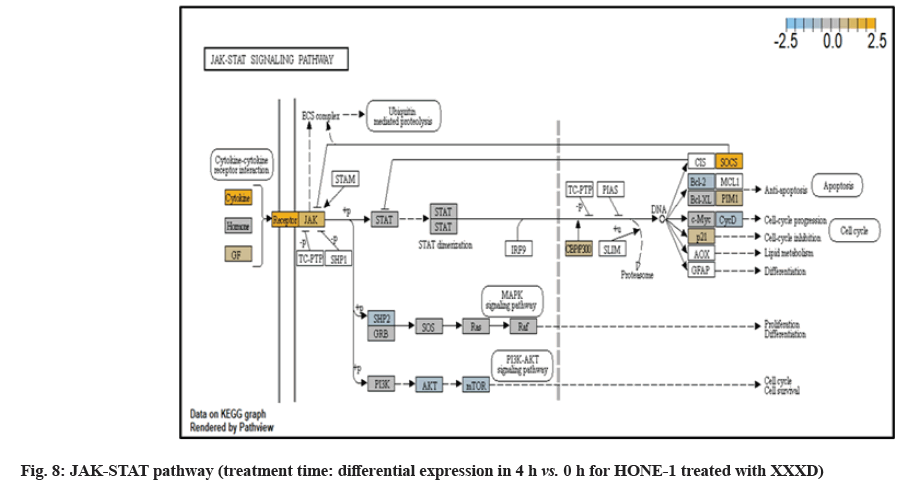
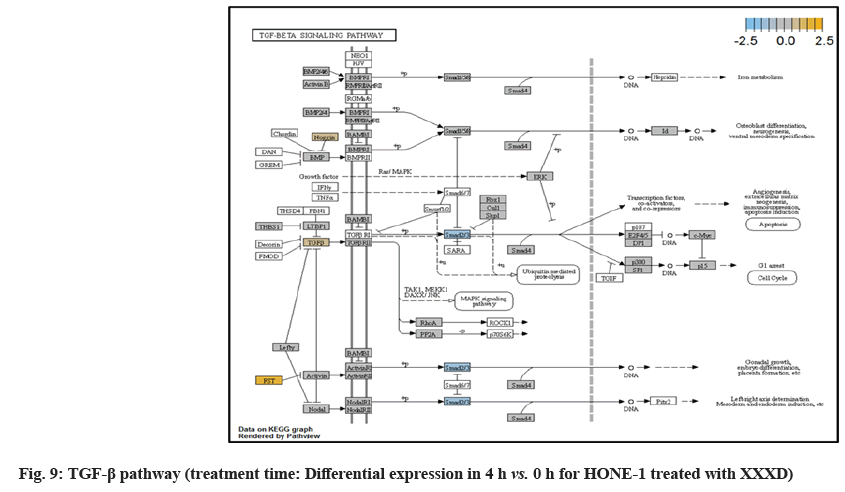
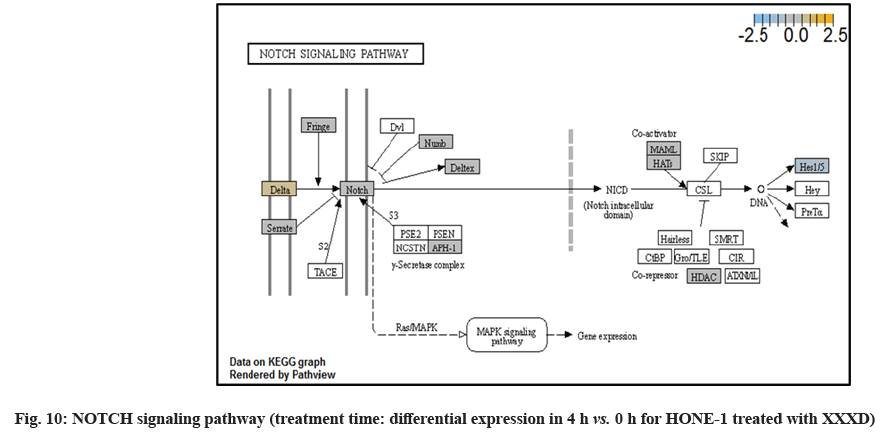
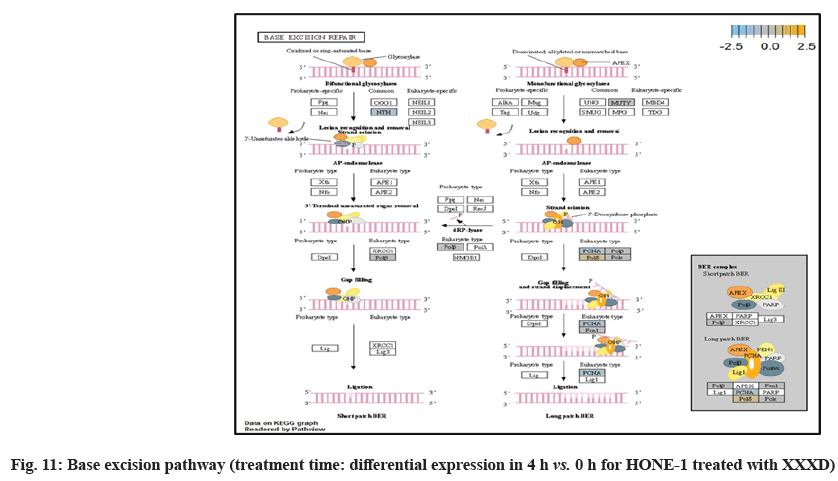
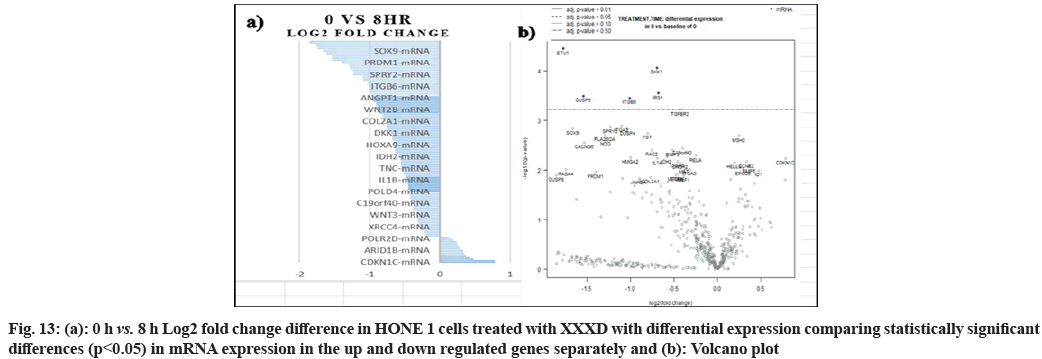
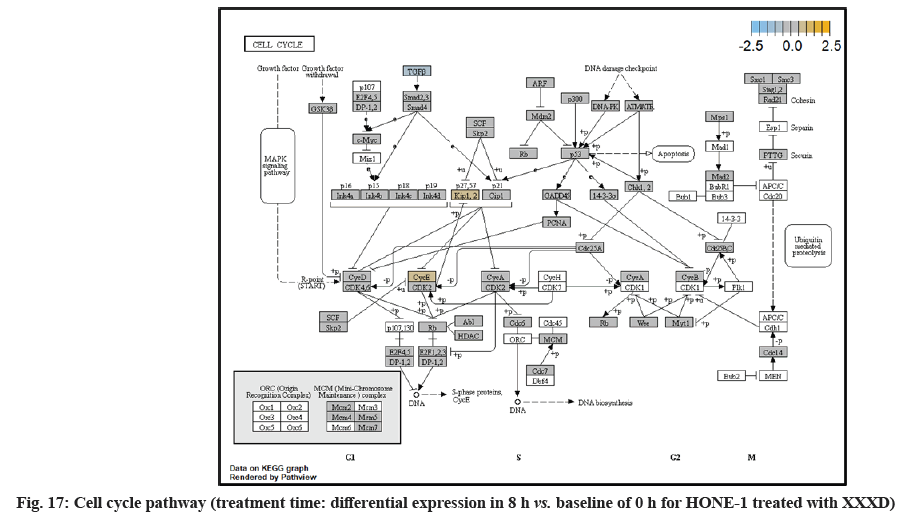
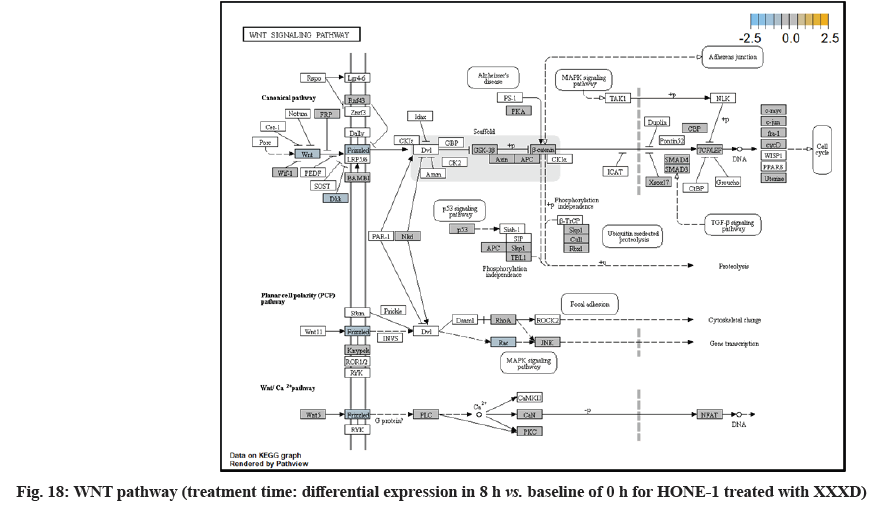
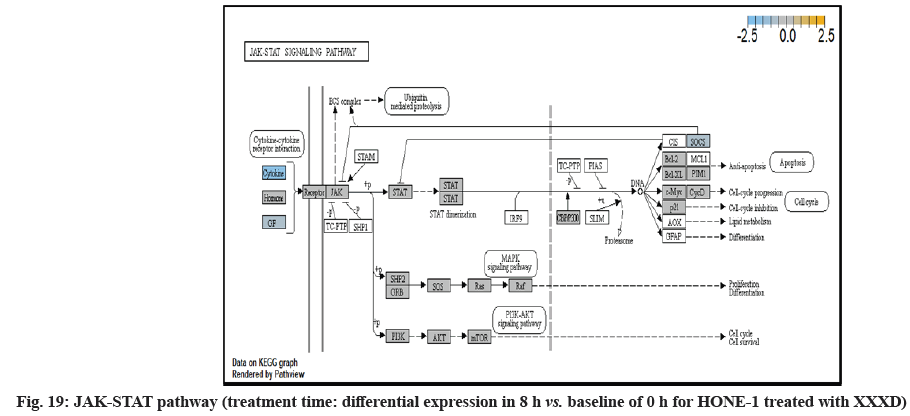
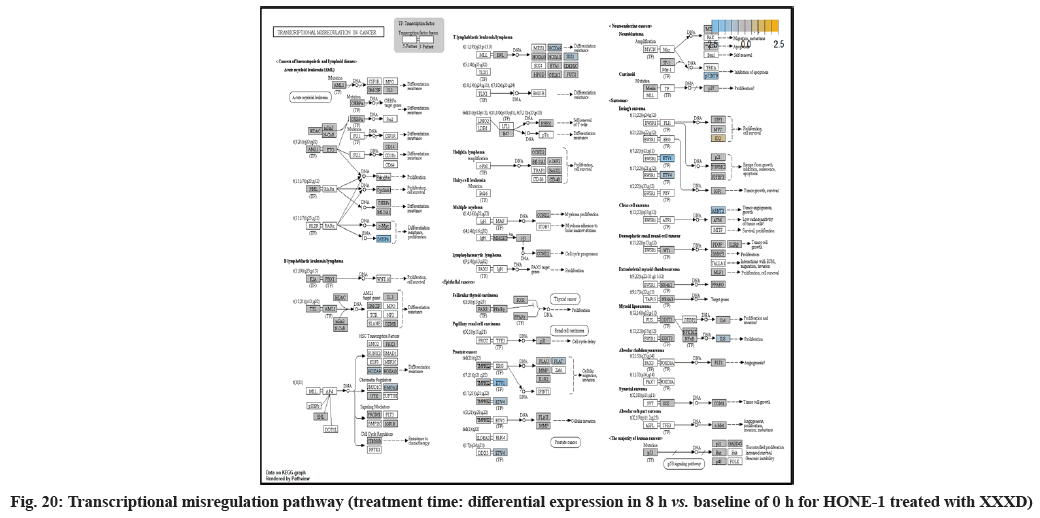
In fig. 1, shows the log2 fold change of regulated genes. Among 621 genes assayed, 119 up regulated genes represented by positive fold change whereas 66 down regulated genes with negative fold change (p<0.05); shows the volcano plot of the top 20 most significant differentially expressed genes. Table 2 shows the top 20 most significant differentially expressed genes (measured in log2 fold change) with the selected covariate at treatment time 4 h vs. 0 h. Regulation of genes at this time point were mainly initiated by signaling pathway such as Mitogen-Activated Protein Kinases (MAPK), Phosphatidylinositol 3-Kinase (PI3K), Ras, cell cycle–apoptosis, Wingless/Integrated (WNT), Janus Kinase/Signal Transducers and Activators of Transcription (JAK/STAT), Neurogenic locus notch homolog protein 1 (Notch 1), transcription misregulation, nucleotide excision and base excision. Gene expression data were mapped onto KEGG pathway graphs by path view function of the pan cancer pathway software, providing intuitive views of both up- and down-regulation at the pathway level. Representative graphs for signaling pathway are shown in fig. 2-fig. 12.
| Genes | Log2 fold change | p value | Gene sets | Biological functions |
|---|---|---|---|---|
| HIST1H3 (H3FK) | -1.35 | 0.00022 | Transcriptional misregulation | Basic nuclear proteins regulate nucleosome structure of the chromosomal fiber in eukaryotes |
| HIST1H3G (H3FH) | -1.26 | 0.00022 | Transcriptional misregulation | Basic nuclear proteins regulate nucleosome structure of the chromosomal fiber in eukaryotes |
| NTHL1 | -0.754 | 0.00022 | DNA damage- repair | Contains oxidized pyrimidine residues and has apurinic/apyrimidinic lyase activity |
| FANC | -0.576 | 5.34E-05 | DNA damage-repair | DNA repair protein for a post replication repair or a cell cycle checkpoint function |
| SMC3 | -0.36 | 0.00013 | Cell cycle-apoptosis | Cohesin complex that holds together sister chromatids during mitosis, enables proper chromosome segregation |
| MSH2 | -0.345 | 0.00028 | Driver gene | Binds to DNA mismatches thereby initiating DNA repair |
| RRAS2 (TC21) | 0.429 | 8.28E-06 | MAPK, Ras | Protein associates with the plasma membrane and may function as a signal transducer |
| ERCC6 | 0.596 | 0.00019 | DNA damage-repair | A DNA-binding protein for transcription-coupled excision repair |
| IL1RAP | 0.69 | 0.00019 | Cell cycle-apoptosis | Initiates signalling events that result in the activation of interleukin 1- responsive genes |
| CASP7 | 0.864 | 0.00011 | Cell cycle-apoptosis | Activates cascade of caspases responsible for apoptosis. |
| CDH1 | 0.915 | 6.41E-05 | Driver gene | Proteolytically generate the mature glycoprotein |
| ETS2 | 0.947 | 0.0001 | Ras | Regulates genes involved in development and apoptosis |
| TNFSF10 | 1.2 | 0.00022 | Cell cycle-apoptosis | Cytokine, induces apoptosis |
| CDKN1C | 1.33 | 0.00028 | Cell cycle- apoptosis | Inhibitor of several G1 cyclin/Cdk complexes negative regulator of cell proliferation |
| MYD88 | 1.38 | 3.18E-05 | Cell cycle-apoptosis, driver gene | Essential signal transducer in the IL-1 and Toll-like receptor signaling pathways |
| WNT10A | 1.52 | 1.23E-05 | Wnt, hedgehog | Implicated in oncogenesis including regulation of cell fate and patterning during embryogenesis |
| ITGB8 | 1.56 | 5.07E-05 | PI3K | Adhesion receptors that function in signaling from the extracellular matrix to the cell |
| ID2 | 1.6 | 0.00029 | TGF-beta, Transcriptional Misregulation | Inhibits the functions of basic helix-loop-helix ranscription factors by suppressing their heterodimerization partners through the HLH domains |
| SOCS3 | 1.76 | 7.52E-05 | JAK-STAT | Cytokine-inducible repressor for cytokine signaling, inhibit the activity of JAK2 kinase |
| IGFBP3 | 2.72 | 6.75E-05 | Transcriptional Misregulation | Changes the interaction of IGFs with their cell surface receptors. Pro- apoptotic effects mediated by its receptor TMEM219/IGFBP-3R |
Table 2: The 20 Most Significant Differentially Expressed Genes (Measured In Log2 Fold Change) at Treatment Time 4 h vs. 0 h with (XXXD)
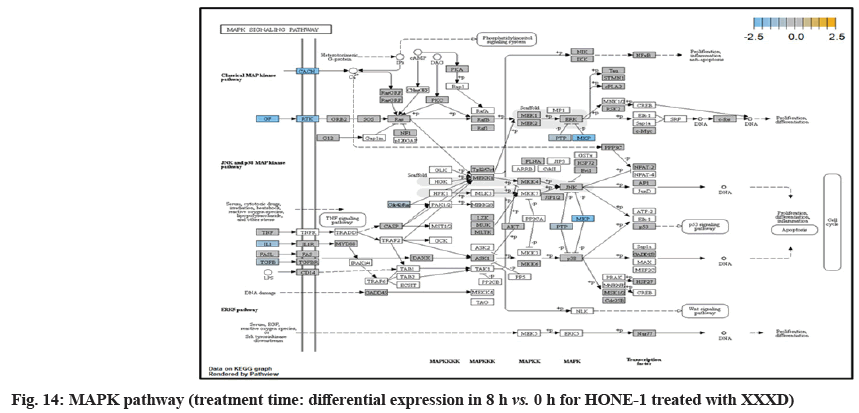
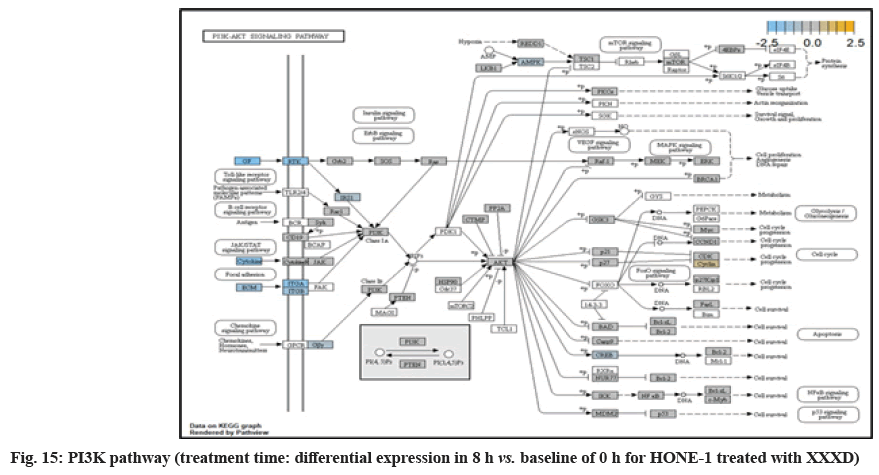
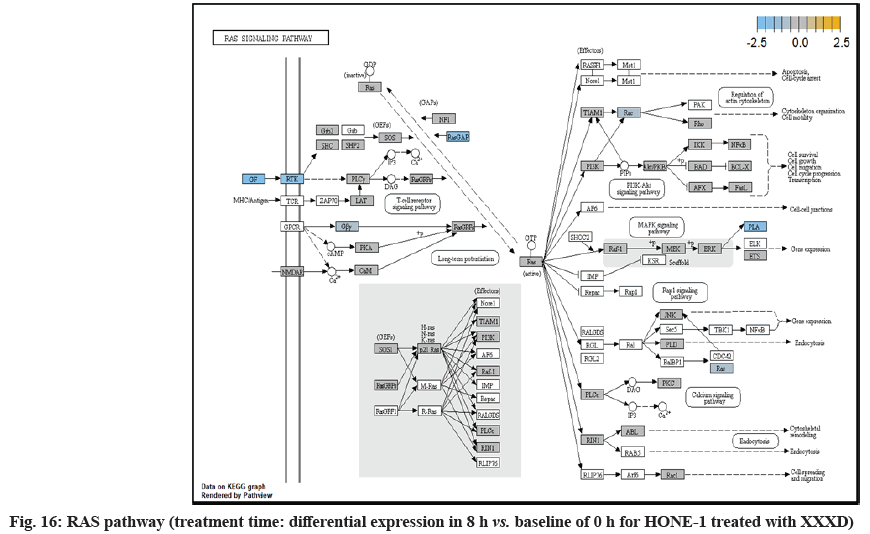
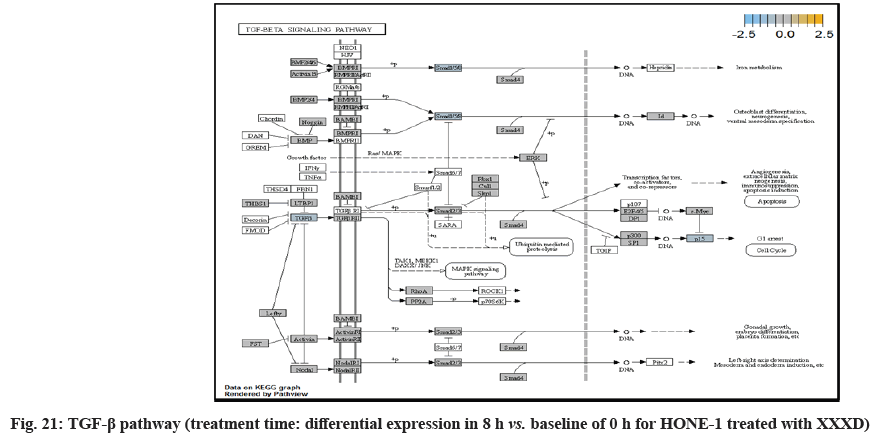
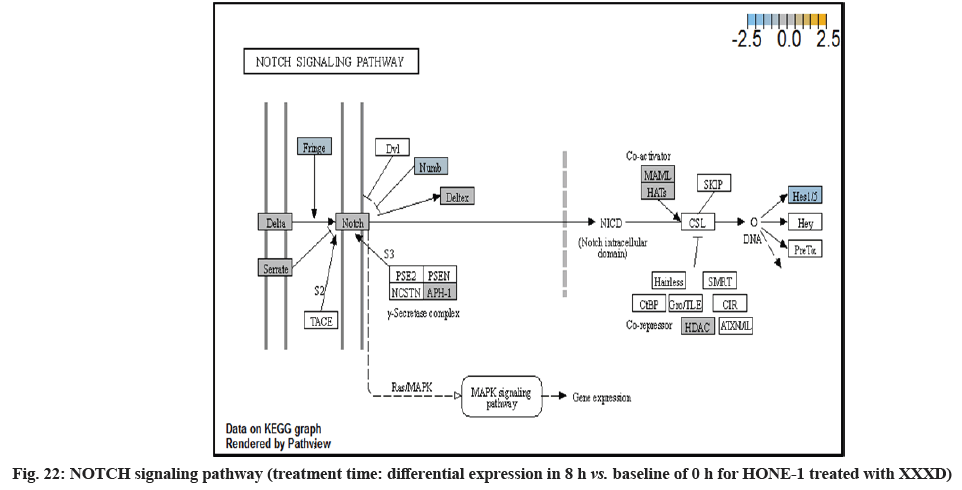
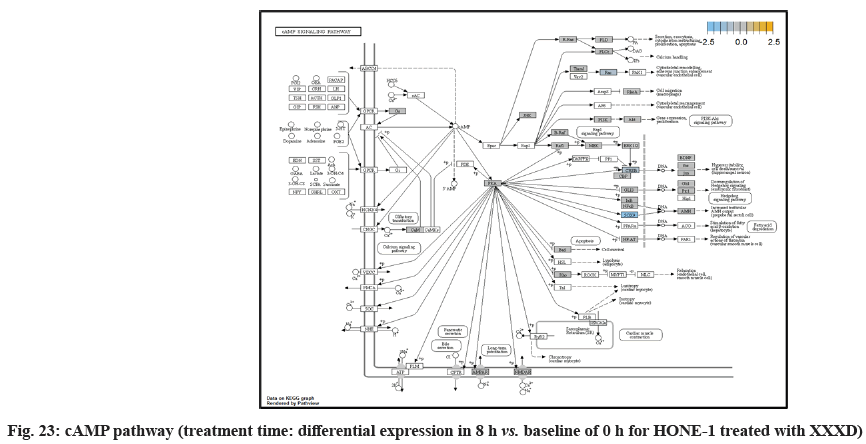
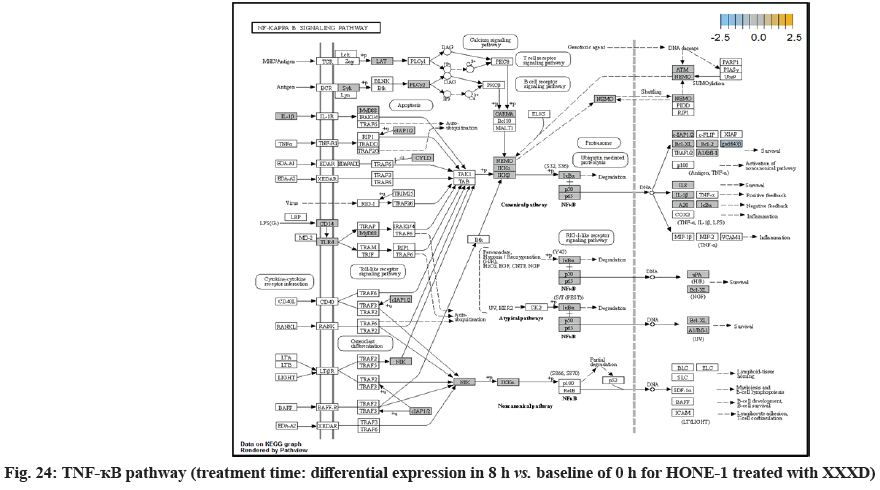
In fig. 13, shows the log2 fold change of regulated genes. Among 621 genes assayed, 18 upregulated genes represented by positive fold change whereas 134 downregulated genes with negative fold change (p<0.05); shows the volcano plot of the top 20 most significant differentially expressed genes. Table 3 shows the 20 most significant differentially expressed genes (measured in log2 fold change) with the selected covariate at treatment time 8 h vs. 0 h. Regulation of genes at this time point were mainly initiated by signaling pathway such as MAPK, PI3K, RAS, cell cycle–apoptosis, WNT, JAK-STAT, Notch, transcription misregulation, Tumor Necrosis Factor-Kappa B (TNF- ?B) and cyclic Adenosine Monophosphate (cAMP). Gene expression data were mapped onto KEGG pathway graphs by path view function of the pan cancer pathway software, providing intuitive views of both up and downregulation at the pathway level. Representative graphs for signaling pathway are shown in fig. 14 to fig. 24.
| Gene | Log2 fold change | p value | Gene set | Biological functions |
|---|---|---|---|---|
| ETV1 | -1.77 | 3.53E-05 | Transcriptional misregulation | Regulates biological processes like cell growth, angiogenesis |
| SOX9 | -1.66 | 0.00144 | Driver gene | Regulates transcription of the Anti-Muellerian Hormone (AMH) gene |
| DUSP6 (MKP3) | -1.54 | 0.00032 | MAPK, transcriptional misregulation | Inactivate their target kinases by dephosphorylating |
| CACNG6 | -1.53 | 0.00279 | MAPK | Membrane protein to stabilize the calcium channel in an inactive state |
| PLA2G2A | -1.29 | 0.00197 | Ras | Involves in the regulation of the phospholipid metabolism in bio membranes |
| NOG | -1.28 | 0.0024 | TGF-β | Secreted polypeptide binds and inactivates TGFβ signaling proteins, such as BMP4 |
| SPRY2 | -1.23 | 0.00135 | JAK-STAT | Inhibitory activity on receptor tyrosine kinase signaling proteins |
| ITGA2 | -1.1 | 0.00128 | PI3K | Adhesion receptors that function in signaling from the extracellular matrix to the cell |
| DUSP4 | -1.03 | 0.0015 | MAPK | Cellular proliferation and differentiation. |
| ITGB6 | -1.01 | 0.00035 | PI3K | Adhesion receptors that function in signaling from the extracellular matrix to the cell |
| FST | -0.801 | 0.00181 | TGF-β | Regulator of pituitary FSH secretion, inhibitor to Activin and BPM TGFβ-related growth factors |
| RAC2 | -0.752 | 0.00396 | MAPK, Ras, Wnt | Located in plasma membrane, regulate secretion, phagocytosis, and cell polarization |
| DKK1 | -0.696 | 8.53E-05 | Wnt | Embryonic, bone development. |
| IRS1 | -0.679 | 0.00028 | PI3K | Phosphorylated by insulin receptor tyrosine kinase. |
| IDH2 | -0.581 | 0.00551 | Driver gene | Enzyme found in the mitochondria, for intermediary metabolism and energy production |
| BNIP3 | -0.515 | 0.004 | Chromatin modification | Mitochondrial protein that contains a BH3 domain and acts as a pro-apoptotic factor |
| TGFBR2 | -0.425 | 0.0006 | MAPK, TGF-β, transcriptional misregulation | Receptor/ligand complex phosphorylates proteins which enter the nucleus and regulate the transcription of genes related to cell proliferation and cell cycle arrest |
| C19ORF40 | -0.403 | 0.00365 | DNA damage-repair | Involves in DNA damage response |
| RELA | -0.248 | 0.00517 | Cell cycle-apoptosis MAPK, PI3K, Ras, transcriptional misregulation | Enzyme found in the mitochondria, for intermediary metabolism and energy production |
| MSH2 | 0.245 | 0.00204 | Driver gene | Forms two different heterodimers, binds to DNA mismatches thereby initiating DNA repair |
Table 3: The 20 Most Significant Differentially Expressed Genes (Measured In Log2 Fold Change) at Treatment Time 8 h vs. 0 h with XXXD
This study was designed to uncover alterations in gene expression patterns and pathway mechanisms involved in NPC and subsequent comparison at different time points upon treated with XXXD.
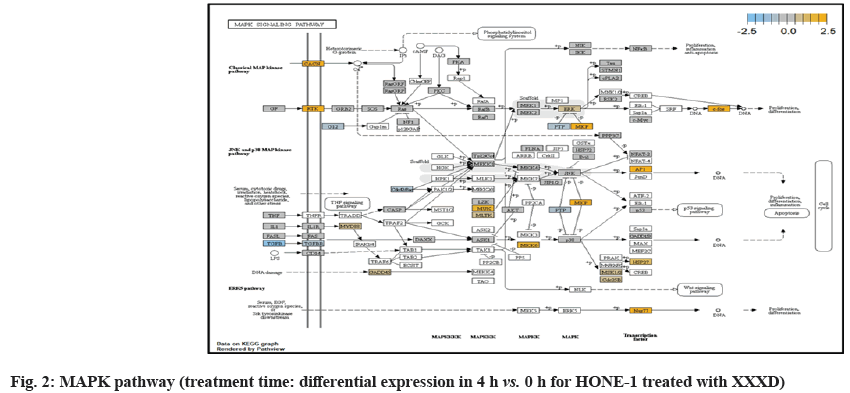
At 4 h vs. 0 h treatment time, transcriptional misregulation pathway initiated the downregulation of HIST1H3 and HIST1F3 as well as upregulation of Inhibitor of DNA Binding 2 (ID2) and Insulin- Like Growth Factor Binding Protein 3 (IGFBP-3). HIST1H3 was the most extenuatingly expressed gene for HONE-1 treated with XXXD. HIST1H3 and HIST1F3 both encoded nuclear protein H3 clustered histone which executed function of organizing of chromatin in eukaryotic cells[11]. As initiated in transcriptional misregulation pathway, it was observed in the molecular pathological pathway of multiple myeloma that H3 was deregulated. In the upstream of H3, MMSET gene formed fusion with Immunoglobulin Heavy (IgH) locus in t (4; 14) (p16; q32). MMSET possesses methyltransferase activity for core histone H3[12]. Enhanced expression of MMSET can be a potential pathogenic factor for multiple myeloma[12]. However it was detected in the path view that MMSET was in a low expression state but IgH was not present. Hence, the fusion was not formed. This may imply that XXXD could have suppressed expression of IgH and induced deregulation of H3 and Cyclin D2 (CCND2). CCND2 was responsible for progress of cell cycle. Diminished expression of CCND2 could deter cell proliferation. The elevated expression of CCND2 was particularly associated with tumorigenesis[13]. Amplified expression of CCND2 had been implicated in gastric cancer[14]. Silencing of CCND2 could decrease the development and metastasis of NSCLC. Hence, the attenuated expression of H3 and CCND2 at this time point correlated to negative regulation of cell growth in HONE-1 and might as well contributed to cell cycle arrest and apoptosis of tumor cells.
The expression of IGFBP3 was most highly amplified at this 4 h vs. 0 h treatment time. It was observed in the path view of transcriptional misregulation pathway. XXXD could have suppressed the expression of Ewing Sarcoma Breakpoint Region 1 (EWRS1) and Fatty Liver Index (FLI) and amplified expression of IGFBP-3, p21 and ID2. IGFBP3 can adjust cell development either with Insulin like Growth Factor 1 (IGF1) or independently[15].
Elevated expression of IGFBP3 induced apoptosis and increased effect of cisplatin in by deactivating IGF1 signaling[16]. The intracellular signaling via Smad2 would be triggered when IGFBP-3 combined with Transforming Growth Factor β-Receptor 2 (TGFBR2) receptors and contributed to the activation of type I TGF-β receptors. Subsequently, this pathway was associated to inhibition of proliferation. Meanwhile, P21 was unregulated. Protein p21 had the function of inhibiting cell cycle progression and often being deregulated in some tumours[17]. As expression of p21 was amplified at 4 h vs. 0 h time point, it could possibly help to suppress cell proliferation of HONE-1.
Moreover, EWRS-FLI-1 fusion will conventionally induce another set of target genes namely ID2, MYC and IGF1 for cell proliferation and survival. It was shown in the extract from fig. 17 that EWRS1 and FLI did not exist. However, ID2 was upregulated while MYC (MYC Proto-Oncogene, BHLH Transcription Factor) and IGF1 were both in quiescence state. Upregulation of ID2 could be initiated in TGFβ pathway. ID2 protein was a negative regulator of basic helix- loop-helix transcription factors. It generally repressed the cell differentiation and promotes cell proliferation in tumor[18]. Deregulation of ID2 attenuated cell invasiveness of human salivary gland cancer cells[18]. Low expression levels of ID2 enabled Glioblastoma (GBM)-derived cell lines to survive glucose deprivation and hence conduced to greater growth opportunity[19]. The elevated expression of ID2 in the 4 h vs. 0 h time point suggested that HONE-1 might struggle for greater cell survival by triggering signaling in TGFβ pathway. Nevertheless, activation of MYC proto-oncogene had been an initiation factor for human cancer occurrence[20].
IGF1 advocated on cell proliferation and differentiation as well as being a strong suppressor for apoptosis[21].
NTHL1 and FANC were deregulated in DNA damage- repair while ERCC6 was elevated in expression. The endogenous and exogenous detrimental factors can contribute Reactive Oxygen Species (ROS) which engender DNA damage[22]. The affected cells would initiate Base Excision Repair (BER) pathway to stabilize genome[23]. NTHL1 encodes DNA N-glycosylase activity to catalyze base excision repair. However, augmented expression of NTHL1 protein levels can contribute to DNA damage that caused genomic imbalance and tumorigenesis[24]. Increased level of NTHL1 protein and were found in various human cancers such as in lung, breast, prostate and pancreas[25]. The elevated expression of NTHL1 gene was noted in NSCLC[24]. The deregulation of NTHL1 in this experiment may ascertain HONE-1 cell growth was under restraint with XXXD treatment.
ERCC6 (known as CSB) encoded a DNA-binding protein which was essential for transcription-coupled excision repair and is responsible for producing Cockayne Syndrome B protein (CSB). CSB repairs damaged DNA so as to remedy with gene transcription. At 4 h vs. 0 h time point, expression of ERCC6 was elevated.
Up-regulation of Myeloid Differentiation Factor 88 (MYD88), Cyclin Dependent Kinase Inhibitor 1C (CDKNIC), Tumor Necrosis Factor Superfamily Member 10 (TNFSF10), Caspase 7 (CASP7), Interleukin-1 Receptor Accessory Protein (IL1RAP) were started off by cell cycle-apoptosis pathway except Structural Maintenance of Chromosomes 3 (SMC3) was deregulated. MYD88 was an important adaptor protein for signal transducer in the Interleukin-1 (IL- 1) and Toll-Like Receptor (TLR) signaling pathways. MYD88 triggers signaling molecules that turn on a group of interacting proteins in NF-κBwhich regulates immunity and pro-inflammation. Conventionally MYD88 will propagate signal to activate downstream Interleukin-1 (IL-1) Receptor-Associated Kinases (IRAKs)-TNF Receptor Associated Factor 6 (TRAF6) and the IKK complex to further trigger Nuclear Factor Kappa B (NF-κB) which controls the production of pro- inflammatory cytokines and type 1 Interferons (IFNs) [26]. However, it was seen in the path view that though the expression of MYD88 was amplified together with IL1R receptor (IL-1, RAP, IL1RAP), the downstream genes such as IRAKs-TRAF6 did not stimulated. The downstream pathway stopped in MYD88. The upregulation of both MYD88 and IL1R receptor in HONE-1 might be due to immunity response from interaction of cytotoxicity of XXXD. Lack of response from downstream genes might cause them to self- destruction or undergo apoptosis.
Expression of CDKNIC (also known as KIP2 or P57) was seen to have elevated as the down-regulation of its inhibitor cyclin E/Cyclin-Dependent Kinase 2 (CDK2) complex which governs the cycle arrest in G1 phase. Kinase Inhibitory Protein (KIP) 1, KIP2 and (CDK) Interacting Protein (CIP) 1 were both upregulated to suppress the amplified expression of cyclin A/CDK2 and also cyclin E/CDK2 complex. CDKNIC was known as tumor suppressor. CDKN1C was normally deregulated in many common human malignancies through several mechanisms[27]. In this experiment, CDKN1C was noted for its up-regulation and its inhibition towards cyclin/CDK complexes. It may suggest that the cell cycle of HONE-1 in G1 phase could encounter DNA damage and cell arrest under the interaction of XXXD treatment.
TNFSF10 (also known as Trail) protein was a cytokine that belongs to the TNF ligand family[28]. At 4 h vs. 0 h, TNFSF10 was highly expressed and bind with D1, D2, D4 and D5 which are called death receptor. The augmented expression of Trail in HONE-1 may due to the irritation of XXXD interaction at the first 4 h. TNFSF10 was well known as tumors suppressor and induces cell apoptosis in cancer[29]. Moreover, TNFSF10 alone or integrated with chemotherapeutics had exhibited favorable anticancer result by mediating apoptosis[30].
The upregulation of caspase-7 envisaged the execution of apoptosis of HONE-1 in mitochondrial pathway. Meanwhile, expression of the anti-apoptosis protein B-cell lymphoma 2 (Bcl-2) was attenuated and caspase-9 stimulated expression of caspase-7.
SMC3 is a component of the multimeric cohesion complex. Its main function is to ensure normal chromosome segregation by gripping sister chromatids during mitosis. At 4 h vs. 0 h time point, SMC3 was deactivated with low expression initiated in cell cycle pathway. This implied an adverse impact for HONE-1 to proliferate through a normal cell cycle progression.
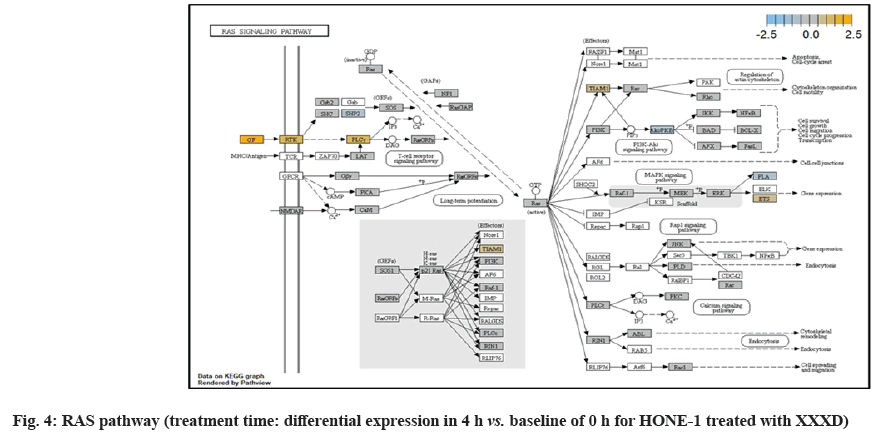
ETS2 was mildly upregulated in RAS pathway. It is a transcriptional factor to translocate signal for cell proliferation, apoptosis and survival in the thymus[31]. As RAS associated with MAPK signaling pathway was often initiated for cellular growth and development, the deactivation of MAPK cascade was seen in fig. 16. This may suggest that HONE-1 could encounter cellular stress from intervention of XXXD treatment. The upregulation of ETS2 could mean to transcribe signal for cell apoptosis.
WNT10A was an important ligand governing adult epithelial development as well as region-specific differentiation, and modulate activation downstream β-catenin pathway[32]. WNT10A protein had been implicated in oncogenesis during developmental processes. Etiologic for Tooth Agenesis (TA) were often been accounted for WNT10A mutations[33]. At 4 h vs. 0 h time point, WNT10A was upregulated but expression of its receptor namely frizzled was attenuated. Subsequently, no downstream genes were stimulated and thus contributed to deactivation of beta (β)-catenin pathway.
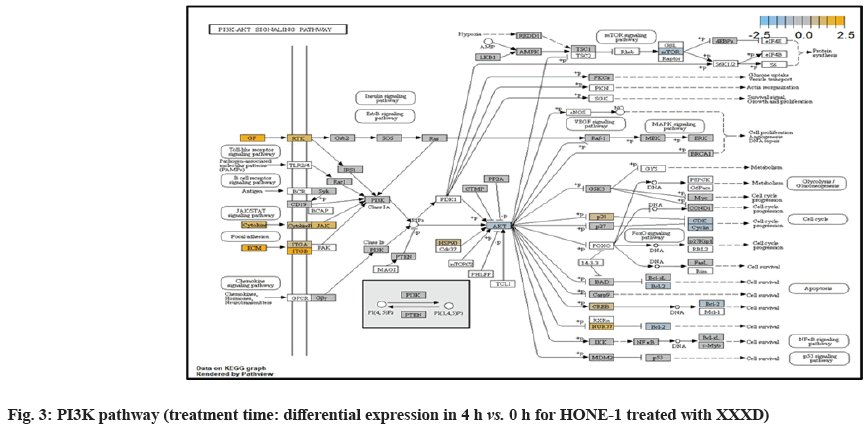
PI3K pathway initiated upregulation of ITGB8 receptor and the signaling molecule Extracellular Matrix (ECM). Nevertheless, the cellular activator molecule was not seen in the path view; resulting in the absence of the signal being triggered towards PI3K and attenuation of cell survival and proliferation of HONE-1. It may suggest that HONE-1 could encounter considerable cellular stress from the treatment of XXXD.
In JAK-STAT pathway, expression of cytokine ligand and the receptor was elevated. Meanwhile, Suppressor of Cytokine Signalling 1 (SOCS1) was in higher level of expression than JAK kinase. SOCS1 inhibited target kinase JAK by recruiting the E3 ubiquitin ligase complex. The E3 complex polyubiquitinated and degraded JAK by proteasome as well as preventing the kinase JAK from phosphorylating its cytokine receptor. The suppressed JAK kinase could not actively transduce signal to its canonical targets STAT. Consequently, STAT dimers translocated signal to the nucleus and induced the attenuated expression of cell cycle progression promoter CCND2 and pro-survival BCL-2, B-cell lymphoma-Extra Large (Bcl-XL) and augmented the expression of cell cycle progression inhibitor p21. It could be postulated that HONE-1 could have adopted an autophagy process.
In summary, as for cellular sustainability of HONE-1 at 4 h vs. 0 h time point, it was observed that HONE- 1 confronted with huge detrimental impact from the XXXD treatment. This can be witnessed from the level of expression in majority genes being induced to perform primarily on inhibition in cell proliferation and to advocate cell differentiation and apoptosis in HONE-1. Particularly in cell cycle pathway, high level expression of CDKN1C, P21 and TNFS10 protein conduced to greater cell cycle arrest and apoptosis whereas deregulation of CCND2 and pro-survival BCL-2, Bcl-XL protein eventually decreased the survival rate of HONE-1. Meanwhile, signaling ligand receptors for promoting growth such as WNT10A and frizzled in WNT pathway as well as ECM and ITGD8 of PI3K pathways, ID2 and MYD88 in MAPK pathway were highly activated, however, the transduction of signal by these genes within cytoplasmic downstream were mostly deregulated. This may suggest that HONE- 1 could endure intensive cellular stress from interaction of XXXD during 4 h vs. 0 h period.
At 8 h vs. 0 h time point, transcriptional misregulation pathway initiated the down-regulation of ETV1, SOX9, TGFBR2 and RELA. ETV1 was the most downregulated gene at 8 h vs. 0 h treatment time for HONE-1 treated with XXXD.
ETV1 belonged to the ETS family of transcription factors. Often associated with MAPK, PI3K pathways, ETV1 protein modulated biological processes like cell growth, angiogenesis; the EWSR1-ETV1 fusion proteins were thought to be the "oncogenic drivers" in Ewing sarcoma. In fig. 20, EWSR1 was detected absent in EWSR1/ETV1 fusion. Hence, this may imply that XXXD could had suppressed expression of EWSR1 and induced low expression of ETV1 and ETV 4. Conventionally, EWSR1/ETV1 and EWSR1/ETV4 in association with EWSR1/FEV tended to exert its influence for neoplasm localization[34]. At 8 h vs. 0 h treatment time, ETV1 as well as ETV4 were deregulated. It could be postulated that the pathogenesis for HONE-1 cell growth and proliferation was reasonably degraded due to the deactivation of biological functions of these two proteins.
Both transcriptional misregulation and MAPK pathway initiated the down-regulation of Dual Specificity Phosphatase 6 (DUSP6), also known as MKP-3 in MAPK pathway. MKP-3 and MKP-2 (DUSPA 4) deactivates its target kinases ERK, JNK and P38 by dephosphorylation which contributed to reduction of cellular proliferation and differentiation of HONE-1.
Calcium Voltage-Gated Channel Auxiliary Subunit Gamma 6 (CACNG6) was an integral membrane protein to stabilize the calcium channel in an inactive (closed) state. The Calcium (Ca2+) concentration inside the cell will influence expression of genes which are important for neuronal development. The inflow of Ca2+ into L-type Calcium Channel (LTC) can effectively activate transcription factors such as cAMP Response Element Binding factor (CREB) and Myocyte-Enhancing Factor 2 (MEF2). Binding of Ca2+-Calmodulin (CaM) was important to transfer the Ca2+ signal to the nucleus and is essential for activating RAS/MAPK pathway and stimulate the genes necessary for neuronal survival and plasticity purposes[35]. At 8 h vs. 0 h treatment time, it was shown that expression of CACNG6 was attenuated in MAPK pathway which regarded signaling to nucleus by an L-type calcium channel–calmodulin complex via RAS and MAPK cascade.
Insulin Receptor Substrate 1 (IRS1) modulated biological processes such as proliferation and survival. IRS1 was an intracellular signaling adaptor protein particularly for insulin hormone ligand. It was envisaged that IRS1 was deregulated together with the growth factor ligand in MAPK pathway which conferred to deactivation of downstream genes.
Down-regulation of PLA2A2G and RAC2 were initiated in RAS pathway. PLA2s are commonly found in nature either as an intracellular or extracellular enzymes[36]. Augmented expression of PLA2G2A has been noted to implicate in various types of malignant tumour such as pancreatic cancer and prostate cancer[37,38]. In RAS pathway, ERK kinase deregulated the transcription factor PLA2 and hence, conferred the degradation of cell proliferation in HONE-1. RAC2 was a member of Rho family of GTPases protein and localized to the plasma membrane. It modulated cell growth, cytoskeletal reorganization, and the activation of protein kinases. The attenuated expression of RAC2 might deactivate RAS as well.
ITGA2 and ITGA6 receptors as well as the signaling molecule ECM were downregulated in the PI3K pathway. It was envisaged that the cellular activator FAK did not respond to receptor and triggered no signal towards PI3K downstream.
Dickkopf-1 (DKK1) was the suppressor for receptor frizzled of WNT pathway. At 8 h vs. 0 h treatment time, WNT pathway initiated the downregulation of DKK1, ligand WNT10A, frizzled receptor together. Subsequently, no downstream genes was stimulated and appeared in the path view conferring to deactivation of β-catenin pathway.
TGFβ pathway initiated attenuation of expression of Noggin (Nog), FST and TGFBR2. NOG and FST are the inhibitor to ligand Bone Morphogenetic Protein (BMP) and activating in TGFβ pathway respectively. As shown in the path view, expression of NOG and FST were elevated in 4 h vs. 0 h time point but attenuated at 8 h vs. 0 h. The attenuated expression might correlate with the frail expression of extracellular signaling molecules during XXXD treatment. Meanwhile, TGFBR2 receptor was deregulated together with the TGFβ ligand too.
The IDH2 encoded protein for producing enzyme called isocitrate dehydrogenase 2 which was found in mitochondria and control intermediary metabolism and energy production. The decreasing expression of IDH2 may implicate that HONE-1 experienced intracellular stress.
BCL-2 Interacting Protein 3 (BNIP3) encoded mitochondrial protein that acted as a pro-apoptotic factor. Expression of BNIP3 was attenuated and expressed with negative log2fold change -0.515. Chromosome 19 Open Reading Frame 40 (C19ORF40) (FA Core Complex Associated Protein 24 (FAAP24)) functioned in DNA repair through recruitment of the Fanconi anemia core complex to damage DNA. The expression was attenuated and expressed with negative log2 fold change -0.403. The downregulation of BNIP3 and C19ORF40 may correlate with the apoptosis resistance of HONE-1 throughout the 8 h treatment time.
RELA (also known as p65, NF-κB) was a transcription factor to regulate mainly for cell development and apoptosis, inflammation and immunity. RELA (p65) formed a heterodimeric complex with the p50 dimers bind at kappa-B sites in the DNA of their target genes and the individual dimers had distinct preferences for different κB sites that they could bind with distinguishable affinity and specificity. Different dimer combinations acted as transcriptional activators or repressors, respectively. RELA (p65) was downregulated and expressed with negative log2fold change -0.248. The deactivation of RELA subsequently did not induce activation of urokinase-type Plasminogen Activator (uPA) and Bcl-XL in downstream. uPA, also known as PLAU, and pro-survival Bcl-XL protein was not observed for being regulated. This could further decrease the viability of HONE-1 due to intervention of XXXD. High level expression of uPA often associates with invasive tumour growth and poor prognosis particularly in breast cancer[39]. MutS Homolog 2 (MSH2) was a tumour suppressor gene and more specifically a caretaker gene that codes for a DNA Mismatch Repair (MMR) protein, MSH2, which forms a heterodimer with MSH6 to make the human MutSα MMR complex. It also dimerized with MSH3 to form the MutSβ DNA repair complex. Recently, there were reports on cases of deficiency of MMR protein, MSH2 correlated with prostate cancer in a Japanese patient[40]. In addition, MSH2 deficiency may cause breast cancer tumorigenesis and accelerate development and progression[41]. Expression of MSH2 was elevated at this time point suggest that XXXD treatment had induced cytotoxicity effect towards HONE-1.
At 8 h vs. 0 h treatment time, it could be summarized that XXXD continued to exert its cytotoxicity effect on HONE-1. This again can be witnessed from down- regulation of a number of genes which could instigate cell growth and proliferation, particularly like ETV1, SOX, DUSPA6, CACNG6, IRS1 and IDH2. In addition, many growth factor ligands and receptors were upregulated but the intracellular downstream genes either being knockdown or not active, resulting in the transduction of growth signal towards transcription factors in vain or frail. Evidence gathered is GF, RTK and IRS1 as well as ECM, ITGA and ITGB which were initiated in PI3K pathway; DKKI and frizzled which were initiated in WNT pathway. This may suggest that XXXD is imposed considerably suppression for cell proliferation in HONE-1 through expression of above mentioned genes associated with their related pathways.
In view of the gene expression of HONE-1 when treated with XXXD at 2 time points within 8 h, majority of genes were induced to inhibit cell proliferation and cause apoptosis in HONE-1. It was observed that expression of ETV1 again was depreciated at 3 different time points such as 4 h vs. 0 h, 8 h vs. 0 h and the log2 fold change value for differential expression were -1.18 (p<0.05), -1.77 (p<0.05) respectively. Thereupon, these findings suggested that ETV1 could be targeted as the core gene for therapeutic purpose in treating NPC particularly HONE-1. ETV1 was often implicated in melanomas, breast and other types of cancer[42]. Overexpression of ETV1 associated with MAPKs instigated invasive tumour growth in OE33 oesophageal squamous cancer[43]. Hence, methods such as regulation of its expression, interaction with cofactors or inhibiting its crucial target gene products could consequently be utilized to treat cancer (including NPC) which was dependent on the PEA3 group of ETS transcription factors[44].
Secondary metabolites abundantly found in XXXD can justify its anti-cancer effect. This study justified the potential effect of XXXD in inducing cytotoxicity on HONE-1 cell line. This is a new attempt to study the effect of XXXD for the total gene expression on HONE-1 cells and demonstrated that both XXXD was able to regulate the expression of the key genes associated with MAPK, PI3K, RAS, WNT, TGFβ, JAK-STAT, transcription misregulation and NF-?B signaling pathway in HONE-1 cells. Nevertheless, XXXD induced attenuation or amplification in expression of the related genes, theoretically, impeded the carcinogenesis progression of HONE-1 cells. In addition, this study suggests that ETV1 could be regarded as the target gene for therapeutic biomarker in treating NPC particularly HONE-1. More functional assay should be conducted to further support this finding.
Funding:
This study “A Multiplexed Gene Expression Study on NPC treated with Xiao Xian Xiong Decoction” in 2017 supported by Universiti Tunku Abdul Rahman Research Fund (UTARRF) (IPSR/RMC/ UTARRF/2017-C1-W01).
Conflict of interests:
The authors declared no conflict of interests.
References
- Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Pineros M, et al. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer 2019;144(8):1941-53.
[Crossref] [Google Scholar] [PubMed]
- Greenwell M, Rahman PK. Medicinal plants: Their use in anticancer treatment. Int J Pharm Sci Res 2015;6(10):4103.
[Crossref] [Google Scholar] [PubMed]
- Liu C, Huang Y. Chinese herbal medicine on cardiovascular diseases and the mechanisms of action. Front Pharmacol 2016;7:469.
[Crossref] [Google Scholar] [PubMed]
- Scheid V, Bensky D, Ellis A, Barolet R. Chinese herbal medicine: Formulas and strategies. 2nd ed. Eastland press; 2009.
- Su T, Tan Y, Tsui MS, Yi H, Fu XQ, Li T, et al. Metabolomics reveals the mechanisms for the cardiotoxicity of Pinelliae Rhizoma and the toxicity-reducing effect of processing. Sci Rep 2016;6(1):34692.
[Crossref] [Google Scholar] [PubMed]
- Yu X, Tang L, Wu H, Zhang X, Luo H, Guo R, et al. Trichosanthes fructus: Botany, traditional uses, phytochemistry and pharmacology. J Ethnopharmacol 2018;224:177-94.
- Pang B, Yu XT, Zhou Q, Zhao TY, Wang H, Gu CJ, et al. Effect of Rhizoma coptidis (Huang Lian) on treating diabetes mellitus. Evid Based Complement Alternat Med 2015;2015:921416.
[Crossref] [Google Scholar] [PubMed]
- Han R, Ye JX, Quan LH, Liu CY, Yang M, Liao YH. Evaluating pulmonary toxicity of Shuang–Huang–Lian in vitro and in vivo. J Ethnopharm 2011;135(2):522-9.
[Crossref] [Google Scholar] [PubMed]
- Chen YH, Liu XW, Huang JL, Baloch S, Xu X, Pei XF. Microbial diversity and chemical analysis of Shuidouchi, traditional Chinese fermented soybean. Food Res Int 2019;116:1289-97.
- Wu B, Liu M, Liu H, Li W, Tan S, Zhang S, et al. Meta-analysis of traditional Chinese patent medicine for ischemic stroke. Stroke 2007;38(6):1973-9.
- Bhasin M, Reinherz EL, Reche PA. Recognition and classification of histones using support vector machine. J Comput Biol 2006;13(1):102-12.
[Crossref] [Google Scholar] [PubMed]
- Marango J, Shimoyama M, Nishio H, Meyer JA, Min DJ, Sirulnik A, et al. The MMSET protein is a histone methyltransferase with characteristics of a transcriptional corepressor. Blood 2008;111(6):3145-54.
[Crossref] [Google Scholar] [PubMed]
- Santarius T, Shipley J, Brewer D, Stratton MR, Cooper CS. A census of amplified and overexpressed human cancer genes. Nat Rev Cancer 2010;10(1):59-64.
[Crossref] [Google Scholar] [PubMed]
- Takano Y, Kato Y, van Diest PJ, Masuda M, Mitomi H, Okayasu I. Cyclin D2 overexpression and lack of p27 correlate positively and cyclin E inversely with a poor prognosis in gastric cancer cases. Am J Pathol 2000;156(2):585-94.
[Crossref] [Google Scholar] [PubMed]
- Varma Shrivastav S, Bhardwaj A, Pathak KA, Shrivastav A. Insulin-like growth factor binding protein-3 (IGFBP-3): Unraveling the role in mediating IGF-independent effects within the cell. Front Cell Dev Biol 2020;8:286.
[Crossref] [Google Scholar] [PubMed]
- Wang YA, Sun Y, Palmer J, Solomides C, Huang LC, Shyr Y, et al. IGFBP3 modulates lung tumorigenesis and cell growth through IGF1 signaling. Mol Cancer Res 2017;15(7):896-904.
- Shamloo B, Usluer S. p21 in cancer research. Cancers 2019;11(8):1178.
[Crossref] [Google Scholar] [PubMed]
- Sumida T, Ishikawa A, Nakano H, Yamada T, Mori Y, Desprez PY. Targeting ID2 expression triggers a more differentiated phenotype and reduces aggressiveness in human salivary gland cancer cells. Genes Cells 2016;21(8):915-20.
[Crossref] [Google Scholar] [PubMed]
- Zhang Z, Rahme GJ, Chatterjee PD, Havrda MC, Israel MA. ID2 promotes survival of glioblastoma cells during metabolic stress by regulating mitochondrial function. Cell Death Dis 2017;8(2):e2615.
[Crossref] [Google Scholar] [PubMed]
- Gabay M, Li Y, Felsher DW. MYC activation is a hallmark of cancer initiation and maintenance. Cold Spring Harb Perspect Med 2014;4(6):a014241.
[Crossref] [Google Scholar] [PubMed]
- Furstenberger G, Senn HJ. Insulin-like growth factors and cancer. Lancet Oncol 2002;3(5):298-302.
[Crossref] [Google Scholar] [PubMed]
- Bauer NC, Corbett AH, Doetsch PW. The current state of eukaryotic DNA base damage and repair. Nucleic Acids Res 2015;43(21):10083-101.
[Crossref] [Google Scholar] [PubMed]
- Maynard S, Schurman SH, Harboe C, de Souza-Pinto NC, Bohr VA. Base excision repair of oxidative DNA damage and association with cancer and aging. Carcinogenesis 2009;30(1):2-10.
[Crossref] [Google Scholar] [PubMed]
- Limpose KL, Trego KS, Li Z, Leung SW, Sarker AH, Shah JA, et al. Overexpression of the base excision repair NTHL1 glycosylase causes genomic instability and early cellular hallmarks of cancer. Nucleic Acids Res 2018;46(9):4515-32.
[Crossref] [Google Scholar] [PubMed]
- Albertson DG. Gene amplification in cancer. Trends Genet 2006;22(8):447-55.
[Crossref] [Google Scholar] [PubMed]
- Saikh KU. MyD88 and beyond: A perspective on MyD88-targeted therapeutic approach for modulation of host immunity. Immunol Res 2021;69:117-28.
[Crossref] [Google Scholar] [PubMed]
- Pateras IS, Apostolopoulou K, Niforou K, Kotsinas A, Gorgoulis VG. p57KIP2: Kiping the cell under control. Mol Cancer Res 2009;7(12):1902-19.
[Crossref] [Google Scholar] [PubMed]
- Starkey MR, Nguyen DH, Essilfie AT, Kim RY, Hatchwell LM, Collison AM, et al. Tumor necrosis factor-related apoptosis-inducing ligand translates neonatal respiratory infection into chronic lung disease. Mucosal Immunol 2014;7(3):478-88.
- He W, Wang Q, Xu J, Xu X, Padilla MT, Ren G, et al. Attenuation of TNFSF10/TRAIL-induced apoptosis by an autophagic survival pathway involving TRAF2-and RIPK1/RIP1-mediated MAPK8/JNK activation. Autophagy 2012;8(12):1811-21.
[Crossref] [Google Scholar] [PubMed]
- Hellwig CT, Rehm M. TRAIL signaling and synergy mechanisms used in TRAIL-based combination therapies. Mol Cancer Ther 2012;11(1):3-13.
[Crossref] [Google Scholar] [PubMed]
- Zaldumbide A, Carlotti F, Pognonec P, Boulukos KE. The role of the Ets2 transcription factor in the proliferation, maturation and survival of mouse thymocytes. J Immunol 2002;169(9):4873-81.
[Crossref] [Google Scholar] [PubMed]
- Xu Z, Yan Y, Xiao L, Dai S, Zeng S, Qian L, et al. Radiosensitizing effect of diosmetin on radioresistant lung cancer cells via Akt signaling pathway. PloS One 2017;12(4):e0175977.
- Zeng Y, Baugh E, Akyalcin S, Letra A. Functional effects of WNT10A rare variants associated with tooth agenesis. J Dental Res 2021;100(3):302-9.
[Crossref] [Google Scholar] [PubMed]
- Im YH, Kim HT, Lee C, Poulin D, Welford S, Sorensen PH, et al. EWS-FLI1, EWS-ERG, and EWS-ETV1 oncoproteins of Ewing tumor family all suppress transcription of transforming growth factor β type II receptor gene. Cancer Res 2000;60(6):1536-40.
[Google Scholar] [PubMed]
- Dolmetsch RE, Pajvani U, Fife K, Spotts JM, Greenberg ME. Signaling to the nucleus by an L-type calcium channel-calmodulin complex through the MAP kinase pathway. Science 2001;294(5541):333-9.
[Crossref] [Google Scholar] [PubMed]
- Six DA, Dennis EA. The expanding superfamily of phospholipase A2 enzymes: Classification and characterization. Biochim Biophysica Acta 2000;1488(1-2):1-9.
[Crossref] [Google Scholar] [PubMed]
- Kashiwagi M, Friess H, Uhl W, Berberat P, Abou-Shady M, Martignoni M, et al. Group II and IV phospholipase A2 are produced in human pancreatic cancer cells and influence prognosis. Gut 1999;45(4):605-12.
- Jiang J, Neubauer BL, Graff JR, Chedid M, Thomas JE, Roehm NW, et al. Expression of group IIA secretory phospholipase A2 is elevated in prostatic intraepithelial neoplasia and adenocarcinoma. Am J Pathol 2002;160(2):667-71.
[Crossref] [Google Scholar] [PubMed]
- Banys-Paluchowski M, Witzel I, Aktas B, Fasching PA, Hartkopf A, Janni W, et al. The prognostic relevance of urokinase-type plasminogen activator (uPA) in the blood of patients with metastatic breast cancer. Sci Rep 2019;9(1):2318.
- Kagawa M, Kawakami S, Yamamoto A, Suzuki O, Eguchi H, Okazaki Y, et al. Prevalence and clinicopathological/molecular characteristics of mismatch repair protein-deficient tumours among surgically treated patients with prostate cancer in a Japanese hospital-based population. Japan J Clin Oncol 2021;51(4):639-45.
[Crossref] [Google Scholar] [PubMed]
- Malik SS, Mubarik S, Aftab A, Khan R, Masood N, Asif M, et al. Correlation of MSH2 exonic deletions and protein downregulation with breast cancer biomarkers and outcome in Pakistani women/patients. Environ Sci Pollut Res 2021;28:3066-77.
[Crossref] [Google Scholar] [PubMed]
- Oh S, Shin S, Janknecht R. ETV1, 4 and 5: An oncogenic subfamily of ETS transcription factors. Biochim Biophys Acta 2012;1826(1):1-2.
[Crossref] [Google Scholar] [PubMed]
- Keld R, Guo B, Downey P, Gulmann C, Ang YS, Sharrocks AD. The ERK MAP kinase-PEA3/ETV4-MMP-1 axis is operative in oesophageal adenocarcinoma. Mol Cancer 2010;9(1):1-4.
- Prucca CG, Racca AC, Velazquez FN, Cardozo Gizzi AM, Rodríguez Berdini L, Caputto BL. Impairing activation of phospholipid synthesis by c-Fos interferes with glioblastoma cell proliferation. Biochem J 2020;477(23):4675-88.
[Crossref] [Google Scholar] [PubMed]