- *Corresponding Author:
- S.M.Chitra
Department of Maruthuvam, Government Siddha Medical College, Arumbakkam, Chennai, Tamil Nadu 600106, India
E-mail: chittu758@gmail.com
| Date of Received | 25 April 2022 |
| Date of Revision | 08 May 2023 |
| Date of Acceptance | 04 January 2024 |
| Indian J Pharm Sci 2024;86(1):45-53 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The pandemic of coronavirus disease-19 involved various traditional medical formulations as therapeutics for treatment along with boosting the immunity to overcome the crisis. Thippili rasayanam was one among the medicines recommended by the Ministry of Ayurveda, Yoga, Unani, Siddha and Homoeopathy for coronavirus disease-19 in Siddha. In spite of the effectiveness, its role against the virus target was not clarified. As a consequence, the present docking study was performed to know its action against inhibition of the target enzyme ribonucleic acid-dependent ribonucleic acid polymerase. The phytocomponents of the herbs present in the formulation were retrieved through literature survey and in silico docking analysis was performed through Auto dock tool. The lead molecules were identified with respect to the target protein and the best dock pose was selected based on the molecular interaction. The phytocomponents epicatechin, myrcene, hyoscyamide, piperic acid, piperine and zingiberene, of the formulation Thippili rasayanam revealed maximum 3 interactions while betulonic acid, eucalyptol, galangin, p-thymol and ellagic acid showed 2 interactions with the active binding site of the target enzyme ribonucleic acid-dependent ribonucleic acid polymerase. Docking results suggest, the formulation Thippili rasayanam has potential inhibiting action against coronavirus disease-19 by reducing the viral replication.
Keywords
Coronavirus disease-19, Siddha, Thippili rasayanam, phyto components, docking analysis, ribonucleic acid-dependent ribonucleic acid polymerase
Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2), the deadly virus disease identified at Wuhan, China on December 2019 became a global pandemic and the world had crossed the two waves on 2020, 2021 respectively. At present the third wave is being accomplished by most of the countries including India, by the emergence of Omicron variant. As on January 19, 2022, India has reported around 317 000 new covid cases[1]. Even though there are vaccines produced for Coronavirus Disease-19 (COVID-19) by different manufacturing industries, people in the community and researchers are trying to find the best way to cure the disease, including herbal medicine. The world was relying on self-care practices that include the use of traditional medicine[2]. World Health Organization (WHO) recognizes that traditional, complementary and alternative medicine of proven quality, safety and efficacy has many benefits[3].
India is renowned for its traditional system of medicine and played a vital role in providing healthcare to the people for centuries. The unique Ayurveda, Yoga, Unani, Siddha and Homoeopathy (AYUSH) systems of India are based on definite medical philosophies and represent a way of achieving a healthy lifestyle with conventional and established ideas on the prevention of diseases and the promotion of health. The basic treatment approach of all these systems is holistic and the pharmacological modalities are based on natural products of plants, animals, or mineral origin. There are many medicinal plants indigenous to India and used in the Indian Systems of Medicine which has been reported as potent antiviral with immuno modulatory, anti-allergic, and anti-asthmatic activities. Many of these medicinal plants are also used as integral part of several traditional formulations that have been in use for a long time[4].
In COVID-19, Ribonucleic Acid (RNA) dependent RNA polymerase (RdRp) is the main enzyme that replicates the viral RNA genome and it is a promising drug target[5,6]. RdRp of the SARS-CoV-2 shares 96 % of the sequence identity with SARS-CoV-19 and hence the compounds or medications that are efficient towards RdRp of SARS-CoV are considered to be effective against the novel CoV. Molecular docking analysis of known RdRp-inhibiting antivirals, other Food and Drug Administration-approved medications and phytochemicals to repurpose SARS-CoV-2 is documented[7]. Three countries including India, China and South Korea, have issued guidelines on traditional regimens for the prevention and management of COVID-19[8]. In India, several clinical trials had been conducted in Siddha and Ayurveda as an integrated approach with modern medicine in the management of COVID-19. Among them, a Randomized controlled trial conducted in a tertiary care hospital at Chennai, with Siddha regimen and modern medicine as an integrated approach during June-July 2020 was proved to be effective. In that trial, Thippili rasayanam given was reported as an effective drug[8].
The Ministry of AYUSH (Government of India) had listed out such formulations and recommended their use from time to time as a prophylactic measure in the management of COVID-19. Moreover, the guidelines given by Ministry of AYUSH for Siddha practitioners, in treating mild to moderate COVID-19 patients also encompasses Thippili rasayanam, indicated for difficulty in breathing which is a prominent symptom of COVID-19[10]. Despite the potency of Thippili rasayanam was proven in the Randomized Controlled Trials (RCT) to combat COVID-19, yet there was a gap regarding the action of the biomolecules of the medicine against disease causing viral protein. One of the easiest and effective ways to identify the action potential of biomolecules of a drug against disease causing viral protein is Molecular docking analysis. Hence, it was aimed to explore the action of the phytocomponents present in the Siddha formulation Thippili rasayanam against SARS-CoV-2 virus through in silico analysis.
Objectives:
The objectives of the present study were to analyze the molecular interaction of the phyto principles with the target RNA dependent RNA polymerase (RdRp) enzyme (PDB)-6NUR which was a nonstructural protein (nsp 12) essential for viral replication in RNA viruses; to identify the potential therapeutic efficacy of the phyto principles in inhibiting the target RdRp of COVID-19 virus.
Materials and Methods
Thippili rasayanam was a polyherbal Siddha formulation quoted in the text, Siddha vaithiya thirattu in page number 235-236, written by Kuppusamy mudaliar, published by The Indian medicine and Homeopathy department, Tamil Nadu[11]. The formulation comprises 18 herbs, Piper longum, Piper nigrum, Zingiber officinale, Cuminum cyminum, Nigella sativa, Carum copticum, Hyoscyamus niger, Alpinia officinarum, Alpinia galangal, Terminalia chebula, Phyllanthus emblica, Terminalia bellirica, Syzygium aromaticum, Cinnamomum tamala, Abies spectabilis, Plumbigo indica, Elettaria cardamomum, Cinnamomum verum. The formulation was prepared with the above raw drugs taken as per ratio, mentioned in the text. Each of them was slightly roasted, powdered separately and mixed together with equal quantity of sugar. The mixture was kneaded with sufficient quantity of honey. The obtained rasayanam was indicated for cough and bronchitis.
Ligand preparation:
As a first step, the herbs present in the formulation Thippili rasayanam were explored for the phytocomponents present in them. An extensive literature search was done through PubMed, Google scholar and a sum of 15 bioactive lead compounds were identified and retrieved from them as listed in Table 1[12-25]. Two dimensional structure of the phytocomponents identified were converted into three dimensional structure through Chem Draw pro-online tool version 12.0. The Ligands were prepared through geometry optimization method and Molecular Mechanics Fore Field (MMFF94) programmer. The phyto components ligand properties selected for docking analysis were described in Table 2.
| S. No | Botanical name | Phytocomponents | Literature survey reference |
|---|---|---|---|
| 1 | Piper nigrum | Piperic acid | [12] |
| 2 | Piper longum | piperine | [13] |
| 3 | Terminalia chebula | Gallic acid | [14] |
| 4 | Terminalia bellirica | Epicatechin | [15] |
| 5 | Cumin cyminum | Cinnamaldehyde | [16] |
| 6 | Nigella sativa | Limonene | [17] |
| 7 | Carum copticum | Thymol | [18] |
| 8 | Hyoscyamus niger | Hyoscyamide | [19] |
| 9 | Alpinia officinarum | Galangin | [20] |
| 10 | Alpinia galangal | Eucalyptol | [21] |
| 11 | Phyllanthu emblica | Betulinic acid | [22] |
| 12 | Cinnamomum tamala | Myrcene | [23] |
| 13 | Zingiber officinale | Zingiberene | [24] |
| 14 | Syzygium aromaticum | Ellagic acid, salicylic acid | [25] |
Table 1: List of PHYTO Components Selected for Docking From Herbs Present in the Formulation Thippili Rasayanam Through Literature Survey
| Compound | Molar weight g/mol | Molecular Formula | H Bond Donor | H Bond Acceptor | Rotatable bonds |
|---|---|---|---|---|---|
| Limonene | 136.23 g/mol | C10H16 | 0 | 0 | 1 |
| Epicatechin | 290.271 g/mol | C15H14O6 | 5 | 6 | 1 |
| Myrcene | 136.238 g/mol | C10H16 | 0 | 0 | 4 |
| Betulonic acid | 454.7 g/mol | C30H46O3 | 1 | 3 | 2 |
| Eucalyptol | 154.25 g/mol | C10H18O | 0 | 1 | 0 |
| Galangin | 270.24 g/mol | C15H10O5 | 3 | 5 | 1 |
| Gallic acid | 170.12 g/mol | C7H6O5 | 4 | 5 | 1 |
| Hyoscyamide | 624.7 g/mol | C36H36N2O | 6 | 8 | 13 |
| Piperic acid | 218.2 g/mol | C12H10O4 | 1 | 4 | 3 |
| Piperine | 285.34 g/mol | C17H19NO3 | 0 | 3 | 3 |
| Cuminaldehyde | 148.205 g/mol | C10H12O | 0 | 1 | 2 |
| p-Thymol | 150.221 g/mol | C10H14O | 1 | 1 | 1 |
| Ellagic acid | 302.19 g/mol | C14H6O8 | 4 | 8 | 0 |
| Salicylic acid | 138.12 g/mol | C7H6O3 | 2 | 3 | 1 |
| Zingiberene | 204.35 g/mol | C15H24 | 0 | 0 | 4 |
Table 2: Ligand Properties of the Phyto Components Selected for Docking Analysis
Protein preparation:
From the online repository of Protein Data Bank (www.rcsb.org/pdb), three dimensional protein structure of the target protein RdRp enzyme (PDB) 6NUR was retrieved as shown in fig. 1. Protein clean-up process with removal of water molecules and addition of essential missing hydrogen atoms were carried out. The above were subjected to Ramchandran plot analysis using RAMPAGE to identify the statistical distribution of the ANALYSIScombinations of the amino acid backbone dihedral Ø (Phi) angles and ψ (Psi) angle and subjected to protein clean geometry prior to docking simulation. By Auto dock program, with respect to the target protein, different orientation of the lead molecules was evaluated and the best dock pose was selected based on the two dimensional interaction plot analysis and hydrogen bond plotting with core amino acid analysis[26,27]. Three dimensional pictures of the selected phyto components are shown in fig. 2.
Docking methodology:
For the retrieved phyto principles, docking scores were carried out against various binding pockets of target protein RdRp with the aid of Auto Dock tools. Essential hydrogen atoms, Kollman united atom type charges, and solvation parameters were added. Using Autogrid program affinity (grid) maps of grid points and 0.375 Å spacing were generated. Auto dock parameter set- and distance-dependent dielectric functions were used in the calculation of the van der Waals and the electrostatic terms, respectively. Docking simulations were performed using the Lamarckian Genetic Algorithm (LGA) and the Solis and Wets local search method[28,29]. Initial position, orientation and torsions of the ligand molecules were set randomly. During docking all rotatable torsions were released. Each docking experiment was derived from 2 different runs that were set to terminate after a maximum of 250 000 energy evaluations. The population size was set to 150. During the search, a translational step of 0.2 Å, and quaternion and torsion steps of 5 were applied.
Results and Discussion
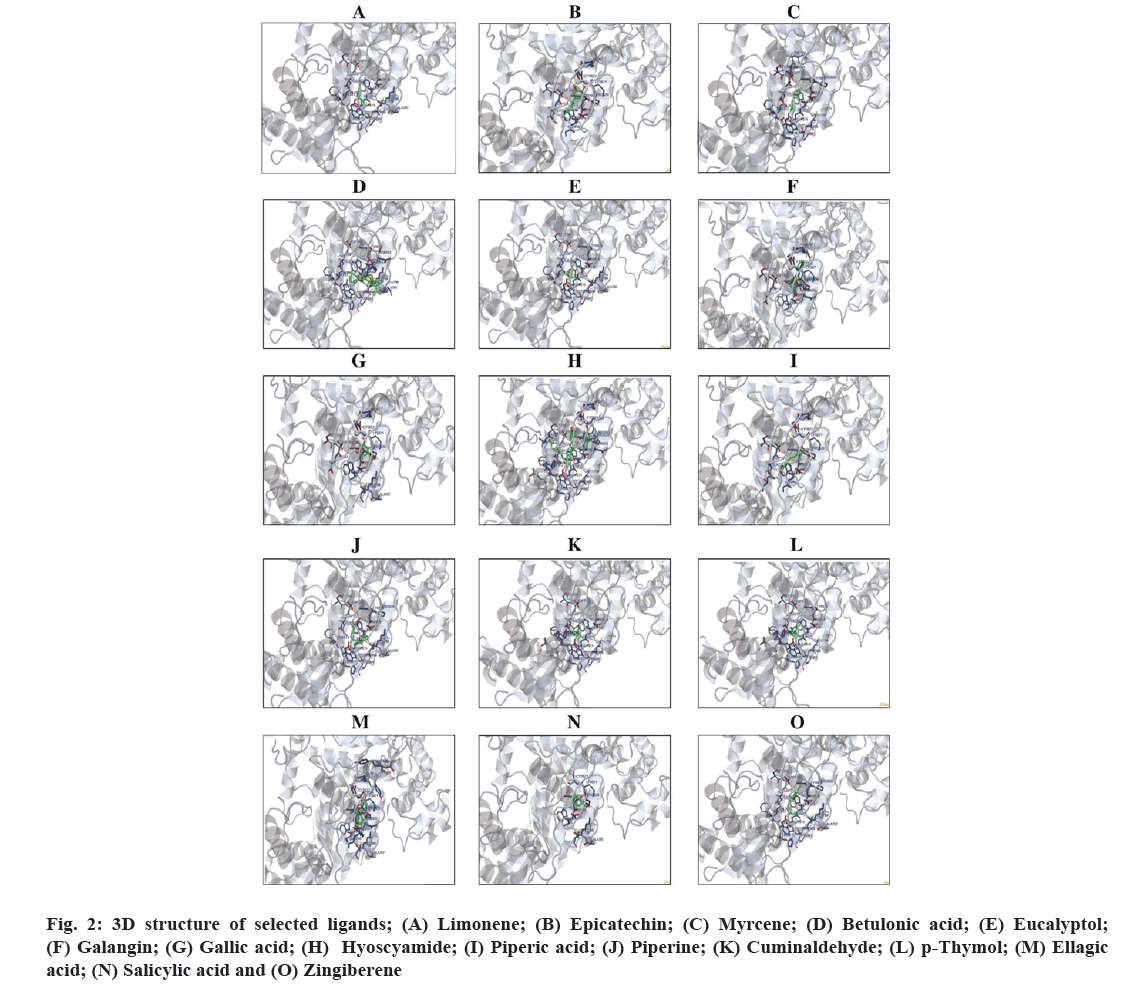
Table 3 shows the summary of the molecular docking studies of phytocomponents like binding free energy, inhibition constant, electrostatic energy, intermolecular energy, total interaction surface and Table 4 shows the amino acid residue interaction of lead phyto compounds against RdRp (PDB)-6NUR. The lead compounds present in them such as epicatechin (-6.27 kcal/mol), myrcene (-4.65 kcal/mol), hyoscyamide (-8.73 kcal/mol), piperic acid (-4.66 kcal/mol), piperine (-6.64 kcal/mol) and zingiberene (-6.24 kcal/mol) revealed a maximum of 3 interactions with all the binding sites on the core active amino acid residues present on the target receptor RdRp and also exhibited lower binding free energy. Followed by this, the compounds like betulonic acid, eucalyptol, galangin, p-thymol and ellagic acid ranked second with the maximum of 2 interactions with the active binding site of the target enzyme RdRp. Among the phytocomponents Hyoscyamide had a highest interaction surface and very lowest free binding energy. Fig. 3 illustrates the Docking Pose of the selected phytocomponents with core amino acid against RdRp (PDB)-6NUR.
| Compounds | Binding free energy Kcal/mol | Inhibition constant KiµM (*mM)(**nM) | Electrostatic energy Kcal/mol | Inter molecular energy Kcal/mol | Total interaction surface |
|---|---|---|---|---|---|
| Limonene | -4.68 kcal/mol | 370.09 µM | -0.03 kcal/mol | -4.98 kcal/mol | 447.17 |
| Epicatechin | -6.27 kcal/mol | 25.49 µM | -0.97 kcal/mol | -6.54 kcal/mol | 572.431 |
| Myrcene | -4.65 kcal/mol | 387.80 µM | -0.01 kcal/mol | -5.76 kcal/mol | 476.388 |
| Betulonic acid | -7.13 kcal/mol | 5.91 µM | -0.77 kcal/mol | -7.69 kcal/mol | 740.74 |
| Eucalyptol | -5.28 kcal/mol | 135.91 µM | -0.01 kcal/mol | -5.28 kcal/mol | 449.175 |
| Galangin | -5.95 kcal/mol | 43.76 µM | -1.80 kcal/mol | -6.83 kcal/mol | 643.94 |
| Gallic acid | -5.73 kcal/mol | 62.85 µM | -1.88 kcal/mol | -5.24 kcal/mol | 399.285 |
| Hyoscyamide | -8.73 kcal/mol | 397.72 µM | -0.81 kcal/mol | -10.09 kcal/mol | 1184.035 |
| Piperic acid | -4.66 kcal/mol | 386.52 µM | -1.67 kcal/mol | -5.55 kcal/mol | 505.323 |
| Piperine | -6.64 kcal/mol | 13.69 µM | -0.09 kcal/mol | -6.43 kcal/mol | 589.153 |
| Cuminaldehye | -4.69 kcal/mol | 362.26 µM | -0.07 kcal/mol | -5.29 kcal/mol | 418.065 |
| p-Thymol | -4.60 kcal/mol | 425.69 µM | -0.15 kcal/mol | -5.20 kcal/mol | 437.566 |
| Ellagic acid | -6.15 kcal/mol | 31.16mM | -1.67 kcal/mol | -5.59 kcal/mol | 620.05 |
| Salicylic acid | -3.52 kcal/mol | 2.61mM | -1.84 kcal/mol | -4.15 kcal/mol | 347.05 |
| Zingiberene | -6.24 kcal/mol | 26.78 mM | -0.01 kcal/mol | -7.21 kcal/mol | 573.56 |
Table 3: Summary of the Molecular Docking Studies of Phyto Components Against RNA Dependent RNA Polymerase (Pdb)-6NUR
| Compounds | Interactions | Amino acid Residues | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Limonene | 1 | 761 ASP* | 800 TRP | 811 GLU | ||||||
| Epicatechin | 3 | 617 TRP | 618 ASP* | 619 TYR | 623 ASP | 760 ASP* | 761 ASP* | |||
| Myrcene | 3 | 617 TRP | 618 ASP* | 619 TYR | 760 ASP* | 761 ASP* | 800 TRP | |||
| Betulonic acid | 2 | 551 LYS | 618 ASP* | 761 ASP* | 800 TRP | 811 GLU | ||||
| Eucalyptol | 2 | 618 ASP* | 761 ASP* | 800 TRP | 811 GLU | |||||
| Galangin | 2 | 618 ASP* | 761 ASP* | 798 LYS | 800 TRP | 811 GLU | ||||
| Gallic acid | 1 | 618 ASP* | 620 PRO | 621 LYS | 623 ASP | 798 LYS | ||||
| Hyoscyamide | 3 | 553 ARG | 618 ASP* | 623 ASP | 758 LEU | 759 SER | 760 ASP* | 761 ASP* | 800 TRP | 811 GLU |
| Piperic acid | 3 | 618 ASP* | 619 TYR | 621 LYS | 623 ASP | 760 ASP* | 761 ASP* | |||
| Piperine | 3 | 618 ASP* | 760 ASP* | 761 ASP* | 800 TRP | 811 GLU | ||||
| Cuminaldehyde | 1 | 761 ASP* | 800 TRP | 811 GLU | 814 SER | |||||
| p-Thymol | 2 | 618 ASP* | 761 ASP* | 800 TRP | 811 GLU | 814 SER | ||||
| Ellagic Acid | 2 | 551 LYS | 618 ASP* | 760 ASP* | 798 LYS | 811 GLU | ||||
| Salicylic acid | 1 | 618 ASP* | 620 PRO | 621 LYS | 798 LYS | |||||
| Zingiberene | 3 | 618 ASP* | 619 TYR | 760 ASP* | 761 ASP* | 800 TRP | 811 GLU | |||
Note: *: Binding interaction of the phyto components with RNA dependent RNA polymerase enzyme
Table 4: Amino Acid Residue Interaction of Lead Phyto Components Against RNA Dependent RNA Polymerase (Pdb)-6NUR
Fig 3: Docking Pose of the selected phytocomponents from the formulation Thippili rasayanam with RNA dependent RNA polymerase- PDB 6NUR; (A) Limonene; (B) Epicatechin; (C) Myrcene; (D) Betulonic acid; (E) Eucalyptol; (F) Galangin; (G) Gallic acid; (H) Hyoscyamide; (I) Piperic acid; (J) Piperine; (K) Cuminaldehyde; (L) p-Thymol; (M) Ellagic acid; (N) Salicylic acid and (O) Zingiberene
Traditional formulations had a pivotal role in treating and preventing chronic as well as novel diseases since centuries. They have been a rich source of bio active medicinal constituents which were not investigated scientifically. Several docking studies had been conducted on individual herbs as well as formulations with respect to traditional system of medicines like Traditional Chinese Medicine (TCM), Ayurvedha and Siddha against COVID-19 virus. Previously, a docking study carried on individual herb Nigella sativa, reported anti-viral potential against SARS-CoV-2 by Ahmad et al.[30]. A study conducted by Alrasheid et al.[31], announced that Gallic acid, Quercetin are important sources for novel anti-viral drug targeting. In the current study also the phytocomponent Gallic acid present in the herb Terminalia chebula of the formulation showed interaction with the RdRp binding site.
Previous studies conducted on siddha formulations Kabasura Kudineer, adathodai kudineer had reported inhibitory action of their phytocomponents aganist Angiotensin-Converting Enzyme 2 (ACE2) receptor spike protein similar to the present study[32,33]. Joshi et al.[34], described some Ayurvedic formulations had anti-thrombotic activity and effective against COVID-19 and suggested that Siddha, Ayurvedha, Unani formulations can be repurposed as they had the potential to prevent and treat COVID-19 that can be tested in future. In this context, the formulation Thippili rasayanam that was selected and repurposed for COVID-19 in Siddha system was found to be effective in a previous study conducted on 2021 and hence as a continuation of that, the present study was done to establish the same.
Individual studies conducted on Thippili rasayanam previously reported about its significant action scientifically. A study conducted by Manoharan et al.[35], revealed that the drug had potent anti-microbial, anti-oxidant, anti-fungal activities. An in vivo study conducted by Vaniswari et al.[36], on Thippili rasayanam reported it possess significant anti-inflammatory and anti-septic activities and it is safe to use in paediatrics. Therefore, it is evident that traditional medicines had its predominance in emerging novel diseases also which has to be studied elaborately in all aspects to prove their therapeutic efficacy.
The computational analysis of the present study concludes that the phyto components present in the siddha formulation Thippili rasayanam has a significant binding efficacy against active amino acid present on the binding sites of target enzyme. Hence, the identified Phyto principles act as potential compounds which exerts promising therapeutic efficacy by inhibiting RdRp enzyme in SARS-CoV-2 RdRp and thereby halting the viral replication. Since the formulation Thippili rasayanam’s effectiveness was proved in clinical trials conducted for COVID-19 and recommended by Ministry of AYUSH, the above study further confirms and establishes it as a strong potential medicine for the management of COVID-19.
Acknowledgements:
The author acknowledges Dr. N. Anbu, HOD, Department of PG Maruthuvam, Government Siddha Medical College, Arumbakkam, Chennai 106, Tamil Nadu, Dr. MGR Medical University, Guindy, Chennai.
Conflict of interest:
The authors declared no conflict of interests.
References
- Third wave of the COVID-19 pandemic in India. Observer Research Foundation; 2022.
- Paudyal V, Sun S, Hussain R, Abutaleb MH, Hedima EW. Complementary and alternative medicines use in COVID-19: A global perspective on practice, policy and research. Res Soc Adm Pharm 2022;18(3):2524-8.
- World Health Organization. WHO traditional medicine strategy: 2014–2023. World Health Organization; 2013.
- Ahmad S, Zahiruddin S, Parveen B, Basist P, Parveen A, Parveen R, et al. Indian medicinal plants and formulations and their potential against COVID-19–preclinical and clinical research. Front Pharmacol 2021;11:2470.
[Crossref] [Google Scholar] [PubMed]
- Li G, De Clercq E. Therapeutic options for the 2019 novel coronavirus (2019-nCoV). Nat Rev Drug Discov 2020;19(3):149-50.
[Crossref] [Google Scholar] [PubMed]
- Morse JS, Lalonde T, Xu S, Liu WR. Learning from the past: Possible urgent prevention and treatment options for severe acute respiratory infections caused by 2019‐nCoV. Chembiochem 2020;21(5):730-8.
[Crossref] [Google Scholar] [PubMed]
- Wu C, Liu Y, Yang Y, Zhang P, Zhong W, Wang Y, et al. Analysis of therapeutic targets for SARS-CoV-2 and discovery of potential drugs by computational methods. Acta Pharm Sin B 2020;10(5):766-88.
[Crossref] [Google Scholar] [PubMed]
- Ang L, Lee HW, Choi JY, Zhang J, Lee MS. Herbal medicine and pattern identification for treating COVID-19: A rapid review of guidelines. Integr Med Res 2020;9(2):100407.
[Crossref] [Google Scholar] [PubMed]
- Chitra SM, Mallika P, Anbu N, NarayanaBabu R, SugunaBai A, Raj RD, et al. An open clinical evaluation of selected siddha regimen in expediting the management of COVID-19–A randomized controlled study. J Ayurveda Integr Med 2022;13(1):100397.
[Crossref] [Google Scholar] [PubMed]
- Guidelines Siddha; Ministry of Ayush; 2021.
- Kuppusamy Mudaliar KN, Uththamarayan KS. Siddha Vaithiya Thirattu. Indian Med Homeopathy Dep 2014;235-236.
- Salehi B, Zakaria ZA, Gyawali R, Ibrahim SA, Rajkovic J, Shinwari ZK, et al. Piper species: A comprehensive review on their phytochemistry, biological activities and applications. Molecules 2019;24(7):1364.
[Crossref] [Google Scholar] [PubMed]
- Tiwari A, Mahadik KR, Gabhe SY. Piperine: A comprehensive review of methods of isolation, purification, and biological properties. Med Drug Discov 2020;7:100027.
- Bag A, Bhattacharyya SK, Chattopadhyay RR. The development of Terminalia chebula Retz. (Combretaceae) in clinical research. Asian Pac J Trop Biomed 2013;3(3):244-52.
[Crossref] [Google Scholar] [PubMed]
- Sobeh M, Mahmoud MF, Hasan RA, Abdelfattah MA, Osman S, Rashid HO, et al. Chemical composition, antioxidant and hepatoprotective activities of methanol extracts from leaves of Terminalia bellirica and Terminalia sericea (Combretaceae). PeerJ 2019;7:e6322.
[Crossref] [Google Scholar] [PubMed]
- Srinivasan K. Cumin (Cuminum cyminum) and black cumin (Nigella sativa) seeds: Traditional uses, chemical constituents, and nutraceutical effects. Food Quality Safety 2018;2(1):1-6.
- Kabir Y, Akasaka-Hashimoto Y, Kubota K, Komai M. Volatile compounds of black cumin (Nigella sativa L.) seeds cultivated in Bangladesh and India. Heliyon 2020;6(10);e05343.
[Crossref] [Google Scholar] [PubMed]
- Boskabady MH, Alitaneh S, Alavinezhad A. Carum copticum L.: A herbal medicine with various pharmacological effects. Biomed Res Int 2014;2014:569087.
[Crossref] [Google Scholar] [PubMed]
- Ma CY, Liu WK, Che CT. Lignanamides and Nonalkaloidal components of Hyoscyamus n iger seeds. J Nat Prod 2002;65(2):206-9.
[Crossref] [Google Scholar] [PubMed]
- Luo H, Cai C, Zhang J, Mo L. Study on the chemical components of Alpinia officinarum. Zhong Yao Cai 1998;21(7):349-51.
[Google Scholar] [PubMed]
- Menon AN. Chemical composition of the volatile oils of Alpinia galanga plant parts from Kerala. J Essential Oil Bearing Plants 2006;9(3):277-82.
- Li B, Huang GQ, Lu RM, Wei JH, Zhong ZG. Study on chemical composition of Phyllanthus emblica. Zhong Yao Cai 2015 Feb 1;38(2):290-3.
[Google Scholar] [PubMed]
- Mir SR, Ali M, Kapoor R. Chemical composition of essential oil of Cinnamomum tamala Nees et Eberm. leaves. Flavour Fragrance J 2004;19(2):112-4.
- Rahmani AH, Aly SM. Active ingredients of ginger as potential candidates in the prevention and treatment of diseases via modulation of biological activities. Int J Physiol Pathophysiol Pharmacol 2014;6(2):125-36.
[Google Scholar] [PubMed]
- Batiha GE, Alkazmi LM, Wasef LG, Beshbishy AM, Nadwa EH, Rashwan EK. Syzygium aromaticum L.(Myrtaceae): Traditional uses, bioactive chemical constituents, pharmacological and toxicological activities. Biomolecules 2020;10(2):202.
[Crossref] [Google Scholar] [PubMed]
- Bikadi Z, Hazai E. Application of the PM6 semi-empirical method to modeling proteins enhances docking accuracy of AutoDock. J Cheminform 2009;1:1-6.
[Crossref] [Google Scholar] [PubMed]
- Halgren TA. Merck molecular force field. I. Basis, form, scope, parameterization, and performance of MMFF94. J Comput Chem 1996;17(5‐6):490-519.
- Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, et al. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J comput Chem 1998;19(14):1639-62.
- Solis FJ, Wets RJ. Minimization by random search techniques. Math Operations Res 1981;6(1):19-30.
- Ahmad S, Abbasi HW, Shahid S, Gul S, Abbasi SW. Molecular docking, simulation and MM-PBSA studies of Nigella sativa compounds: A computational quest to identify potential natural antiviral for COVID-19 treatment. J Biomol Struct Dyn 2021;39(12):4225-33.
[Crossref] [Google Scholar] [PubMed]
- Alrasheid AA, Babiker MY, Awad TA. Evaluation of certain medicinal plants compounds as new potential inhibitors of novel corona virus (COVID-19) using molecular docking analysis. In Silico Pharmacol 2021;9:1-7.
- Vincent S, Arokiyaraj S, Saravanan M, Dhanraj M. Molecular docking studies on the anti-viral effects of compounds from Kabasura Kudineer on SARS-CoV-2 3CLpro. Front Mol Biosci 2020;7:613401.
[Crossref] [Google Scholar] [PubMed]
- Christian GJ, Meenakumari R, Rajamaheswari K, Sekaran P, Gajalakshmi G, Anand T. Phytochemical screening of Adathodai kudineer A Siddha herbal concoction and evaluation of its binding affinity with SARS-CoV-2 spike protein and ACE2 receptor spike protein complex through molecular docking in silico approach. Int J Ayurvedic Med 2021;12(2):366-74.
- Joshi C, Chaudhari A, Joshi C, Joshi M, Bagatharia S. Repurposing of the herbal formulations: molecular docking and molecular dynamics simulation studies to validate the efficacy of phytocompounds against SARS-CoV-2 proteins. J Biomol Struct Dyn 2022;40(18):8405-19.
[Crossref] [Google Scholar] [PubMed]
- Manoharan A. Novel action of Thippili rasayanam- Siddha drug against respiratory pathogens. Int J Recent Sci Res 2016;7(5);10842-5.
- Vaniswari DS, Arunachalam K. Anti-inflammatory and Antiseptic activity of ancient traditional Siddha drug Thippili rasayanam in Pediatric care. Int J Adv Res 2019;7:1035-9.