- *Corresponding Author:
- Jing Liang
Laboratory Department, The Sixth School of Clinical Medicine, Xinjiang Medical University, Urumqi, Xinjiang 830000, China
E-mail: lj95890000@163.com
| This article was originally published in a special issue, “Innovations in Biomedical Research and Drug Development” |
| Indian J Pharm Sci 2023:85(3) Spl Issue “47-54” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The main objective of this study is to analyze the expression levels of messenger ribonucleic acid and long non-coding ribonucleic acid in patients with platelet transfusion refractoriness and reveal the mechanism of T lymphocytes in immune platelet transfusion refractoriness. The Agilent expression profile chip was used to detect the expression levels; gene ontology and Kyoto encyclopedia of genes and genomes enrichment analysis were performed on the differential genes to determine their main biological functions. Unsupervised hierarchical clustering was used to process differential genes and the differential genes among samples were represented in a heat map. The differentially expressed messenger ribonucleic acids and long non-coding ribonucleic acids in different groups were found as 720 and 1719 in normal control group vs. platelet transfusion effective group; 4254 and 12491 in normal control group vs. platelet transfusion ineffective group and 1806 and 6216 in platelet transfusion effective group vs. platelet transfusion ineffective group. Gene ontology and Kyoto encyclopedia of genes and genomes enrichment analysis were performed on differentially expressed genes, and the results were annotated to be related to T cells. The co-expression of the target gene Ras-related protein 1A and long non-coding transactive response deoxyribonucleic acid binding protein 2 was determined through the national center for biotechnology information database and the interaction between micro ribonucleic acid-4739 and Ras-related protein 1A was predicted using the starBase and TargetScan databases. T lymphocytes play an important role in immune platelet transfusion refractoriness and long non-coding transactive response deoxyribonucleic acid binding protein 2 may affect the differentiation of T lymphocytes and promote the occurrence of immune platelet transfusion refractoriness through micro ribonucleic acid-4739 targeting Rasrelated protein 1A gene regulation.
Keywords
Platelet transfusion refractoriness, T lymphocytes, gene chip, long non-coding transactive response deoxyribonucleic acid binding protein 2, micro ribonucleic acid-4739
Platelet Transfusion Refractoriness (PTR) has been a common clinical problem in the treatment of bleeding disorders[1,2], which can cause serious consequences for patients and additional economic burdens for families and society. In recent years, it has been recognized that the mechanism of immune PTR is caused by B cells producing platelet antibodies through humoral immune response[3]. But new research has found that about 17 % of patients with PTR and 40 % of patients with Immune Thrombocytopenia (ITP) had no significant platelet antibody[4], indicating that the humoral immune clearance mechanism cannot fully explain the occurrence of PTR. Studies have found that both Cluster of Differentiation 8+ (CD8+) T cells and Cluster of Differentiation 4+ (CD4+) T cells with antiplatelet reactivity are found in peripheral blood. CD8+ T cells can inhibit B lymphocytes from producing platelet antibodies, while the decrease in the number of CD4+ T cells can make auto reactive B lymphocytes proliferate accordingly, thereby producing some antibodies that promote platelet destruction[5-7], indicating that T lymphocytes can regulate platelet antibody production. The immune response of T lymphocytes may be another mechanism that causes immune PTR, but there are few studies on the regulation mechanism of T lymphocytes on immune PTR. We analyzed the expression levels of messenger Ribonucleic Acid (mRNA) and long noncoding RNA (lncRNA) in patients with PTR by gene chip technology, screened the relevant target genes that regulate T lymphocytes in immune PTR, and explored its possible mechanism of action, in order to further explore the role of T lymphocytes in mediating immune PTR. The development mechanism provides a theoretical basis.
Materials and Methods
Research objects and specimen collection:
A total of 12 patients admitted to the Sixth Affiliated Hospital of Xinjiang Medical University from August 2021 to March 2022 were selected and their Peripheral Blood Mononuclear Cell (PBMC) samples were included in this study, of which 4 PBMC samples from volunteers included in normal control group, 4 patients included in the platelet transfusion effective group and 4 patients included in the platelet transfusion ineffective (PTR) group. The three groups include 1 male and 3 females, and their ages were 40.25±1.708, 41±2.582 and 41.75±1.708 y. All patients in this study gave informed consent and this study was approved by the Ethics Committee of our hospital.
Detection of differential expression of lncRNA and mRNA in PBMC:
Agilent Human lncRNA V6 (4×180K, Design ID: 084410) chips were used to detect the expression levels of mRNA and lncRNA. Agilent Human lncRNA V6 (4×180K, Design ID: 084410) chip was used, NanoDrop® ND-2000 (Thermo Scientific) was used for quantitative analysis of sample RNA and RNA integrity was determined by Agilent Bioanalyzer 2100 (Agilent Technologies). After the qualification of RNA in the quality inspection, the labeling of the sample, hybridization of the chip and the elution refer to the chip standard process was initiated. After the total RNA was reverse-transcribed into double-stranded complementary Deoxyribonucleic Acid (cDNA), Cyanine-3-CTP (Cy3) was used to label the synthesized complementary RNA (cRNA), the labeled cRNA was hybridized with the chip and after elution, the original image was obtained by scanning with Agilent Scanner G2505C (Agilent Technologies).
Data analysis:
The original image was processed by feature extraction software (version 10.7.1.1, Agilent Technologies) to obtain the original data. The data were then subjected to quantile normalization and subsequent processing using GeneSpring software (version 14.8, Agilent Technologies). Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were performed on the differential genes to determine the biological functions and pathways in which the differential genes were mainly involved. Further combine the queries of National Center for Biotechnology Information (NCBI), StarBase and TargetScan databases to screen out key target genes and target pathways. Unsupervised hierarchical clustering is used to process differential genes and the expression status of differential genes between different samples is displayed in a heat map.
Results and Discussion
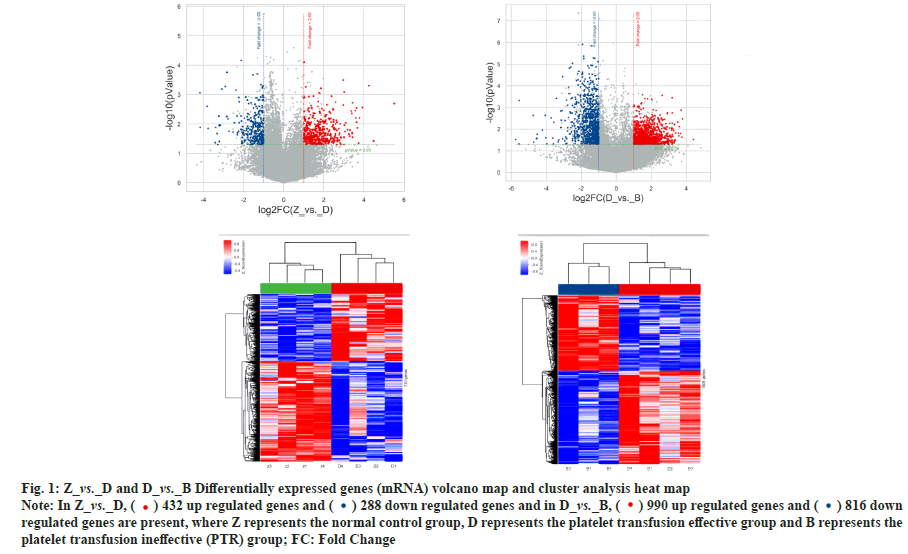
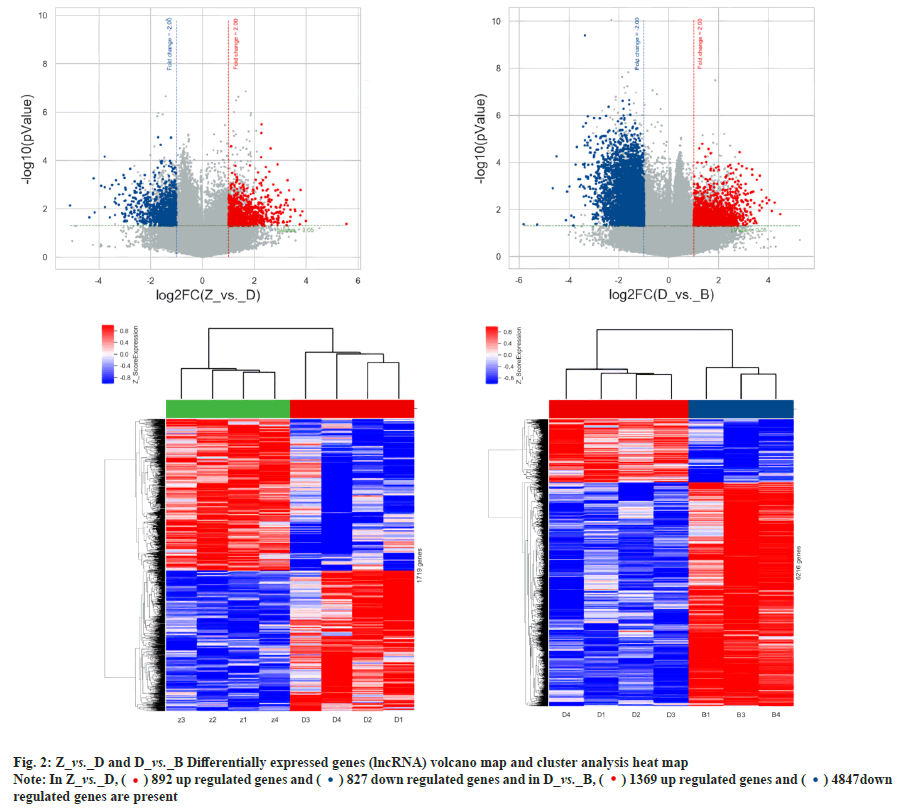
Differential expression and co-expression analysis of lncRNA and mRNA in peripheral blood PBMC of patients with PTR was explained here. Agilent expression profiling chips were used in order to detect the changes in expression levels of mRNA and lncRNA in T lymphocytes of patients with PTR. The results showed that there were a total of 720 differentially expressed mRNAs and 1719 differentially expressed lncRNAs in the normal control group vs. platelet transfusion effective group, a total of 4254 differentially expressed mRNAs and 12491 differentially expressed lncRNAs in the normal control group vs. platelet transfusion ineffective group. A total of 1806 differentially expressed mRNAs and 6216 differentially expressed lncRNAs were found in the platelet transfusion effective group vs. platelet transfusion ineffective group. The analysis results are shown in Table 1, fig. 1 and fig. 2.
| Group name | Type | Up-regulated | Down-regulated | Total |
|---|---|---|---|---|
| comp1_Z_vs._D_FC2P0.05 | Coding | 432 | 288 | 720 |
| comp1_Z_vs._D_FC2P0.05 | lncRNA | 892 | 827 | 1719 |
| comp2_Z_vs._B_FC2P0.05 | Coding | 2501 | 1753 | 4254 |
| comp2_Z_vs._B_FC2P0.05 | lncRNA | 5059 | 7432 | 12491 |
| comp3_D_vs._B_FC2P0.05 | Coding | 990 | 816 | 1806 |
| comp3_D_vs._B_FC2P0.05 | lncRNA | 1369 | 4847 | 6216 |
Note: Z represents the normal control group, D represents the platelet transfusion effective group; B represents the platelet transfusion ineffective group (PTR) and FC: Fold Change
Table 1: Statistics of Differentially Expressed Genes
Fig. 1: Z_vs._D and D_vs._B Differentially expressed genes (mRNA) volcano map and cluster analysis heat map
Note: In Z_vs._D, ( ) 432 up regulated genes and (
) 432 up regulated genes and ( ) 288 down regulated genes and in D_vs._B, (
) 288 down regulated genes and in D_vs._B, ( ) 990 up regulated genes and (
) 990 up regulated genes and ( ) 816 down
) 816 down
regulated genes are present, where Z represents the normal control group, D represents the platelet transfusion effective group and B represents the
platelet transfusion ineffective (PTR) group; FC: Fold Change
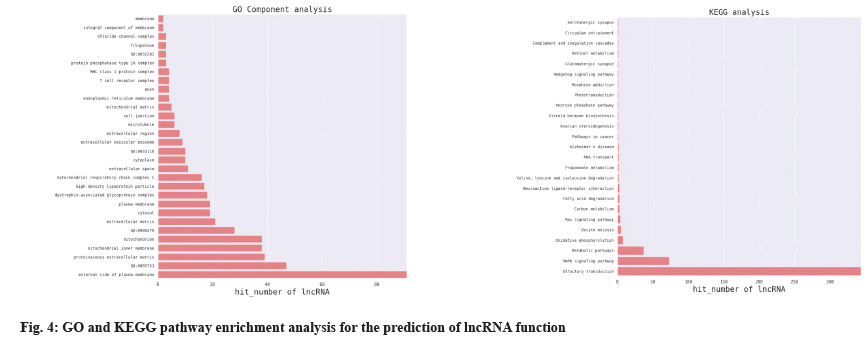
Through GO enrichment analysis, the differentially expressed genes related to the regulation of T lymphocyte differentiation and platelet apoptosis were annotated in three aspects of biological process, cellular components, molecular functions, p21- Activated Kinase 2 (PAK2), Ras Protein Specific Guanine Nucleotide Releasing Factor 2 (RASGRF2), Colony-Stimulating Factor-1 (CSF1), Ras-related protein 1A (Rap1A), Arrestin Beta 1 (ARRB1) and Mitogen-Activated Protein Kinase Kinase Kinase 7 (MAP3K7), etc. were initially identified. The gene was used as a candidate gene and the co-expression relationship between the target gene Rap1A and lnc-Transactive Response Deoxyribonucleic Acid Binding Protein 2 (lnc-TARDBP-2) was determined by combining the NCBI database gene function query, literature query and lncRNA-mRNA coexpression analysis. The analysis results are shown in Table 2 and Table 3.
| Differentially expressed genes | Gene ID | p-value (D_vs._B) | FC (D_vs._B) | p-value (Z_vs._B) | FC (Z_vs._B) |
|---|---|---|---|---|---|
| PAK2 | 5062 | 0.00987 | 2.59682 | 0.00119 | 3.21288 |
| RASGRF2 | 5924 | 0.00698 | -5.1452 | 0.02928 | -2.1382 |
| CSF1 | 1435 | 0.00136 | -8.4982 | 9E-05 | -9.7822 |
| RAP1A | 5906 | 0.03472 | 2.89331 | 0.00234 | 3.45761 |
| ARRB1 | 408 | 0.0111 | 2.83555 | 0.02006 | 2.155 |
| MAP3K7 | 6885 | 0.00441 | 2.89151 | 0.00018 | 3.40379 |
Table 2: Differentially Expressed mRNA Screening
| LncRNA | mRNA | Co-expression | p value | Primary ID |
|---|---|---|---|---|
| lnc-TARDBP-2 | Rap1A | 0.918 | 0.0035 | lnc-TARDBP-2 |
Table 3: LncRNA and mRNA Co-Expression Analysis
GO enrichment analysis was performed on the differentially expressed genes in terms of biological process, cellular components and molecular functions. The Z_vs._D differentially expressed gene GO enrichment analysis results annotated T Cell Receptor (TCR) complexes, negative regulation of TCR signaling pathways, TCR Variable, Diversity and Joining (VDJ) recombination, alpha-beta T cell differentiation and modulation of the immune response, etc. where Z represents the normal control group and D represents the platelet transfusion effective group, as shown in fig. 3.
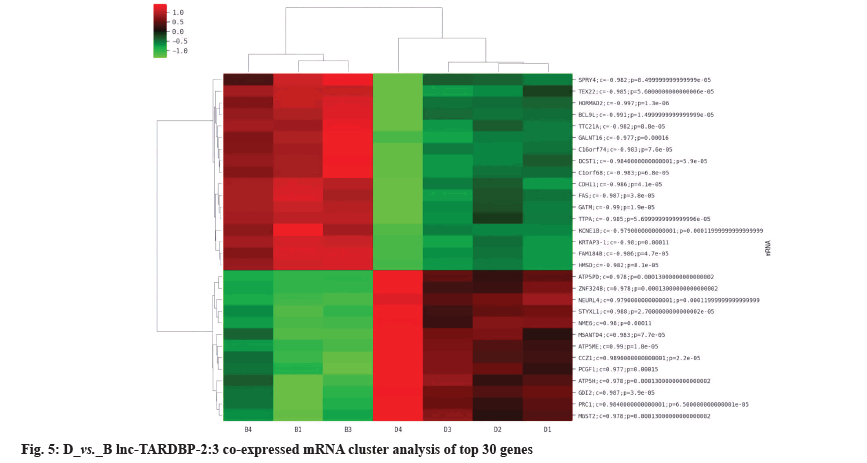
The differentially expressed lncRNAs and mRNAs were analyzed for co-expression. If the p value of the correlation coefficient of the expression values of lncRNAs and mRNAs was p≤0.05 and the absolute value of the correlation was ≥0.8, it was considered to be correlated. The expression values were used for unsupervised cluster analysis to draw heat map. For each differentially expressed lncRNAs, the co-expressed coding genes were calculated and according to GO and KEGG pathway enrichment and the application of NCBI database, it was predicted that Rap1A and lnc-TARDBP-2 had a co-expression relationship, as shown in fig. 4 and fig. 5.
Prediction analysis of the micro Ribonucleic Acid (miRNA) was reported here. To predict the miRNA interacting with Rap1A StarBase database and TargetScan database was used and to predict the miRNA that can interact with lnc-TARDBP-2 RegRNA 2.0 database was used, and further querying the references corresponding to the miRNA to confirm that miR-4739 is the predicted miRNA.
This study was the first to elucidate the differences in gene expression profiles between PBMCs of patients with immune PTR, healthy people and people with effective platelet transfusion through gene chips and found that there were significant gene differences between patients with immune PTR and the other two groups. Through GO enrichment analysis, the results showed that T cells were closely related to immune PTR. At the same time, PAK2, RASGRF2, CSF1, Rap1A, ARRB1, MAP3K7 and other genes were initially identified as candidate genes and further combined with NCBI database gene function query, literature query and lncRNA-mRNA co-expression analysis to determine the co-expression of target genes Rap1A and lnc-TARDBP-2 relation.
Rap1 plays a key role in TCR signal transduction and is expressed in cells by one of its two isoforms: Rap1A and Ras-related protein 1B (Rap1B)[8]. The expression of Rap1A is required for the fine-tuning of the immune response[9]. Su et al. studied the effect of Rap1A deficiency on the number of T cells[10]. When Rap1 was deficient, T cells in Peripheral Lymph Nodes (PLN) decreased significantly and T cells in blood increased. In addition, studies have found that the low expression of Rap1A activity was detected in resting T lymphocytes. When T lymphocyte surface receptors are activated by antigens, the enhancement of Rap1A activity can be clearly detected and Rap1A activation enhances TCR acceptance. Signaling and transduction capabilities further promote the ability of T lymphocyte proliferation and differentiation[11]. In summary, Rap1A has been confirmed to be closely related to the activation of T lymphocytes. At the same time, we found that the expression of Rap1A increased in patients with immune PTR, indicating that Rap1A may play a role in immune PTR by regulating T lymphocytes.
LncRNA plays an important role and has multiple functions in biological processes such as cell function, growth, development and disease development[12], including the regulation of T lymphocyte immunity and platelet biological processes[13]. Ghafouri-Fard et al. found that in stage III immature T cells, Thymic T lymphocyte-natural cytotoxicity triggering Receptor 1 (Thy-ncR1) activates the adjacent surface differentiation antigen CD gene cluster by degrading mRNA-Microfibril Associated Protein 4 (MFAP4), indicating that lncRNAs are involved in the differentiation and maturation of T cells[14]. Liu et al. found that lncRNA may affect the activation of T cells and the proliferation of CD4+ and CD8+ cells by inhibiting miRNA[15,16]. Another study found that silencing lncRNA-Antisense Non-coding RNA in the Inhibitor of the cyclin D-dependent kinases (INK4) Locus (ANRIL) can inhibit the activation of T cells[17]. These studies suggest that lncRNAs play an important role in the differentiation and maturation of immune T lymphocytes in the body. Different lncRNAs can regulate the biological process of T lymphocytes through the corresponding target gene mRNA. It is closely related to death and has a regulatory effect on platelets. Vesicle-Associated Membrane Protein 8 (VAMP8) has an important function in the platelet activation pathway and lnc- Ubiquitin Specific Peptidase 39 (lnc-USP39-1) can regulate the protein-coding gene VAMP8 in cis, thereby regulating platelet activation[18]. Moreover, platelets cannot perform physiological functions without the aggregation of actin filaments lnc- Neuroblastoma Breakpoint Family 9-5:1 (lnc-NBPF 9-5:1), lnc-Spindle Pole body Component 25-6:1 (lnc-SPC25-6:1), lnc-Nuclear factor-kappa-BRepressing Factor-1 (lnc-NKRF-1) and lnc-High Mobility Group Nucleosome binding domain 5-3:1 (lnc-HMGN 5-3:1) closely interact with platelet actin protein filament aggregation and they can delay the activation of platelets and thus help to prolong the life of platelets[19]. As a kind of lncRNA, lnc- TARDBP-2 has been detected to be up-regulated in immune PTR, but its regulatory relationship with T cells and its role in immune PTR needed further study.
In addition, we combined StarBase database and TargetScan database to predict miRNAs interacting with Rap1A. It was determined that miR-4739 may participate in the occurrence of immune PTR by regulating T lymphocytes. A variety of miRNAs have been identified to play key roles in T lymphocyte development, differentiation and function[20]. Dolz et al. found that miR-17-92 is closely related to the proliferation of CD4+ T cells[21]. Veiga et al. found that miR-155 is a key positive regulator of regulatory T cells (Tregs)[22]. At the same time, studies have shown that miRNAs play an important role in hematopoietic differentiation, which is related to the cellular immune response leading to ITP[23]. Zuo et al. found that miR-4739 significantly increased in ITP[24]. Therefore, miR-4739 may participate in the occurrence of PTR by regulating T cell immunity and miR-4739 may become a new target of PTR biomarkers in the future.
To sum up, this study analyzed the gene expression differences between PBMC of immune PTR population, healthy population and platelet transfusion effective population by gene chip technology, selected a batch of valuable target genes and clarified their related functions. Molecular functions found that T lymphocytes may play an important role in the occurrence of immune PTR. Moreover, in PTR patients, lnc-TARDBP-2 may affect the proliferation and differentiation of T lymphocytes through miR-4739 targeting Rap1A gene regulation to promote disease occurrence. Our study provides a theoretical basis for revealing the mechanism of T lymphocytes in the development of immune PTR and also provides valuable genes and directions for further research on the markers of immune PTR. However, the conclusions proposed in this paper need to be confirmed by further molecular biology experiments.
Acknowledgments:
This work was supported by Natural Science Foundation of Xinjiang Uygur Autonomous Region (Fund name: Mechanism of ASICs regulation of NFATinduced PTR platelet/T cell abnormalities through Wnt/Ca2+ pathway, Project No: 2022D01C585).
Conflict of interests:
The authors declared no conflict of interest.
References
- Yang XL, Yu Z. Research progress on causes and countermeasures of ineffective platelet transfusion. Lab Med Clin 2019;16(7):127-30.
- Liang J, Kaiserjiang D, Liu W. Study on expression levels of platelet membrane glycoproteins and T/B lymphocytes based on immune PTR. Mod Biomed Adv 2022;22(4):5.
- Sai J, Tian L. The mechanism and solution of immune platelet transfusion ineffectiveness. Chin J Blood Transfus 2022;35(6):677-82.
- Liu Y, Zhang Y, Chen D, Fu Y. Current status of and global trends in platelet transfusion refractoriness from 2004 to 2021: A bibliometric analysis. Front Med 2022;9:1-17.
[Crossref] [Google scholar] [PubMed]
- Tărniceriu CC, Hurjui LL, Florea ID, Hurjui I, Gradinaru I, Tanase DM, et al. Immune Thrombocytopenic purpura as a hemorrhagic versus thrombotic disease: An updated insight into pathophysiological mechanisms. Medicina 2022;58(2):1-19.
[Crossref] [Google scholar] [PubMed]
- Mingot-Castellano ME, Bastida JM, Caballero-Navarro G, Entrena Ureña L, González-López TJ, González-Porras JR, et al. Novel therapies to address unmet needs in ITP. Pharmaceuticals 2022;15(7):1-24.
[Crossref] [Google scholar] [PubMed]
- Audia S, Mahévas M, Nivet M, Ouandji S, Ciudad M, Bonnotte B. Immune thrombocytopenia: Recent advances in pathogenesis and treatments. Hemasphere 2021;5(6):e574.
[Crossref] [Google scholar] [PubMed]
- Zhang T, Jiang K, Zhu X, Zhao G, Wu H, Deng G, et al. MiR-433 inhibits breast cancer cell growth via the MAPK signaling pathway by targeting Rap1A. Int J Biol Sci 2018;14(6):622-32.
[Crossref] [Google scholar] [PubMed]
- Sun H, Lagarrigue F, Ginsberg MH. The connection between Rap1 and Talin1 in the activation of integrins in blood cells. Front Cell Dev Biol 2022;10:1-8.
[Crossref] [Google scholar] [PubMed]
- Su W, Wynne J, Pinheiro EM, Strazza M, Mor A, Montenont E, et al. Rap1 and its effector RIAM are required for lymphocyte trafficking. Blood 2015;126(25):2695-703.
[Crossref] [Google scholar] [PubMed]
- Golec DP, Hoeppli RE, Henao LMC, McCann J, Levings MK, Baldwin TA. Thymic progenitors of TCRαβ+ CD8αα intestinal intraepithelial lymphocytes require RasGRP1 for development. J Exp Med 2017;214(8):2421-35.
[Crossref] [Google scholar] [PubMed]
- Ren Y, Su Q. Research progress of long non-coding RNA in ischemic heart disease. J PLA Med Coll 2023:1-7.
- Gasic V, Karan-Djurasevic T, Pavlovic D, Zukic B, Pavlovic S, Tosic N. Diagnostic and therapeutic implications of long non-coding RNAs in leukemia. Life 2022;12(11):1-30.
[Crossref] [Google scholar] [PubMed]
- Ghafouri-Fard S, Niazi V, Taheri M. Role of miRNAs and lncRNAs in hematopoietic stem cell differentiation. Noncoding RNA Res 2021;6(1):8-14.
[Crossref] [Google scholar] [PubMed]
- Liu X, Li Y, Jiang X, Ma C, Yu Q, Gao D. Long non-coding RNA: Multiple effects on the differentiation, maturity and cell function of dendritic cells. Clin Immunol 2022;245:109167.
[Crossref] [Google scholar] [PubMed]
- Cheng Q, Chen X, Xu J, Chen M, Zhang P, Wu H, et al. LncRNA X-inactive-specific transcript is a potential biomarker related to changes in CD4+ T cell levels in systemic lupus erythematosus. Rheumatol Autoimmun 2022;2(03):159-74.
- Xu Y, Cao L, Ji S, Shen W. LncRNA ANRIL-mediated miR-181b-5p/S1PR1 axis is involved in the progression of uremic cardiomyopathy through activating T cells. Sci Rep 2022;12(1):1-11.
[Crossref] [Google scholar] [PubMed]
- Ren Q, Barber HK, Crawford GL, Karim ZA, Zhao C, Choi W, et al. Endobrevin/VAMP-8 is the primary v-SNARE for the platelet release reaction. Mol Biol Cell 2007;18(1):24-33.
[Crossref] [Google scholar] [PubMed]
- Litvinov RI, Weisel JW. Blood clot contraction: Mechanisms, pathophysiology and disease. Res Pract Thromb Haemost 2023;7(1):100023.
[Crossref] [Google scholar] [PubMed]
- Naqvi RA, Datta M, Khan SH, Naqvi AR. Regulatory roles of MicroRNA in shaping T cell function, differentiation and polarization. Semin Cell Dev Biol 2022;124:34-47.
[Crossref] [Google scholar] [PubMed]
- Dolz M, Hasiuk M, Gagnon JD, Kornete M, Marone R, Bantug G, et al. Forced expression of the non-coding RNA miR-17-92 restores activation and function in CD28-deficient CD4+ T cells. Iscience 2022;25(11):105372.
[Crossref] [Google scholar] [PubMed]
- Veiga RN, Zambalde ÉP, Cox L, Jucoski TS, Kohler AF, Carvalho TM, et al. Regulation of immune cells by microRNAs and microRNA-based cancer immunotherapy. Adv Exp Med Biol 2022;1385:75-108.
[Crossref] [Google scholar] [PubMed]
- Zhao Y, Cui S, Wang Y, Xu R. The extensive regulation of microRNA in immune thrombocytopenia. Clin Appl Thromb Hemost 2022;28:1-19.
[Crossref] [Google scholar] [PubMed]
- Zuo B, Zhai J, You L, Zhao Y, Yang J, Weng Z, et al. Plasma microRNAs characterising patients with immune thrombo cytopenic purpura. Thromb Haemost 2017;117(7):1420-31.
[Crossref] [Google scholar] [PubMed]
 ) Function; (
) Function; ( ) Process and (
) Process and ( ) Component
) Component