- *Corresponding Author:
- N. Shivaiah
Department of Studies and Research in Biochemistry
Tumkur University
Tumakuru 572103, India
E-mail: nagarajubiochem@gmail.com
| Date of Received | 11 December 2019 |
| Date of Revision | 21 July 2021 |
| Date of Acceptance | 06 September 2021 |
| Indian J Pharm Sci 2021;83(5):918-924 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The objective of this study was to identify and evaluate for the first time the anticoagulant and fibrinogenolytic protease from Psidium guajava seed extract. The Psidium guajava seed extract in sodium dodecyl sulfate polyacrylamide gel electrophoresis showed protein bands in high molecular weight region between 122 kDa and 47 kDa and two distinct bands in the lower molecular weight region at ~16.6 kDa and ~14.3 kDa. The protein content in Psidium guajava seed extract was found to be 1 mg/ml with protease specific activity 0.8 U. The proteolytic activity was supported by zymography studies using casein/gelatin as substrates. Psidium guajava seed extract hydrolysed all the subunits of fibrinogen and alpha-polymer and alpha-chain subunits of fibrin at a concentration of 20 μg and 120 μg, respectively. Psidium guajava seed extract prolonged the clotting time in recalcification time from control 161 s to 487 s, increasing by ~3 folds. This indicates the anticoagulant nature associated with Psidium guajava seed extract. Our study provides for the first time, the scientific validation for the presence of a protease with anticoagulant and fibrinogenolytic property from Psidium guajava seed extract.
Keywords
Psidium guajava, fibrinogen, hypertension, diabetes, anticoagulant
Stroke is an immediate pause in the supply of blood to the brain due to the blockage of cerebral artery with thromboembolic occlusion[1]. The causes of thrombosis are multifactorial. The thrombus formation occurs by the activation of blood coagulation factors during vascular injuries or imbalance in endogenous anticoagulants[2–5]. Stroke can be divided into two categories: ischemic and hemorrhagic. Majority of the stroke caused leads to cerebral infarction, venous thrombosis and embolism[6,7]. The most common causes of cerebral infarction are hypertension, diabetes, hyperlipidaemia, smoking and genetic diseases[8].
The possible contributors for these vascular complications during these disease conditions are found to be majorly platelet hypersensitivity, endothelial cell dysfunction and imbalance in blood coagulation mechanisms[9–11]. Blood coagulation is a complex cascade of enzymatic reactions, which arrest the blood from bleeding; while, thrombolysis/ fibrinolysis is the process of dissolution of clot by the degradation of fibrin into soluble fragments[12]. Under normal hemostasis condition, there is balance between blood coagulation and fibrinolysis. But, when there is pathological abnormality under disease condition the imbalance is observed, which may lead to thrombus formation[13].
Antithrombotic drugs such as anticoagulants, antiplatelet and fibrinolytic drugs either target procoagulant factors or dissolve the thrombus formed. These are used as prophylaxis or treatment of arterial and venous thrombosis[14-16]. Currently used anticoagulants are heparin, warfarin and their derivatives. Antiplatelet drugs that are currently used are aspirin, clopidogrel and abciximab[16-19]. Fibrinolytic agents are proteases which have potential to degrade thrombi directly or indirectly. Proteases acting on thrombi directly are plasmin, brinase and trypsin; while, protease dissolving thrombi indirectly is Tissue Plasminogen Activator (t-PA)[20–23]. The major concerns against using these thrombolytic drugs are excessive bleeding, anaphylactic reactions, heparin-induced thrombocytopenia and toxicity[24–28]. Hence, currently there is lot of interest in the use of natural products as the new thrombolytic drugs with more potency, safety, chemical stability which is devoid of risk of hemorrhage and toxicity from natural sources.
Medicinal plants are enormous and important source for the indigenous medical systems[29]. Psidium guajava (P. guajava), belongs to family Myrtaceae, commonly called as ‘guava’, is a small medicinal tree native to South America but extends throughout the European, African and Asian continents[30]. P. guajava is used as both nutraceutical (fruit) and medicinal (leaves, bark, seeds, roots) agents in traditional formulations[31]. P. guajava is mostly grown for the food industry to produce candies, juices, jams and frozen pulp[32]. The decoctions or pastes of whole plant or shoots are used to treat skin ailments and as astringents in dysmenorrhea, miscarriages, uterine bleeding and premature labor. The bark is used as an astringent to treat of ulcers wounds and diarrhea. The leaf extracts are used as febrifuge, antispasmodic, to treat viral disease and for rheumatism[33]. The seeds are associated with antimicrobial, gastrointestinal and anticarcinogenic activities. The different extracts of the parts of the tree are known to possess antimicrobial, hypoglycaemic, anti-inflammatory, anti-hypertensive, gastroenteritis, dysentery, conjunctivitis, constipation and antitumour effect[31,34-36]. In addition, it is used to treat hyperlipidemia, diabetes mellitus, cardiovascular diseases, liver disorders and parasitic infection[37]. Previous studies have shown that P. guajava inhibits cholesterol levels and incidence of stroke[32]. But the mechanism of inhibition of stroke is not clearly depicted. Different parts of the plants have been used in traditional system of medicines. But there is no report for the presence of protease in the seeds of P. guajava fruit. This is the first report on protease associated with anticoagulant and fibrinolytic activity from the seeds of P. guajava fruit.
Materials and Methods
Materials:
Casein, gelatin and human fibrinogen were purchased from Sigma-Aldrich chemicals, St. Louis, MO, USA. Liquicellin and uniplastin was procured from Tulip Diagnostics Pvt. Ltd, Goa, India. All other reagents were of analytical grade.
Sample collection:
Pink P. guajava fruits were purchased from local market. The fruits were cut and the seeds were taken out of the fruit part. The collected seeds were washed with distilled water, wiped with filter paper and then shade dried at room temperature for 3 to 4 d. The dried seeds were then pulverized into fine powder.
Preparation of extract:
The finely powdered seeds were weighed using digital balance and extracted with sodium phosphate buffer (10 mM, pH 7.0) in the ratio 1:25 on a magnetic stirrer at 500 rpm for about 24 h at room temperature. The P. guajava Seed Extract (PgSE) was collected and stored at -20° until further use.
Electrophoresis:
Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE) was performed as described by Laemmli[38],. Briefly, PgSE (160 µg) was treated with non-reducing sample buffer and incubated in boiling water bath for 3 min. The sample was then loaded onto 12 % SDS-PAGE along with standard molecular weight markers ranging from 7 to 175 kDa and electrophoresis was performed. The gel was stained with 0.25 % Coomassie brilliant blue R-250 to visualize the protein bands.
Zymography:
Zymography was carried out according to the method of Laemmli[38]. Briefly, the 12 % gel was incorporated with 0.2 % casein and gelatin separately as substrate for the detection of proteolytic activity. The PgSE (80 µg) in 10 mM sodium phosphate buffer (pH 7.0) was incubated with non-reducing sample buffer at 37° for 30 min and electrophoresis was carried out. After electrophoresis, the gels were washed with 2.5 % of Triton X-100 for 1 h to remove SDS. The gels were incubated overnight in incubation buffer containing Tris–HCl (50 mM, pH 7.6, 10 mM calcium chloride (CaCl2) and 150 mM Sodium Chloride (NaCl)). The gels were then stained with 0.25 % Coomassie brilliant blue R-250 to observe the activity bands.
Protease activity:
Protease activity was performed as described by Satake et al.[39], using casein (2 % in 200 mM Tris–HCl buffer, pH 7.0) as substrate. Briefly, 0.4 ml of casein was incubated with different concentration of PgSE (0-300 µg) in 10 mM sodium phosphate buffer (pH 7.0) at 37º for 2.5 h. About 1.5 ml of Trichloroacetic acid (TCA) (0.44 M) was added to terminate the reaction and allowed to stand for 30 min at room temperature. The mixture was centrifuged at 3000 rpm for 5 min and the supernatant (1 ml) was mixed with 0.4 M sodium carbonate (2.5 ml) and 1:2 diluted Folin reagent (0.5 ml). The colour developed was read at 660 nm. One unit of enzyme activity was defined as the amount of enzyme required to increase in the absorbance of 0.01 at 660 nm. Protease activity was expressed as units/min/mg.
Fibrinogenolytic activity:
Fibrinogenolytic activity was performed as described by Gubbiveeranna et al.[13]. Briefly, human fibrinogen (50 µg) was treated with different concentration of PgSE (0.5–20 µg) in 25 µl of 10 mM sodium phosphate buffer (pH 7.0) and incubated at 37° for 2.5 h. Reducing sample buffer (10 µl) containing 1M urea, 4 % SDS and 4 % beta (β)-mercaptoethanol was added to terminate the reaction and kept in boiling water bath for 3 min. The hydrolyzed products were analyzed in 12 % SDS- PAGE and visualized by staining with Coomassie brilliant blue R-250.
Fibrinolytic activity:
Fibrinolytic activity was carried out according to the method of Shivaiah and Kempaiah[40]. Briefly, trisodium citrate (3.2 %) treated blood in the ratio 1:9 was centrifuged at 3000 rpm for 5-10 min. The supernatant obtained was separated and used as Platelet Poor Plasma (PPP). Equal volume of PPP (100 µl) and 25 mM CaCl2 (100 µl) was incubated at 37º to get fibrin clot. The clot formed was thoroughly washed with 10 mM sodium phosphate buffer (pH 7.0) for 5-6 times. The washed fibrin clot was incubated with different concentration of PgSE (15 to 120 µg) in a total reaction volume of 40 μl of 10 mM sodium phosphate buffer (pH 7.0) at 37º for 2.5 h. Reducing sample buffer (20 µl) containing 1M urea, 4 % SDS and 4 % β-mercaptoethanol was added to terminate the reaction and kept in boiling water bath for 3 min. An aliquot (20 μl) of the supernatant was subjected to 10 % SDS-PAGE to analyze fibrin hydrolyzing pattern.
Recalcification time (RT):
Plasma recalcification time was determined according to the method described by Gubbiveeranna et al.[13]. Briefly, PPP (100 µl) was pre-warmed to 37º before use and incubated with different concentration of PgSE (0-80 µg) in 10 mM sodium phosphate buffer (pH 7.0) at 37° for 5 min. Later, 100 µl of 25 mM CaCl2 was added and the clotting time was recorded. The 10 mM sodium phosphate buffer (pH 7.0) alone without PgSE was considered as negative control.
Protein estimation:
The protein concentration was estimated as described by Lowry et al.[41]. Briefly, bovine serum albumin (BSA) was used as standard and the protein concentration of PgSE was determined by comparing with known concentration of BSA.
Statistical analysis:
The experiments were performed in triplicates and the data obtained from the experiments were expressed as mean±standard error of mean (SEM). The results were statistically analysed using one–way analysis of variance (ANOVA) followed by Tukey’s Multiple Comparison Test. The data were considered significant at p<0.05.
Results and Discussion
The PgSE (160 µg) was treated with non-reducing sample buffer and incubated in boiling water bath for 3 min. The sample was then loaded onto 12 % SDS-PAGE under non-reducing condition. After electrophoresis, the gel was stained with Coomassie Brilliant Blue R-250 to visualise the protein bands. PgSE exhibited dense bands in the high molecular weight region between 122 kDa and 47 kDa. Two distinct bands were seen in the lower molecular weight region at ~16.6 kDa and ~14.3 kDa (fig. 1).
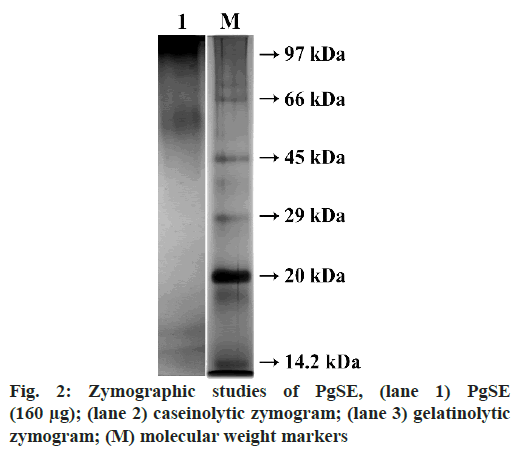
PgSE was evaluated for proteolytic activity using 2 % casein as substrate. It showed a specific activity of 0.8 U/mg/ml. Casein (0.2 %) and gelatin (0.2 %) were copolymerized with the polyacrylamide gel separately for the detection of proteolytic activity. PgSE (80 µg) was incubated with non-reducing sample buffer at 37º for 1.5 h and loaded onto 12 % SDS-PAGE under non- reducing condition. After electrophoresis, gels were washed with 2.5 % of Triton X-100 for 1 h to remove SDS. The gels were incubated overnight in incubation buffer containing Tris–HCl (50 mM, pH 7.6, 10 mM Cacl2 and 150 mM NaCl). Gels were then stained with 0.25 % Coomassie brilliant blue R-250 to visualize the activity bands. The bands were observed at molecular weight region ~97 kDa and between 34 kDa and 18.2 kDa for caseinolytic activity. Similarly, translucent activity bands in the molecular weight region ~96.7 kDa and between 52.3 kDa and 24.9 kDa were seen for the gelatinolytic activity (fig. 2).
PgSE was studied for fibrinogenolytic activity using human fibrinogen, which is a 340 kDa soluble plasma glycoprotein, composed of three subunits (Aα, Bβ and γ). Fibrinogen plays a crucial role in arresting blood during vascular injury by getting converted to fibrin upon action of thrombin. The fibrin is subsequently converted to fibrin-based blood clot. PgSE hydrolysed all the chains (Aα, Bβ and λ) of fibrinogen in a dose dependent manner. The fibrinogen (50 µg) was incubated with different concentration of PgSE (0.5 µg to 20 μg) in 10 mM sodium phosphate buffer (pH 7.0) at 37° for 2.5 h. The reaction was terminated by adding denaturing sample buffer containing 1 M urea, 4 % SDS and 4 % β-mercaptoethanol and kept in boiling water bath for 3 min. SDS-PAGE (12 %) was performed in reducing condition to visualize degradation pattern. After electrophoresis, the gel was stained with 0.25 % Coomassie brilliant blue R-250 (fig. 3).
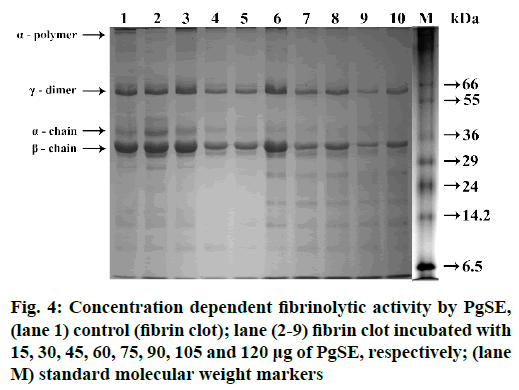
The fibrinolytic activity of PgSE was studied using human fibrin. PgSE hydrolyzed fibrin clot in a dose dependent manner. α-polymer and α-chain subunits of fibrin were hydrolysed at 90 µg followed by partial degradation of λ-dimer and β-chain subunits at a concentration of 120 µg. Equal volumes of PPP (100 µl) and 25 mM Cacl2 (100 µl) were mixed and incubated to get fibrin clot. The fibrin clot was washed in 10 mM sodium phosphate buffer (pH 7.0) and incubated with different concentration of PgSE (15 µg to 120 µg) in 10 mM sodium phosphate buffer (pH 7.0) at 37° for 2.5 h. The reaction was terminated by adding denaturing sample buffer containing 1 M urea, 4 % SDS and 4 % β-mercaptoethanol and kept in boiling water bath for 3 min. The sample was loaded onto 10 % SDS-PAGE in reducing condition. After electrophoresis, the gel was stained with Coomassie Brilliant blue R-250 to visualize the bands (fig. 4).
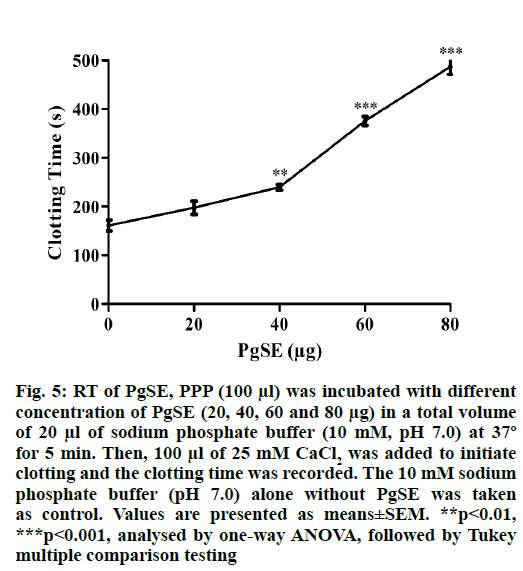
PgSE which was found to contain protease associated with fibrinogenolytic and fibrinolytic activity showed anticoagulant activity upon incubation with PPP. PgSE increased the recalcification time in a dose dependent manner indicating its anticoagulant property. PgSE increased the clotting time from control 161 s to 487 s at a concentration of 80 µg. The PgSE increased the clotting time by 3.03-folds (fig. 5).
Fig. 5: RT of PgSE, PPP (100 μl) was incubated with different concentration of PgSE (20, 40, 60 and 80 μg) in a total volume of 20 μl of sodium phosphate buffer (10 mM, pH 7.0) at 37º for 5 min. Then, 100 μl of 25 mM CaCl2 was added to initiate clotting and the clotting time was recorded. The 10 mM sodium phosphate buffer (pH 7.0) alone without PgSE was taken as control. Values are presented as means±SEM. **p<0.01, ***p<0.001, analysed by one-way ANOVA, followed by Tukey multiple comparison testing
Pathophysiological associated with hypertension, diabetes, inflammation, hypercholesterolemia, genetic diseases and metabolic syndrome reported to disturb the hemostatic and fibrinolytic mechanisms by abruptly activating blood coagulation, hypofibrinolysis and platelet hyperaggregation[42]. With the new research area focusing on identifying new anticoagulants, much attention has been focused on potent anticoagulants from the natural source particularly from medicinal plant extracts with more efficiency, safety, chemical stability and devoid of side effects.
Several medicinal plant extracts have been reported to exhibit anticoagulant activity[10]. Compounds such as chitosan, glucan, polysaccharides, proteins, sulfated flavonoids, tannins, tea catechins and triterpenes have been reported to show anticoagulant activity and/or antithrombotic effects[43]. P. guajava is traditionally used for various therapeutic purposes. Studies have reported that P. guajava leaf extract showed significant anticoagulant activity associated with protective effect on antithrombin III activity[44]. The leaf extract has been shown to contain three phenolic compounds gallic acid, ferulic acid and quercetin. The anticoagulant nature of the leaf extract has been attributed to these phenolic compounds[45]. Hydroalcoholic leaves extract of P. guajava and its bioactive fractions showed antithrombocytopenic activity. The analysis showed the presence of phenolic and flavonoid compounds such as quercetin, ellagic acid and kaempferol[37].
In this study, we have analysed PgSE for the presence of protease and its effect on fibrinogen, fibrin and blood coagulation cascade to evaluate its anticoagulant property. PgSE showed protein bands in high molecular weight region between 122 kDa and 47 kDa and two distinct bands in the lower molecular weight region at ~16.6 kDa and ~14.3 kDa as analysed by SDS-PAGE. The protein concentration in PgSE was found to be 1 mg/ml. PgSE showed a specific activity of 0.8 U/mg/ ml using casein as substrate. The proteolytic activity was supported by zymography studies using casein and gelatin as substrates. In zymography studies, PgSE hydrolyzed casein at molecular weight region ~97 kDa and between 34 kDa and 18.2 kDa. The degradation of gelatin was observed in the molecular weight region ~96.7 kDa and between 52.3 kDa and 24.9 kDa.
Plant proteases are involved in variety of physiological processes in plant life such as regulated cell death, photosynthesis, immune response, embryogenesis, remodelling of extracellular matrix and regulation of signal transduction[46-48]. Plant proteases have been extracted in aqueous solution from different parts of the plants such as flowers, seeds, roots and leaves[5]. Plant proteases have high stability, unique substrate specificity and wide pH range for enzyme activity[49-51]. Hence, proteases isolated from plants have been used to treat diverse diseases. Some of them exhibit antitumor, antimicrobial and antifungal activities[50]. The plant proteases have been widely used in wound healing, digestion disorders, burn debridement and blood coagulation, thrombolysis process and oral healthcare[52].
PgSE showed fibrinogenolytic activity and degraded the subunits of fibrinogen and fibrin. The degradation is dose dependent manner. PgSE hydrolyzed Aα, Bβ and γ chains of fibrinogen at a concentration of 20 µg while the fibrin subunits (α-polymer and α-chain) were degraded at a comparatively high concentration of 120 µg. γ-dimer and β-chains of fibrin were resistant for hydrolysis. The α-polymer and α-chain are more susceptible for degradation compared to the γ-dimer and β-chains of fibrin. PgSE hydrolyzed the cross-linked fibrin subunits. Fibrin together with platelet aggregate stabilizes the platelet plug, which stops bleeding from vascular injuries. Anticoagulant plant proteases have been studied which increases the plasma coagulation time. These proteases digest fibrinogen in different pattern; some hydrolyse fibrinogen completely while others show partial hydrolysis. Most of the proteases also showed fibrinolytic activity and ability to lyse blood clots. Some of the proteases are Kitamase from Aster yomena, protease from P. japonicas, Hirtin from Euphornia hirta, Codiase from Codium fragile and protease from Costaria costata[52].
PgSE interfered in the blood coagulation cascade as analysed by RT. PgSE, at a concentration of 80 µg, prolonged the clotting time in RT from control 161 s to 487 s, increasing the RT by ~3.03 folds. This clearly indicates the anticoagulant nature associated with PgSE. The increase in clotting time of plasma in RT method indicates the interference of PgSE in common pathway of blood coagulation.
In conclusion, PgSE exhibited proteolytic activity degrades fibrinogen, fibrin and inhibited blood coagulation cascade indicating its interference in thrombus formation. Our study provides for the first time, the scientific validation for the presence of a protease with anticoagulant property from PgSE. This study provides an opportunity to identify, isolate and characterize the active principle from PgSE having clinical significance and decipher its molecular mechanism.
Conflicts of interest:
There is no conflict of interest to disclose.
References
- Levine SR, Brott TG. Thrombolytic therapy in cerebrovascular disorders. Prog Cardiovasc Dis 1992;34(4):235-62.
- Davie EW, Fujikawa K, Kisiel W. The coagulation cascade: initiation, maintenance and regulation. Biochemistry 1991;30(43):10363-70.
- Huisman MV, Barco S, Cannegieter SC, Gal G Le, Konstantinides SV, Reitsma PH, et al. Pulmonary embolism. Nat Rev Dis Primers 2018;4:18028.
- Osman AA, Ju W, Sun D, Qi B. Deep venous thrombosis: a literature review. Int J Clin Exp Med 2018;11:1551-61.
- Gubbiveeranna V, Kusuma CG, Bhavana S, Sumachirayu CK, Ravikumar H, Nagaraju S. Potent procoagulant and platelet aggregation inducing serine protease from Tridax procumbens extract. Pharmacogn Res 2019;11(4):363-70.
- Min SK, Park JS, Luo L, Kwon YS, Lee HC, Jung H, et al. Assessment of C-phycocyanin effect on astrocytes-mediated neuroprotection against oxidative brain injury using 2D and 3D astrocyte tissue model. Sci Rep 2015;5:14418.
- Mackman N. Triggers, targets and treatments for thrombosis. Nature 2008;451(7181):914-8.
- Yoo J, Chung CS, Kang SS. Relation of plasma homocyst(e)ine to cerebral infarction and cerebral atherosclerosis. Stroke 1998;29(12):2478-83.
- Rajendran P, Rengarajan T, Thangavel J, Nishigaki Y, Sakthisekaran DP, Sethi G, et al. The vascular endothelium and human diseases. Int J Biol Sci 2013;9(10):1057-69.
- Kusuma CG, Gubbiveeranna V, Sumachirayu CK, Bhavana S, Nagaraju S. In vitro anti-oxidant and anti-platelet activity of Manilkara zapota (L.) P Royen latex. Int J Botany Stud 2020;5(5):405-8.
- Xie P, Zhang Y, Wang X, Wei J, Kang W. Antithrombotic effect and mechanism of Rubus spp. Blackberry. Food Funct 2017;8:2000-12.
- Gubbiveeranna V, Pavan KMA, Kusuma CG, Bhavana S, Sumachirayu CK, Sneharani AH, et al. Role of Tridax procumbens in green synthesis of cerium oxide nanoparticles and its effect on blood coagulation cascade and platelet aggregation. Int J Botany Stud 2020;5(3):79-86.
- Gubbiveeranna V, Kusuma CG, Bhavana S, Sumachirayu CK, Nagaraju S. Anti-hemostatic protease from Jatropha curcas latex with fibrinogen lytic activity. J Pharmacogn Phytochem 2019;8:1303-10.
- Simkhada JR, Cho SS, Mander P, Choi YH, Yoo JC. Purification, biochemical properties and antithrombotic effect of a novel Streptomyces enzyme on carrageenan-induced mice tail thrombosis model. Thromb Res 2012;129(2):176-82.
- Cakarer S, Eyupoglu E, Gunes CO, Kuseoglu BG, Berberoglu HK, Keskin C. Evaluation of the hemostatic effects of ankaferd blood stopper during dental extractions in patients on antithrombotic therapy. Clin Appl Thromb Hemost 2013;19(1):96-9.
- Chen C, Yang FQ, Zhang Q, Wang FQ, Hu YJ, Xia ZN. Natural products for antithrombosis. Evid Based Complement Alternat Med 2015;2015:876426.
- Becattini C, Agnelli G. Aspirin for prevention and treatment of venous thromboembolism. Blood Rev 2014;28(3):103–8.
- Fuentes E, Caballero J, Alarcon M, Rojas A, Palomo I. Chlorogenic acid inhibits human platelet activation and thrombus formation. Plos One 2014;9(3):e90699.
- Wang TY, Magid DJ, Ting HH, Li S, Alexander KP, Roe MT, et al. The quality of antiplatelet and anticoagulant medication administration among ST-segment elevation myocardial infarction patients transferred for primary percutaneous coronary intervention. Am Heart J 2014;167(6):833-9.
- Butcher K, Shuaib A, Saver J, Donnan G, Davis SM, Norrving B, et al. Thrombolysis in the developing world: is there a role for streptokinase? Int J Stroke 2013;8:560-5.
- Gomaraschi M, Ossoli A, Vitali C, Pozzi S, Serdoz VL, Pitzorno C, et al. Off-target effects of thrombolytic drugs: apolipoprotein A-I proteolysis by alteplase and tenecteplase. Biochem Pharmacol 2013;85:525-30.
- Alkjaersig N, Fletcher AP, Sherry S. The mechanism of clot dissolution by plasmin. J Clin Invest 1959;38(7):1086–95.
- Collen D. The plasminogen (fibrinolytic) system. Thromb Haemost 1999;82(2):259-70.
- Beijering RJR, Cate H ten, Cate JW ten. Clinical applications of new antithrombotic agents. Ann Hematol 1996;72(4):177-83.
- Risch L, Huber AR. Improving the definition of heparin-induced thrombocytopenia. Arch Intern Med 2004;164(15):1699.
- Bhakuni T, Ali MF, Ahmad I, Bano S, Ansari S, Jairajpuri MA. Role of heparin and non-heparin binding serpins in coagulation and angiogenesis: A complex interplay. Arch Biochem Biophys 2016;604:128-42.
- Baruah DB, Dash RN, Chaudhari MR, Kadam SS. Plasminogen activators: A comparison. Vascul Pharmacol 2006;44(1):1–9.
- Wiman B, Collen D. Molecular mechanism of physiological fibrinolysis. Nature 1978;272(5653):549-50.
- Gubbiveeranna V, Nagaraju S. Ethnomedicinal, phytochemical constituents and pharmacological activities of Tridax procumbens: a review. Int J Pharm Pharm Sci 2016;8:1-7.
- Kaneria M, Chanda S. Phytochemical and pharmacognostic evaluation of leaves of Psidium guajava L. (Myrtaceae). Pharmacogn J 2011;3(23):41-5.
- Gutierrez RMP, Mitchell S, Solis RV. Psidium guajava: A review of its traditional uses, phytochemistry and pharmacology. J Ethnopharmacol 2008;117(1):1-27.
- Barbalho SM, Farinazzi-Machado FM V, Goulart R de A, Brunnati ACS, Ottoboni AMMB, Nicolau CCT. Psidium guajava (Guava): A plant of multipurpose medicinal applications. Med Aromat Plants 2012;1:1000104.
- Ravi K, Divyashree P. Psidium guajava: A review on its potential as an adjunct in treating periodontal disease. Pharmacogn Rev 2014;8(16):96–100.
- Rishika D, Sharma R. An update of pharmacological activity of Psidium guajava in the management of various disorders. Int J Pharm Sci Res 2012;3:3577–84.
- Chandra RGS, Ramesh TN, Veeranna VG, Maiya PS. Antimicrobial activity studies of zinc oxide, zinc acetate and layered zinc hydroxysalt. Int J Sci Res 2013;01:524-30.
- Nagaraju S, Gubbiveeranna V, Mudili V, Dhananjaya MC, Rangappa S, Ramachandrappa LT, et al. Microbial Catalysts. Nova Science Publishers; 2019.
- Ahmad A, Ibrahim M, Chester K, Khan W, Ahmad S, Ansari S. Antithrombocytopenic potential of bioactivity guided fractions of traditionally used Psidium guajava Linn. leaves in busulfan induced-thrombocytopenic rats. Pharmacogn Mag 2019;15:S440-8.
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970;227(5259):680–5.
- Satake K, Okuyama T, Ohashi M, Shinoda T. The spectrophotometric determination of amine, amino acid and peptide with 2,4,6-trinitrobenzene 1-sulfonic acid. J Biochem 1960;47:654-60.
- Shivaiah N, Kempaiah K. ‘Partitagin’, a unique β, γ-fibrinogenase that inhibits platelet aggregation from Hippasa partita spider venom. Blood Coagul Fibrinolysis 2011;22:24-8.
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin-phenol reagent. J Biol Chem 1951;193:265–75.
- Fritsma GA, Walenga JM. Thrombotic Disorders and Laboratory Assessment. Rodak’s hematology: clinical principles and applications. 6th ed. Canada: Elsevier; 2020.
- Akram M, Rashid A. Anti-coagulant activity of plants: mini review. J Thromb Thrombolysis 1234;44:406-11.
- Nazeruddin GM, Pingale SS, Shaikh SS. Pharmacological review of Tridax procumbens L. Der Pharma Sinica 2011;2:172–5.
- Hsieh CL, Lin YC, Yen GC, Chen HY. Preventive effects of guava (Psidium guajava L.) leaves and its active compounds against α-dicarbonyl compounds-induced blood coagulation. Food Chem 2007;103:528–35.
- Schaller A. A cut above the rest: the regulatory function of plant proteases. Planta 2004:183-97.
- Ahmad Shah M, Ahmad Mir S, Amir Paray M. Plant proteases as milk-clotting enzymes in cheesemaking: a review. Dairy Sci Technol 2014;94:5-16.
- Cui L, Xing M, Xu L, Wang J, Zhang X, Ma C, et al. Antithrombotic components of Malus halliana Koehne flowers. Food Chem Toxicol 2018;119:326-33.
- Gubbiveeranna V, Kusuma C, Sumachirayu C, Bhavana S, Ravikumar H, Nagaraju S. Anticoagulant and fibrino(geno)lytic protease from Leucas aspera. Int J Pharm Sci Res 2021;12:2470-6.
- Gubbiveeranna V, Shivaiah N. Ethnomedicinal, phytochemical constituents and pharmacological activities of Tridax procumbens: A review. Int J Pharm Pharm Sci 2016;8:1-7.
- Thoyajakshi RS, Poornima D. Anticoagulant, fibrinogenolytic and anti-platelet aggregation activities of Lablab purpureus (L.) Sweet seed radicle aqueous extract. Plant Sci Today 2021;8:89-94.
- Anastasia VB, Natalia VK, Anastasiia IP, Lyudmila VS, Andrey AZJ. Trends and Prospects of Plant Proteases in Therapeutics. Curr Med Chem 2019;26:465-86.