- *Corresponding Author:
- Yanyun Zhu
Department of Medical Oncology,
The Fifth Medical Center of People's Liberation Army General Hospital,
Haidian,
Beijing
100071,
China
E-mail: zhuyymm06@163.com
| This article was originally published in a special issue, “Modern Applications in Biomedical Research and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2022:84(3) Spl Issue “136-143” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
To investigate the association between tumor protein 53 polymorphism and esophageal cancer cases using systematic review and meta-analysis, this provides a basis for future researches. All studies on tumor protein 53 mutations in patients with esophageal cancer and its influencing factors, which published before May 2021, were collected by searching Cochrane Library, PubMed, Embase, China National Knowledge Infrastructure and Wanfang data. Meta-analysis was performed using RevMan 5.3. Ten eligible studies (Chinese n=7, English n=3) involving 1379 cases were included in this analysis. Meta-analysis showed that the odds ratio value (95 % confidence interval) of tumor protein 53 mutations in Caucasian patients with esophageal cancer was 70.51 (9.77, 508.88), z=4.22, p<0.0001; the odds ratio value (95 % confidence interval) of tumor protein 53 mutations in Chinese Han patients with esophageal cancer was 1.06 (0.63, 1.79), z=0.22, p=0.83; the odds ratio value (95 % confidence interval) of tumor protein 53 mutations in Chinese Uyghur patients with esophageal cancer was 18.48 (4.44, 76.90), z=4.01, p<0.0001. The odds ratio value (95 % confidence interval) of tumor protein 53 mutations in patients with esophageal cancer was 1.01 (0.60, 1.72), z=0.05, p=0.96; the odds ratio value (95 % confidence interval) of tumor protein 53 deletions in patients with esophageal cancer was 23.02 (9.04, 58.63), z=6.57, p≤0.00001. There was no significant difference in tumor protein 53 mutations between patients with esophageal cancer and the control group. There is a certain correlation between tumor protein 53 polymorphism and the incidence of esophageal cancer and the change of tumor protein 53 gene will affect the risk and prognosis of esophageal cancer.
Keywords
Esophageal cancer, tumor protein p53, gene polymorphism, meta-analysis
Esophageal Cancer (EC) is one of the most common malignancies of the digestive tract in the world, mainly divided into Esophageal Squamous Cell Carcinoma (ESCC) and Esophageal Adenocarcinoma (EAC)[1]. The incidence of EC varies in different regions and races worldwide. China is a region with a high incidence of EC. Different from Europe and the United States, squamous cell carcinoma have majority of cases in China[2] and some ethnic minorities have higher risk of EC[3].
Activation of proto-oncogenes and inhibition of tumor suppressor genes are currently recognized molecular mechanisms of cancer. EC has been proved to be related to activation of multiple genes. A study has firstly performed a comprehensive analysis of EC using Whole Genome Sequencing (WGS), Whole Exome Sequencing (WES) and array Comparative Genomic Hybridization (aCGH), which showed that more than 83 % of ESCC contained mutations and polymorphisms of Tumor Protein 53 (TP53) gene and the abnormal expression of TP53 in Barrett's Esophagus (BE) was also one of the risk factors for EAC[4]. TP53 gene is an important tumor suppressor gene located on human chromosome 17p13.1. It mainly affects the function of downstream genes through transcription, which mainly includes induction of cell cycle arrest, Deoxyribonucleic Acid (DNA) repair, changes in cell metabolism, cell senescence, cell apoptosis, etc.[5]. It is considered that high frequency mutations or gene deletions of TP53 gene are common in various progressive and metastatic tumors of cancer and the mutant TP53 not only no longer has the original tumor inhibition function of wild-type p53 gene, but also can promote tumor formation[6]. Moreover, TP53 gene has a variety of polymorphisms, which plays an important role in the prognosis, disease development and inheritance of tumors. In this study, we performed a metaanalysis of foreign and domestic studies to estimate the association between TP53 polymorphism and the cases of EC, which provides a medical basis for the risk analysis of EC.
Materials and Methods
Publication search:
We searched the articles using the terms “EC”, “ESCC”, “TP53”, “Single Nucleotide Polymorphisms (SNPs)” in Cochrane Library, PubMed, Embase, China National Knowledge Infrastructure (CNKI) and Wanfang database. Among them, the Chinese search terms were “TP53 gene” and “EC”. The above terms were searched by combining subject words and free words.
Inclusion criteria:
Research type contains domestic and overseas studies on the correlation between TP53 mutations and polymorphism and patients with EC published in English and Chinese; research object has tissues of patients diagnosed with EC were taken for Fluorescence In Situ Hybridization (FISH) test to detect TP53 mutations and deletion and outcome indicator was to detect the TP53 polymorphism.
Exclusion criteria:
Duplicate publications; abstracts, reviews, case reports, conference papers; Non-Chinese and English literature; the full-text article was not available and the study that can’t be extracted or converted into required data.
Literature screening and data:
Two researchers independently screened the literature and extracted data according to the inclusion and exclusion criteria and cross-checked them. If there is any inconsistency, a decision will be made through discussion or consultation with a third researcher. The data extracted included the first author, year of publication, country, number of patients, age, body weight, gender, ethnicity, race, test method and genetic mutation, etc.
Quality evaluation of the included literature:
The quality of the included literature was evaluated using the Strengthening the Reporting of Genetic Association Studies (STREGA) tool, including, whether the research background, purpose and assumptions were clearly stated; whether the criteria of inclusion and exclusion is clear; whether variables are clearly defined; whether the concentration detection method is reliable; whether it conforms to Hardy-Weinberg balance; whether to provide demographics; whether statistical methods can be repeated and whether there is fund support[7]. Each item scored 1 point, the total score is 9 points and 6 points are for research with reliable quality[8].
Statistical analysis:
Meta-analysis was performed using RevMan 5.3. Measurement data were presented as “z” and its 95 % Confidence Interval (CI). The heterogeneity of the results was determined by χ2 test and the test level was alpha (α)=0.1. If statistical heterogeneity exists between studies (p<0.1, Chinese Journal of Hospital Pharmacy I2≥50 %), random effects model was used for analysis; instead, the fixed effects model was used for analysis. The publication bias was evaluated by Begg’s test and Egger’s test.
Results and Discussion
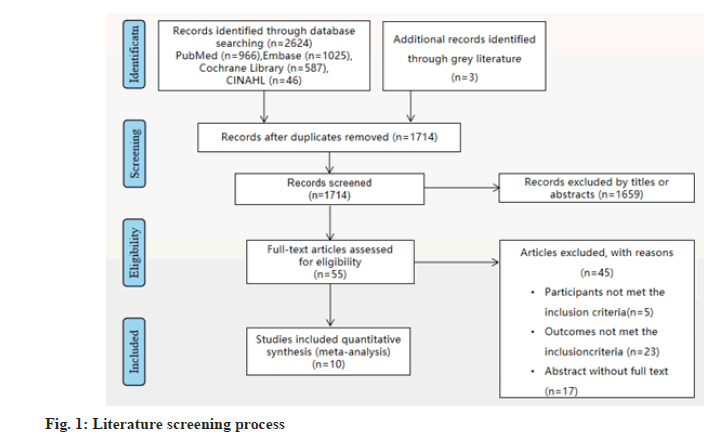
A total of 2624 related literatures were obtained in the preliminary examination. After deletions of duplicates and reading the titles, abstracts and full texts according to the inclusion and exclusion criteria, 10 literatures were finally included[3,9-17], involving a total of 1397 patients. The literature screening process was shown in fig. 1. The basic characteristics of the included literatures were shown in Table 1.
| Country | Mutation assessment methods | Nationality | Cases | Control | Histological classification of EC | |||
|---|---|---|---|---|---|---|---|---|
| First author and year of publication | Positive | Negative | Positive | Negative | ||||
| China | FISH | Uyghur | 22 | 67 | 1 | 67 | ESCC | |
| Abudoureheman A 2018 | ||||||||
| China | Niyaz M 2020 | FISH | Uyghur | 68 | 214 | 2 | 24 | ESCC (patients) |
| China | Xifeng T 2020 | FISH | Han | 6 | 19 | 0 | 10 | ESCC |
| Germany | Melling N 2019 | FISH | Caucasian | 110 | 269 | 0 | 129 | EAC |
| Germany | Melling N 2019 | FISH | Caucasian | 46 | 237 | 0 | 56 | ESCC |
| China | Niyaz M 2013 | FISH | Kazak | 22 | 40 | 0 | 10 | ESCC |
| China | Niyaz M 2020 | FISH | Uyghur | 56 | 117 | 2 | 97 | ESCC (tissues) |
| China | Shiping G 2002 | PCR | Han | 6 | 22 | 12 | 22 | ESCC |
| China | Lei Z 2002 | PCR | Han | 71 | 91 | 153 | 204 | ESCC |
| China | Shuying L 2017 | PCR | Han | 55 | 118 | 63 | 118 | ESCC |
| China | Ye J 2016 | PCR | Han | 2 | 30 | 0 | 30 | ESCC |
| China | Jianhui Z 2003 | PCR | Han | 136 | 173 | 96 | 136 | ESCC |
Note: PCR: Polymer Chain Reaction
Table 1: Basic characteristics of the included literatures
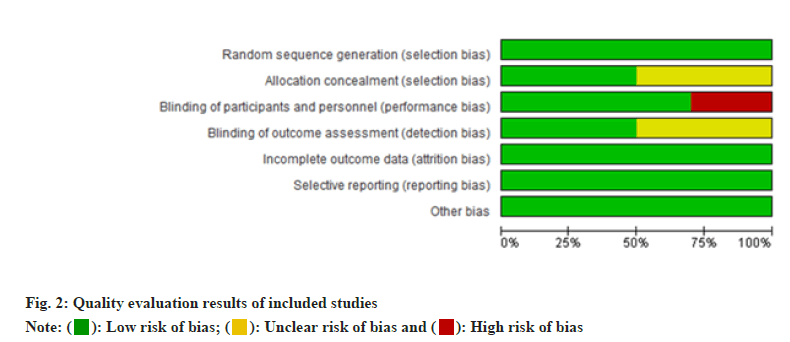
The results showed that the quality of 10 literatures[3,8-17] was reliable. The quality evaluation results of the included studies were shown in fig. 2.
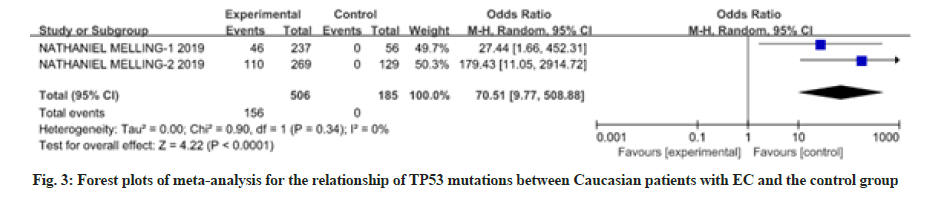
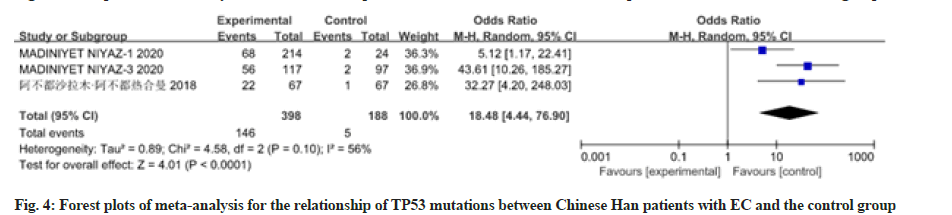
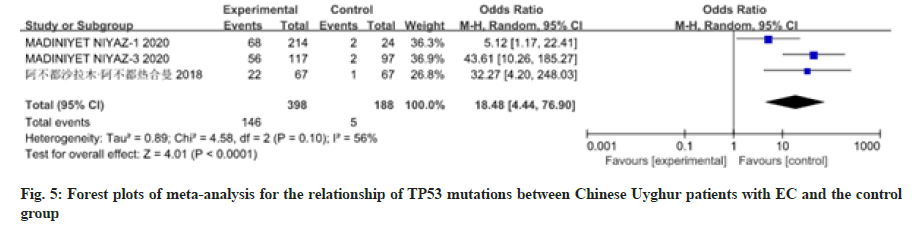
12 studies have reported the relationship between patients with EC of different races and nationalities and TP53 mutations. There was heterogeneity among the studies, so the random effects model was used for analysis, as shown in fig. 3-fig. 5. Meta-analysis results showed that the Odds Ratio (OR) value (95 % CI) of TP53 mutations in Caucasian patients with EC was 70.51 (9.77, 508.88), z=4.22, p<0.0001; the OR value (95 % CI) of TP53 mutations in Chinese Han patients with EC was 1.06 (0.63, 1.79), z=0.22, p=0.83; the OR value (95 % CI) of TP53 mutations in Chinese Uyghur patients with EC was 18.48 (4.44, 76.90), z=4.01, p<0.0001. There was no significant difference in TP53 mutations between Chinese Han patients with EC and the control group, while TP53 polymorphism in Caucasians and Chinese Uyghur patients with EC were statistically significantly different with the control group.
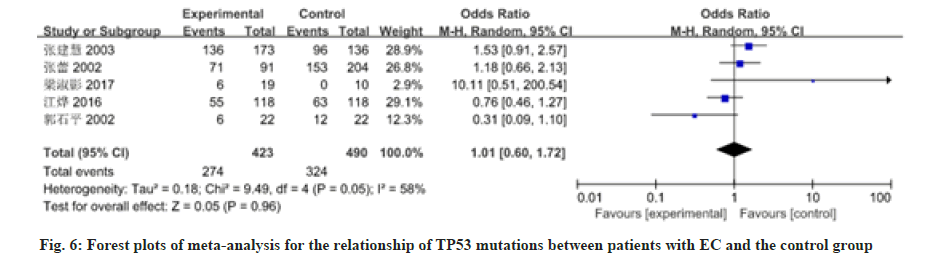
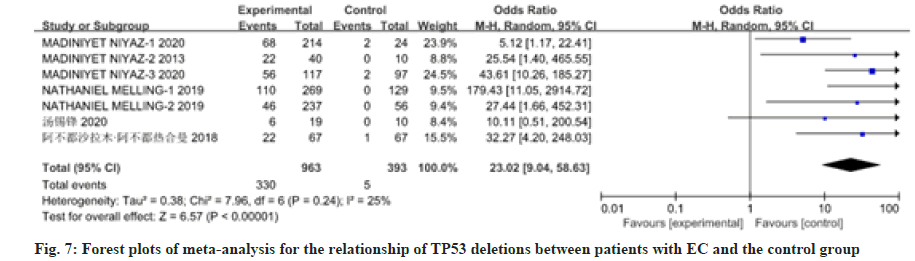
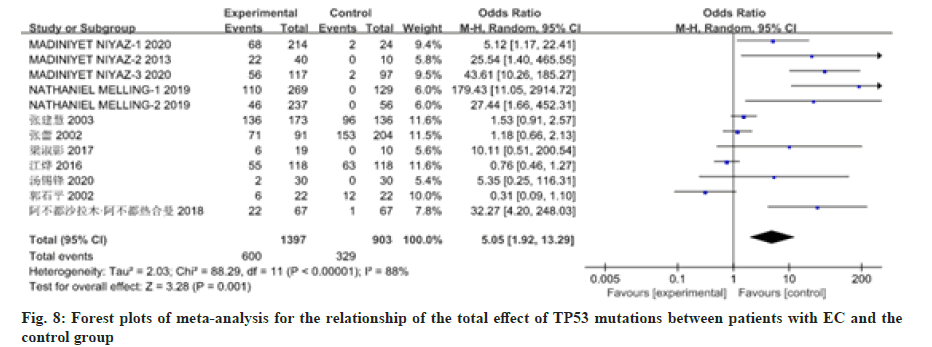
12 studies have reported on the association between patients with EC and mutations in the TP53 gene or deletion of the gene. There was heterogeneity among the studies, so the random effects model was used for analysis, as shown in fig. 6 and fig. 7. Meta-analysis results showed that the OR value (95 % CI) of TP53 mutations in patients with EC was 1.01 (0.60, 1.72), z=0.05, p=0.96; the OR value (95 % CI) of TP53 deletions in patients with EC was 23.02 (9.04, 58.63), z=6.57, p≤0.00001. There was no significant difference in TP53 mutations between patients with EC and the control group. The total effect of TP53 mutations in patients with EC was analyzed in the 12 studies included in the analysis. There was heterogeneity among studies (p≤0.00001, I2=0.88 %), so the random effects model was used for analysis, as shown in fig. 8. The result showed the OR value (95 % CI) was 5.05 (1.92, 13.29). Compared with the control group, the total effect of TP53 gene methylation in patients with EC was statistically significant (z=3.28, p=0.001).
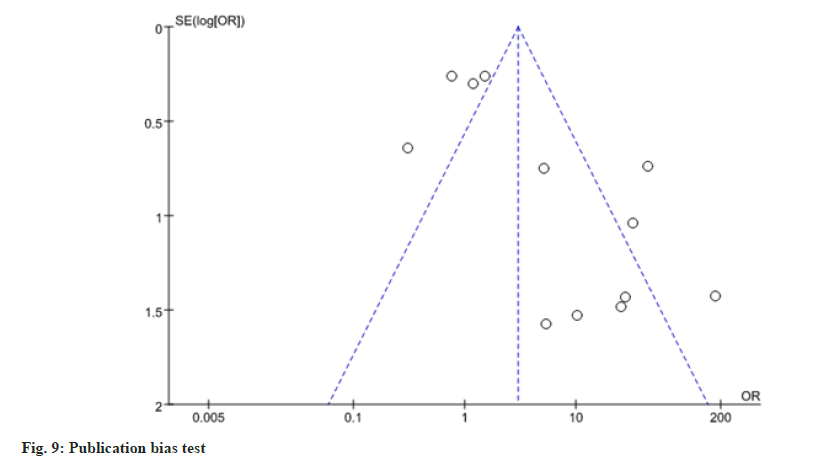
Stata 14.0 was used to conduct publication bias analysis based on whether TP53 gene methylation occurred in patients with EC. The results showed that p values of both Begg’ s test and Egger’s test were greater than 0.05 and the distribution on both sides of the funnel plot was roughly symmetrical, suggesting that the publication bias of this study was small, as shown in fig. 9. EC has a high morbidity in China and its related mortality ranks the top 5 among tumor diseases. However, the early diagnosis and treatment of EC is greatly hindered due to the lack of obvious early symptoms. 70 %-80 % of patients with EC can only be diagnosed after the disease progresses to the middle or advanced stage and the prognosis is very poor. Therefore, early diagnosis is of great clinical significance to improve the prognosis of patients with EC. Currently, the main treatment for EC is surgical resection, but some studies have shown that the overall survival rate of EC has not been significantly improved[18]. TP53 is the gene with the highest correlation with cancer among known tumor suppressor genes[19]. Mutation or polymorphism of TP53 is present in almost all types of cancer and TP53 mutations and polymorphism can be detected at the early stage of cancer, even before cell cancerization[20,21]. Therefore, TP53 plays a pivotal role in the early diagnosis and progression of EC. Studies have shown that patients with TP53 mutations have a higher degree of malignancy and worse prognosis[22], so TP53 polymorphism is of great significance for the early diagnosis of EC.
SNPs refer to the polymorphism of DNA sequence caused by the variation of single nucleotide at the gene level, which is the most common human heritable variation. The polymorphic structure of TP53 gene is very complex. At present, more than 80 TP53 polymorphisms have been identified and verified, most of which are located outside intron splicing sites or on non-coding exons. Most of the polymorphisms may be associated with cancer risk, but they have not been confirmed yet.
The TP53 72 codon is the most widely studied for TP53 polymorphism and it was first found by Dumont et al. that the TP53 72 codon Arg can strongly induce apoptosis[23]. Currently, the TP53 Arg72Pro is associated with an increased risk of EC, with significant ethnic differences. A large number of epidemiological evidence supports that Arg72Pro polymorphisms significantly increase the risk of EC in Asian populations[24]. In addition, TP53 gene also has other cancer-related polymorphisms.
V217M is the only non-silent polymorphism in the DNA binding domain, which can increase the reverse transcriptional activity of part of the TP53 response domain, such as Cyclin Dependent Kinase Inhibitor 1A (CDKN1A) and BCL2 Associated X (BAX) and has the possibility of anticancer effect. However, the activity of G360a is slightly reduced, which may increase the risk of cancer[25], but this has not been confirmed by epidemiological and clinical trials.
P47S polymorphism may reduce the risk of cancer by inducing phosphorylation at S46 and enhancing TP53-mediated apoptosis[26]. However, in animal experiments, this polymorphism only showed a mild phenotypic effect and its effect on cancer susceptibility remains to be determined.
This result of this study showed that, ethnic Uyghur was correlated with the prevalence of EC (OR value was 70.51, 18.48, 1.01 and 23.02 respectively), which was similar to the results of most current epidemiological surveys. This may be due to the inherent differences in cell rate[27] and drug sensitivity[28] caused by the natural single nucleic acid polymorphism of TP53 gene in people of different races and ethnicities, which lead to differences in the incidence and risk of EC. Therefore, we conclude that there are significant differences in the incidence of EC among different ethnic groups, which may be related to the naturally occurring polymorphism of TP53. The mutation and deletion of TP53 gene were correlated with the incidence of EC (OR value was 1.01, 23.02 respectively), which was consistent with the results of most current epidemiological surveys. The mutation or deletion of TP53 gene may directly affect the expression of p53 protein. Studies have shown[29] that p53 protein overexpression can be detected in precancerous cells of EC and the amount of p53 protein increases with the increase of the severity of the disease. During the development of esophageal squamous epithelial disease, more than 50 % of patients have p53 protein overexpression and the accumulation percentage of p53 protein in different types of lesions is very close to the corresponding mutation percentage of TP53 gene[30]. Therefore, we concluded that the risk and prognosis of EC are significantly related to gene mutations and gene deletions, which may be caused by gene deletions and mutations affecting the expression of p53 protein.
In conclusion, TP53 mutations are correlated with the incidence of EC to a certain extent and TP53 mutations will affect the risk and prognosis of EC. However, there are still some shortcomings in this study, the data of different ethnic groups and races included in this metaanalysis did not control the factors such as their living areas or bad living habits. TP53 mutations are not only related to genetics and genes, but also related to other non-genetic factors, such as diet, living environment, living habits and other factors[31]. The included literature in this study could not exclude mutations of TP53 gene caused by living habits in different regions, such as smoking and drinking, or environmental carcinogens in different environments. The number of Han cases included was small, because the TP53 mutations frequency of EC patients in different regions of China was greatly different and it was necessary to increase the number of cases of Han patients with EC in different regions for research. Only 9 literatures were included in this study, with a small number of literatures and small sample size. Therefore, further studies with larger samples and multiple regions are needed to draw more reliable conclusions, so as to provide ideas for reducing the risk of EC and improving the survival rate and prognosis of patients.
Acknowledgements
The work was supported by the Senior Department of Oncology, the Fifth Medical Center of People's Liberation Army General Hospital.
Conflict of Interests
The authors declare that they have no competing interests.
References
- Wang WP, Chen LQ. Current situation and prospective development of surgical treatment for esophageal carcinoma. Chin J Clin Thorac Cardiovasc Surg 2011;18(1):58-65.
- Niyaz M, Turghun A, Ping ZH, Zhu Z, Sheyhedin I, Ren C, et al. TP53 gene deletion in esophageal cancer tissues of patients and its clinical significance. Mol Med Rep 2013;7(1):122-6.
[Crossref] [Google Scholar] [Pub Med]
- Abudoureheman A, Niyaz M, Zhang HP. The gene phenotype, protein expression of TP53 gene in Uyghur’s esophageal carcinoma and its correlation with prognosis. Chin Med Herald 2018;15(17):17-20.
- Mayne ST, Risch HA, Dubrow R, Chow WH, Gammon MD, Vaughan TL, et al. Nutrient intake and risk of subtypes of esophageal and gastric cancer. Cancer Epidemiol Prev Biomarkers 2001;10(10):1055-62.
- Zhou JL. Research progress on molecular mechanism of p53 inactivation in cancers and drugs targeting p53. Life Sci Res 2010;2:1-11.
- Lu QR, Liu JX, Yu EQ. Detection and clinical significance of PIK3CA, TP53, PTEN genes mutation in breast cancer on next-generation sequencing. Chin J Curr Adv Gen Sur 2019;22(7):530-5.
- Little J, Higgins JP, Ioannidis JP, Moher D, Gagnon F, von Elm E, et al. Strengthening the Rtporting of Genetic Association Studies (STREGA)-An extension of the STROBE statement. Genet Epidemiol 2009;33(7):581-98.
[Crossref] [Google Scholar] [Pub Med]
- Liu M. Design and implementation of systematic evaluation: Meta-analysis. Beijing: People's Medical Publishing House; 2011. p. 72.
- Niyaz M, Ainiwaer J, Abudureheman A, Zhang L, Sheyhidin I, Turhong A, et al. Association between TP53 gene deletion and protein expression in esophageal squamous cell carcinoma and its prognostic significance. Oncol Lett 2020;20(2):1855-65.
[Crossref] [Google Scholar] [Pub Med]
- Zhang L. The relationship between genetic and epigenetic alterations in MGMT and TP53 genes and ESCC. Peking Union Medical College, China; 2002.
- Melling N, Norrenbrock S, Kluth M, Simon R, Hube-Magg C, Steurer S, et al. p53 overexpression is a prognosticator of poor outcome in esophageal cancer. Oncol Lett 2019;17(4):3826-34.
[Crossref] [Google Scholar] [Pub Med]
- Niyaz M, Turghun A, Ping ZH, Zhu Z, Sheyhedin I, Ren C, et al. TP53 gene deletion in esophageal cancer tissues of patients and its clinical significance. Mol Med Rep 2013;7(1):122-6.
[Crossref] [Google Scholar] [Pub Med]
- Tang XF, Ji XZ. Correlation of TP53 gene phenotype and protein expression with 19 cases of esophageal cancer in children in Sanya. Chin J Birth Health Heredity 2020;28(7):123-5.
- Zhang JH, Li Y, Wang R, Wen DG, Wu ML, He M. p53 gene polymorphism with susceptibility to esophageal cancer and lung cancer in Chinese population. Zhonghua Zhong Liu Za Zhi 2003;25(4):365-7.
- Guo SP. Loss of heterozygosity on p53 in human esophageal cancer. Shanxi Medical University, China; 2002.
- Liang SY. Study of TP53 mutation in esophageal squamous cell carcinoma. Zhengzhou Univ J 2008;5:26-9.
- Jiang Y. Analysis of TP53 gene mutation and its intratumor heterogeneity in esophageal squamous cell carcinoma. Anhui Med Univ 2002;24(3):55-9.
- Wang S, Zhao Y, Bernard D, Aguilar A, Kumar S. Targeting the MDM2-p53 protein-protein interaction for new cancer therapeutics. Protein Protein Interactions 2012;8:57-79.
- Lakin ND, Jackson SP. Regulation of p53 in response to DNA damage. Oncogene 1999;18(53):7644-55.
[Crossref] [Google Scholar] [Pub Med]
- Niyaz M, Turghun A, Ping ZH, Zhu Z, Sheyhedin I, Ren C, et al. TP53 gene deletion in esophageal cancer tissues of patients and its clinical significance. Mol Med Rep 2013;7(1):122-6.
[Crossref] [Google Scholar] [Pub Med]
- Chen TC, Wu CT, Wang CP, Lou PJ, Ko JY, Chang YL. The differences of immunologic and TP53 mutant phenotypes between synchronous and metachronous head and neck cancer and esophageal cancer. Oral Oncol 2020;111:104945.
[Crossref] [Google Scholar] [Pub Med]
- Uchino S, Saito T, Inomata M, Osawa N, Chikuba K, Etoh K, et al. Prognostic significance of the p53 mutation in esophageal cancer. Jpn J Clin Oncol 1996;26(5):287-92.
[Crossref] [Google Scholar] [Pub Med]
- Dumont P, Leu JL, Della Pietra AC, George DL, Murphy M. The codon 72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet 2003;33(3):357-65.
[Crossref] [Google Scholar] [Pub Med]
- Zhao Y, Wang F, Shan S, Zhao Y, Qiu X, Li X, et al. Genetic polymorphism of p53, but not GSTP1, is association with susceptibility to esophageal cancer risk-a meta-analysis. Int J Med Sci 2010;7(5):300.
[Crossref] [Google Scholar] [Pub Med]
- Olivier M, Hollstein M, Hainaut P. TP53 mutations in human cancers: Origins, consequences and clinical use. Cold Spring Harb Perspect Biol 2010;2(1):a001008.
[Crossref] [Google Scholar] [Pub Med]
- Pistritto G, Puca R, Nardinocchi L, Sacchi A, D'Orazi G. HIPK2-induced p53Ser46 phosphorylation activates the KILLER/DR5-mediated caspase-8 extrinsic apoptotic pathway. Cell Death Differ 2007;14(10):1837-9.
[Crossref] [Google Scholar] [Pub Med]
- Dahabreh IJ, Linardou H, Bouzika P, Varvarigou V, Murray S. TP53 Arg72Pro polymorphism and colorectal cancer risk: A systematic review and meta-analysis. Cancer Epidemiol Prev Biomarkers 2010;19(7):1840-7.
[Crossref] [Google Scholar] [Pub Med]
- Gianfagna F, Feo ED, van Duijn CM, Ricciardi G, Boccia S. A systematic review of meta-analyses on gene polymorphisms and gastric cancer risk. Curr Genomics 2008;9(6):361-74.
[Crossref] [Google Scholar] [Pub Med]
- Bellini MF, Leite KR, Cury PM, Silva AE. p53, p16 and Fhit proteins expressions in chronic esophagitis and Chagas disease. Anticancer Res 2008;28(6A):3793-9.
- Han U, Can OI, Han S, Kayhan B, Onal BU. Expressions of p53, VEGF C, p21: Could they be used in preoperative evaluation of lymph node metastasis of esophageal squamous cell carcinoma? Dis Esophagus 2007;20(5):379-85.
[Crossref] [Google Scholar] [Pub Med]
- Oki E, Zhao Y, Yoshida R, Egashira A, Ohgaki K, Morita M, et al. The difference in p53 mutations between cancers of the upper and lower gastrointestinal tract. Digestion 2009;79(1):33-9.
[Crossref] [Google Scholar] [Pub Med]