- *Corresponding Authors:
- A. L. Iglesias
Escuela de Ciencias de la Ingeniería y Tecnología, Universidad Autónoma de Baja California, No. 1000. Blvd. Universitario 21500
E-mail: aiglesias@uabc.edu.mx - l. J. Villarreal-Gómez
Facultad de Ciencias Químicas e Ingeniería, Universidad Autónoma de Baja California, Tijuana, Baja California, México
| Date of Submission | 02 June 2018 |
| Date of Revision | 30 September 2018 |
| Date of Acceptance | 23 February 2019 |
| Indian J Pharm Sci 2019;81(2):333-343 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
A series of new Schiff base compounds derived from substituted 3-aminopyrazoles and dialdehydes were synthesized and characterized by 1H, 13C nuclear magnetic resonance, Fourier-transform infrared, ultraviolet-visible, gas chromatography-mass spectrometry and high resolution mass spectrometry. The antimicrobial activity of the ligands was screened against the bacterial Gram-negative species Escherichia coli, Pseudomonas aeruginosa and the Gram-positive species Staphylococcus aureus, using gentamycin as a control. Minimal inhibitory concentration was determined by the microdilution method. The anticancer activity was evaluated against HCT116 colorectal cancer cells, with etoposide as a control, using MTS-PMS assay and expressed as IC50 values. The pharmacological studies showed that in general ligands exhibited broad-spectrum antibacterial activity, which decreased in the following order Escherichia coli>Staphylococcus aureus>Pseudomonas aeruginosa. Bis(imino)pyridine Schiff bases (2a-e) have excellent activity towards Staphylococcus aureus; compounds 2a and 2d (3.125 µg/ml) are several times more potent than the control drug, whilst bis(imino)benzene compounds (3a-e) showed significant pathogenic activity toward Pseudomonas aeruginosa with MIC values of 6.25 µg/ml for 3c and 3e. Compound 2c showed higher cytotoxicity (0.40 µM) than etoposide. The results suggest that pyrazole ring as well as the substitution pattern on the heterocyclic moiety have effect on bioactivity.
Keywords
Pyrazole, bioactivity, Schiff base, antibacterial activity, cytotoxic activity, bis(imino)pyridine, bis(imino)benzene
Treatment of infectious diseases is an ever-increasing problem owing to the growing number of multi-drug resistant pathogens. Nosocomial infections are on the rise due to antibiotic-resistant microorganisms, which have led to increased morbidity and mortality in many hospitals. Cancer is a major disease worldwide [1]; colorectal cancer particularly, is the second leading cause of death by cancer in Western countries and has one of the highest mortality rates in both men and women. These cancer patients are especially susceptible to nosocomial infections; as a result of their immunosuppressed systems (due to the treatments associated with illness); which in turn leads to prolonged stays, disability and economic burden [2,3]. Therefore, the search for novel compounds that displayed a broad range of therapeutic activities that can address these major health problems is of outmost importance [4]. New therapeutic drugs that contain a heterocyclic core are on the forefront of pharmaceutical research [5,6]. Pyrazoles, (1,2-azoles) have proven to be a remarkable scaffold for the synthesis of biologically active compounds [7-9] thanks to their wide range of medical applications that center mainly on antimicrobial [10-12], antifungal [8,12,13], antiinflammatory [7], analgesic, and anxiolytic activities [13,14]. Some pyrazole derivatives have also shown antitumor activities towards several cancer cell lines, including leukemia (K562), HeLa cervix adenocarcinoma and Fem-x melanoma [14]. Consequently, in recent years pyrazole-base compounds have become a target of great interest.
Schiff bases or imines, products of the condensation of carbonyl compound and primary amines, are important molecules that have been extensively studied owing to their broad range of industrial and biomedical applications [15]. The relative ease of their preparation, as well as the facile modification of the electronic and steric factor of the ligands; together with their chelating properties toward different metals, have made them attractive targets in the field of medicinal chemistry. Their growing importance stems from their diverse pharmacological properties: antibacterial, antifungal, antimalarial, antiinflammatory and antiviral [16-18]. Imines have also been found to possess cytotoxic and antiproliferative activity towards several cancer cell lines like leukemia, colorectal adenocarcinoma (Caco-2), and pancreatic cancer (Panc-1) [19-21], where the presence of the azomethine bond (CH=N) is believe to be critical to biological activity [15,16,22].
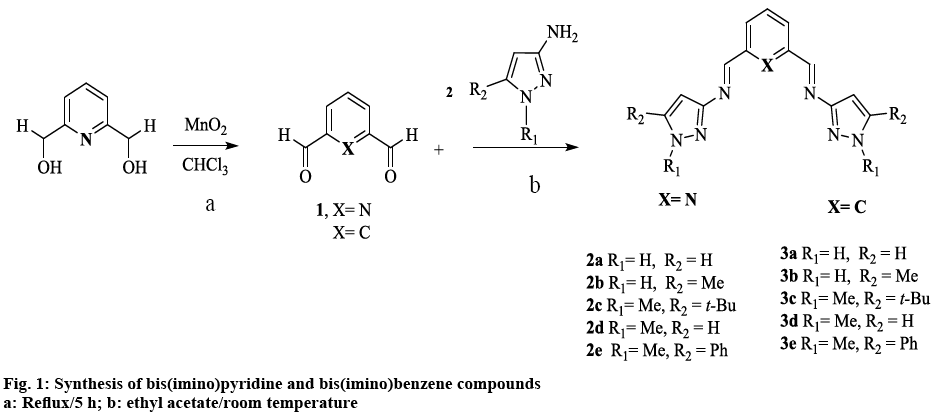
Inspired by the aforementioned and continuing our work on Schiff base compounds and their applications [23-26], the pyrazole nucleus was incorporated into the design and architecture of the imine ligand, with the goal of finding compounds that elicit and enhance bioactivity. The synthesis of two homologous families of NNN-bis(imino)pyridine and NN-bis(imino) benzene compounds derived from 3-aminopyrazole was reported, as well as their biological activity was evaluated. We expect that the presence of the different substituents on the pyrazole ring, together with the change in the aromatic system (pyridine, benzene) of the Schiff bases; may have an effect in the in vitro antibacterial and anticancer activity of these potential chemotherapeutics. Finally, despite the fact that transition metal complexes with bis(imino)pyridine ligands, have been extensively used for last two decades as highly active catalyst; especially towards the polymerization of numerous and diverse olefins [27], few reports have been devoted to explore their potential as chemotherapeutics. To the best of our knowledge, this is the first report of pyrazole containing bis(imino)- pyridine or benzene compounds, coupled with the evaluation of their pharmacological activity.
Materials and Methods
Reactions were carried out under an inert atmosphere or using standard Schlenk techniques. Solvents were dried according to standard techniques. 1,3-benzenedicarboxaldehyde, 2,6-pyridinedi methanol, 2,6-dimethylanline and 3-aminopyrazoles were purchased from Aldrich and used as received without further purification. Melting point determinations were made on a Stuart SMP10 and are uncorrected. UV/Vis spectra were acquired at room temperature (Beckman Coulter model DU730). Fourier-transform infrared (FTIR) spectra were recorded on a Perkin-Elmer FTIR 1605 spectrophotometer (ATR mode). 1H, 13C nuclear magnetic resonance (NMR) spectra were acquired at 400 MHz with Bruker Avance III spectrometer at 30°, chemical shifts were reported in ppm and referenced to residual solvent resonance. Gas chromatography-mass spectrometry (GC-MS) spectra were done by direct insertion on an Agilent Technologies 5975C. High resolution mass spectrometry (HRMS) was collected on a micrOTOF-Q III MS instrument with electrospray ionization using sodium formate as calibrant.
Synthesis of 2,6-pyridinedicarboxaldehyde (1):
To a solution of 2,6 pyridinedimethanol (5 g, 36 mmol) in 250 ml of CHCl3, MnO2 (48.98 g, 576 mmol) was added. The black slurry solution was refluxed for 5 h. After the reaction was allowed to cool down, the solution was filtered through a double layer celite bed. The yellow solution was evaporated under reduced pressure, washed (3×50 ml) with hexane and dried in vacuo to afford a white crystalline solid [28]. Yield: (2.68 g, 55 %), melting point (mp) 120-121°, IR (ATR): 1715 cm−1. 1H NMR (400 MHz, CDCl3): δ 10.18 (s, 2H), 8.18 (d, J=8 Hz, 2H), 8.09 (t, J=8 Hz, 1H).
2,6-Bis(2,6-dimethyl-phenyl-iminomethyl)pyridine (2):
Compounds 2-3 were synthesized with the following general procedure: 2,6 dimethylaniline (0.361 g, 2.984 mmol) and isophtaldehyde (0.100 g, 1.492 mmol), were dissolved in 25 ml of dry methanol, and reflux for 24 h; upon cooling a yellow precipitate was formed, filtered, dried in vacuo and used without further purification. Yellow bright solid, yield: (0.203 g, 80 %), mp 340-341°. IR (cm-1, ATR): 3307, 2180, 1637, 1471, 1030, 761, 581. 1H NMR (400 MHz, CDCl3): δ 8.33 (s, 2H, CH=N), 8.32 (d, J=7.6, 2H, Hb), 7.90 (t, J=7.6, 1H, Ha), 7.01 (d, 4H, Hf), 6.902 (t, 2H, Hg), 2.098 (s, 12H, He). 13C{1H} NMR (101 MHz, CDCl3): δ 163.17 (CH=N), 154.46, 150.27, 137.34, 128.16, 126.80, 124.18, 122.72, 18.32 (-CH3); UV/ Vis (CH3OH, 1×10-4 M): λ 244 nm, 2653 l cm-1 mol-1. MS (m/z, %), 341.2 (M+, 100). HRMS (ESI-TOF) m/z: [M+Na]+ calcd. for C23H23N3Na 364.1783, found: 364.1784.
2,6-Bis(pyrazolyl-1H-iminomethyl)pyridine (2a):
Schiff bases were synthesized in a similar fashion using ligand 2a as an example: to a solution of 3-aminepyrazole (0.062 g, 0.740 mmol) in dry ethyl acetate, 2,6-pyridinedicarbaldehyde (0.050 g, 0.370 mmol) was added and stirred at room temperature. The reaction was followed by the disappearance of the aldehyde by thinlayer chromatography (hexane:ethyl acetate 1:9), and after 96 h the solution was evaporated under reduced pressure, washed with acetone (3×5 ml) and dried in vacuo to give a light yellow solid. Yield: (0.070 g, 35.0 %), mp 259°. IR (cm−1, ATR): 3217, 2946, 1649, 1574, 1409, 1109, 1017, 664. 1H NMR (400 MHz, dimethyl sulfoxide (DMSO)-d6): δ 12.92 (s, 2H, N-H), 8.93 (s, 2H, HC=N), 8.24 (d, J=8 Hz, 2H, Hb), 8.10 (t, J=7.6 Hz, 1H, Ha), 7.44 (d, J=2 Hz, 2H, Hf), 6.60 (d, J=2.4 Hz, 2H, He). 13C{1H} NMR (101 MHz, DMSO-d6): δ 159.4 (HC=N), 155.0 138.5, 131.3, 130.9, 122.9, 96.8 (Ce). UV/Vis (DMSO, 1×10−4 M): λ 290 nm ε= 1650 l cm−1mol−1, λ 299 nm ε=1360 l cm−1mol−1. MS (m/z, %), 263.1 (M+-2, 100).
2,6-Bis(5-methyl-pyrazolyl-1H-iminomethyl) pyridine (2b):
Light beige solid, yield: (0.065 g, 60.6 %), mp >300°. IR (cm−1, ATR): 3213, 2920, 1636, 1578, 1554, 1517, 1452, 1318, 1080, 995, 759, 597. 1H NMR (400 MHz, DMSO-d6): δ 12.58 (s, 2H, N-H), 8.84 (s, 2H, HC=N), 8.21 (d, J=7.6 Hz, 2H, Hb), 8.04 (t, J=7.6 Hz, 1H, Ha), 6.33 (s, 2H, He), 2.24 (s, 6H, -CH3). 13C{1H} NMR (101 MHz, DMSO-d6): δ 158.7 (HC=N), 121.9, 96.1 (Ce), 11.20 (CH3). UV/Vis (DMSO, 1×10−4 M): λ 303 nm ε=3029 l cm−1mol−1, λ 350 nm ε=2291 l cm−1 mol−1. MS (m/z, %), 293.1 (M+, 100). HRMS (ESI-TOF) m/z: [M+Na]+ calcd. for C15H15N7Na 316.1281, found: 316.1286.
2,6-Bis(5-t-butyl-pyrazolyl-1H-iminomethyl) pyridine (2c):
Orange solid, yield: (0.056 g, 85.0%), mp 278-280° with decomp, IR (cm−1, ATR): 3205, 2962, 1619, 1567, 1484, 1260, 1089, 1015, 794, 692. 1H NMR (400 MHz, DMSO-d6): δ 12.56 (s, 2H, N-H), 8.94 (s, 2H, HC=N), 8.27 (d, J=7.8 Hz, 2H, Hb), 8.01 (t, J=7.8 Hz, 1H, Ha), 6.35 (s, 2H, He), 1.38 (s, 18H, -C(CH3)3). 13C{1H} NMR (101 MHz, DMSO-d6): 158.8 (HC=N), 153.9, 140.0, 139.0, 125.9, 123.1, 94.0 (Ce), 31.9 -C(CH3)3, 30.4 -C(CH3)3. UV/Vis (DMSO, 1×10−4 M): λ 266 nm ε=4657 l cm−1 mol−1, λ 329 nm ε=3423 l cm−1mol−1. MS (m/z, %), 377.2 (M+, 10), 139.1 (50), 124.1(100).
2,6-Bis(pyrazolyl-1methyl-iminomethyl)pyridine (2d):
Light yellow solid, yield: (0.058 g, 79.9 %), mp 142- 143°. IR (cm−1, ATR): 3099, 2961, 1612, 1581, 1562. 1511, 1463, 1419, 1073, 1016, 748, 701. 1H NMR (400 MHz, CDCl3): δ 8.92 (s, 2H, CH=N), 8.34 (d, J=8 Hz, 2H, Hb), 7.90 (t, J=7.6 Hz, 1H, Ha), 7.36 (d, J=2.4 Hz, 2H, Hf), 6.42 (d, J=2.4 Hz, 2H, He), 3.93 (s, 6H, -CH3). 13C{1H} NMR (101 MHz, CDCl3): δ 158.7 (HC=N), 154.2, 136.5, 131.2, 122.1, 119.6, 97.5 (Ce), 38.4 (NCH 3). UV/Vis (CHCl3, 1×10−4 M): λ 322 nm ε=640 l cm−1mol−1, λ 376 ε=523 l cm−1 mol−1. MS (m/z, %), 293.1 (M+, 100). HRMS (ESI-TOF) m/z: [M+Na]+ calcd for C15H15N7Na 316.1281, found: 316.1279.
2,6-Bis(5-phenyl-pyrazolyl-1H-iminomethyl) pyridine (2e):
Light beige solid, yield: (0.240 g, 86.4 %), mp >300°. IR (cm−1, ATR): 3224, 1653, 1570, 1504, 1450, 760, 696. 1H NMR (400 MHz, DMSO-d6): δ 8.98 (s, 2H, CH=N), 8.46 (dd, J=7.90, 1.1 Hz, 2H, Hb), 8.34-8.27 (m, 3H), 8.21 (dd, J=7.6, 0.8 Hz, 4H), 8.16 (t, J= 7.7 Hz, 2H), 8.02 (dd, J=7.6, 1.2 Hz, 2H), 6.39 (s, 2H, He). UV/Vis (DMSO, 1×10−4 M): λ 372 nm, ε= 7880 l cm−1mol−1.
1,3-Bis(2,6-dimethyl-phenyl-iminomethyl)benzene (3):
Yellow solid, yield (0.192 g, 76.2%), mp 143-144°. IR (cm−1, ATR): 2919, 2852, 1635, 1469, 1191, 1088, 767, 760. 1H-NMR (400 MHz, CDCl3): δ 8.29 (s, 1H, d), 8.22 (s, 2H, CH=N), 8.02 (dd, J=7.6,1.6, 2H, Hb), 7.54 (t, J=7.6, 1H, Hc), 6.99 (d, 4H, Hf), 6.88 (t, 2H, Hg), 3.03 (s, 12H, He); 13C{1H} NMR (101 MHz, CDCl3): δ 160.70 (CH=N), 149.93, 135.76, 129.90, 128.49, 127.90, 126.97, 126.15, 122.87, 17.34. UV/Vis (CH2Cl2, 1×10-4M ): λ 335 nm, 3550 ε= l cm-1mol-1; 287 nm, 1850 ε= l cm-1mol-1. HRMS (ESI-TOF) m/z: [M+H]+ calcd. for C24H25N2 341.2012, found: 341.2003.
1,3-Bis(pyrazolyl-1H-iminomethyl)benzene (3a):
Beige solid, yield: (0.020 g, 50.1 %), mp 228-231°. IR (cm−1, ATR): 3209, 3100, 1622, 1601, 1548, 1494, 1384, 1288, 1261, 1156, 797, 694. 1H NMR (400MHz, DMSO-d6): δ 12.77 (s, 2H, N-H), 8.98 (s, 2H, HC=N), 8.52 (s, 1H, Hc), 8.04 (d, J=8 Hz, 2H, Hb), 7.75 (d, J=2 Hz, 2H, Hf), 7.66 (t, J=7.6 Hz, 1H, Ha), 6.49 (d, J=2 Hz, 2H, He). 13C{1H} NMR (101 MHz, DMSO-d6): δ 159.3 (CH=N), 158.9, 137.3, 131.6, 130.5, 129.9, 128.0, 96.8 (Ce). UV/Vis (DMSO, 1×10−4 M): λ 307.0 nm, ε=2713 l cm−1mol−1, λ 346.0 nm, ε=1726 l cm−1mol−1. MS (m/z, %), 263 (M+ -1, 100). HRMS (ESI-TOF) m/z: [M+Na]+ calcd for C14H12N6Na 287.1015, found: 287.1007.
1,3-Bis(5-methylpyrazolyl-1H-iminomethyl) benzene (3b):
Beige solid, yield: (0.144 g, 63.8 %), mp with decomp. >300°. IR (cm−1, ATR): 3239, 3199, 2960, 1618, 1589, 1578, 1507, 1488, 1453, 1152, 1025, 1000. 1H NMR (400 MHz, DMSO-d6): δ 12.50 (s, 2H, N-H), 8.95 (s, 2H, HC=N), 8.52 (s, 1H, Hc), 8.05 (dd, J=7.7, 1.7 Hz, 2H, Hb), 7.68 (t, J=7.7 Hz, 1H, Ha), 6.27 (s, 2H, He), 2.29 (s, 6H, -CH3). 13C{1H} NMR (101 MHz, DMSO-d6): δ 159.1 (CH=N), 158.8, 140.1, 137.3, 131.4, 129.8, 95.7 (Ce) 11.27 (-CH3). UV/Vis (DMSO, 1×10−4 M): λ 304 nm ε=2768 l cm−1mol−1, λ 351 nm ε=2251 l cm−1mol−1. MS (m/z, %), 291.1 (M-1, 100). HRMS (ESI-TOF) m/z: [M+Na]+ calcd for C14H12N6Na 315.1328, found: 315.1327.
1,3-Bis(5-t-butylpyrazolyl-1H-iminomethyl) benzene (3c):
Orange solid, yield: (0.048 g, 96.0 %), mp with decomp. 194°. IR (cm−1, ATR): 3205, 2961, 1618, 1567, 1484, 1259, 1086, 1014, 793. ]H NMR (400 MHz, DMSO-d6): δ 12.53 (s, 2H, N-H), 8.86 (s, 2H, CH=N), 8.43 (d, J=1.7 Hz, 1H, Hc), 7.93 (dd, J=7.7, 1.7 Hz, 2H, Hb), 7.57 (t, J=7.6 Hz, 1H, Ha), 6.22 (s, 2H, He), 1.18 (s, 18H, -C(CH3)3). 13C{1H} NMR (101 MHz, DMSO-d6): δ 157.8 (CH=N), 136.7, 134.6, 130.1, 129.7, 128.8, 91.6, 30.9 (-C(CH3)3), 29.9 (-C(CH3)3). UV/Vis (DMSO, 1×10−4 M): λ 327 nm ε=8380 l cm−1mol−1, λ 364 nm ε=9930 l cm−1mol−1. MS (m/z, %), 375.2 (M+ -1, 10), 124(100), 139(50). HRMS (ESI-TOF) m/z: [M+Na]+ calcd for C22H28N6Na 399.2268, found: 399.2271.
1,3-Bis(pyrazolyl-1-methyl-1H-iminomethyl) benzene (3d):
Light green solid, yield: (0.114 g, 94.6 %), mp 112°. IR (cm-1, ATR): 3096, 2902, 1620, 1590, 1547, 1511, 1487, 1302, 1057, 998, 737. 1H NMR (400 MHz, CDCl3): δ 8.86 (s, 2H, CH=N), 8.39 (t, J=1.7 Hz, 1H, Hc), 8.07 (dd, J=7.7, 1.7 Hz, 2H, Hb), 7.53 (t, J=7.7 Hz, 1H, Ha), 7.32 (d, J=2.3 Hz, 2H, Hf), 6.32 (d, J=2.4 Hz, 2H, He), 3.90 (s, 6H, N-CH3). 13C{1H} NMR (101 MHz, CDCl3): δ (ppm) 158.9 (CH=N), 158.5, 136.7, 131.4, 130.9, 129.7, 129.0, 97.7 (Ce), 39.2 (N-CH3). UV/Vis (CHCl3 1×10−4 M): λ=343 nm, ε=1690 l cm−1mol−1 λ=364 nm, ε=2320 l cm−1mol−1. MS (m/z, %), 292.1 (M+, 100). HRMS (ESI-TOF) m/z: [M+Na]+ calcd for C16H16N6Na 315.1329, found: 315.1330.
1,3-Bis(5-phenylpyrazolyl-1H-iminomethyl) benzene (3e):
Beige solid, yield: (0.217 g, 66.4 %), mp >300°. IR (cm−1, ATR): 3206, 1629, 1582, 1472, 1157, 1070, 797, 756, 693. 1H NMR (400 MHz, DMSO-d6): δ 13.30 (s, 2H, N-H), 9.05 (s, 2H, CH=N), 8.59 (s, 1H, Hc), 8.08 (dd, J=7.7, 1.6 Hz, 2H, b), 7.82 (d, J=7.5 Hz, 5H, Ar), 7.48 (t, J=7.5 Hz, 1H, Ha), 6.97 (s, 2H, He). 13C{1H} NMR (101 MHz, DMSO-d6): δ 160.8 (CH=N), 159.8, 143.7, 137.3, 129.4, 128.8, 127.7, 125.5, 125.2, 95.0 (Ce). UV/Vis (DMSO, 1×10−4 M): λ 383 nm, ε=1,844 l cm−1mol−1. MS (m/z, %), 415.1(M+ -1, 12), 159 (100). HRMS (ESI-TOF) m/z: [M+Na] calcd for C26H20N6Na 439.1642, found: 439.1652.
Antimicrobial assay:
The antimicrobial activity test was performed using the serial dilution method [26] and carried out by inoculating the pathogenic strains Escherichia coli (ATCC 25922), Pseudomonas aeruginosa (ATCC 27853) and Staphylococcus aureus (ATCC 25923), previously incubated for 24 h at 35° in Mueller-Hinton medium (MHB at 23 g/l) in 96 microwell plates, where the inoculated medium was placed, adding the sample at a concentration of 10 mg/ml in DMSO.
Controls (gentamicin as a positive control and inoculated MHB culture medium and DMSO as negative controls) were handled in parallel to verify the inhibition and normal growth of the pathogen. After the inoculation was completed, the absorbance of each well was read at 492 nm using a microplate photometer (Thermo Scientific Multiskan). Subsequently, the microplates were incubated at 35° for 24 h, in order to observe the inhibitory effect on the strains of the samples.
A volume of 96 μl of MHB medium was inoculated with the pathogen strain to be tested and deposited in row A of the microplate, to which 4 μl of the sample (at a concentration of 10 mg/ml in DMSO) was added and homogenized. From row B to H, a volume of 50 μl of inoculated MHB medium was deposited. A volume of 50 μl of the homogenate from row A was taken and transferred to row B by homogenizing and completing a total volume of 100 μl, of which a volume of 50 μl was taken, repeating this procedure consecutively until reaching the wells of row H, from which the remaining 50 μl were discarded. Eight dilutions of the sample of interest were obtained from the dilution process, and incubated at 35° for 24, 48 and 72 h, after which the absorbance of the microwell plate was measured at 492 nm, to calculate the inhibitory effect for each of the established concentrations.
An inhibition curve was constructed at 24, 48 and 72 h of exposure, separately calculating dose-response at these times through a Probit analysis, which would establish the minimum inhibitory concentration (MIC) for each sample. For this Probit analysis, Minitab 17 Statistical Software® package was used. All experiments were done in triplicate.
Anticancer assay:
HCT-116 (ATCC® CCL-247™) colorectal cancer cells were used to evaluate the anticancer activity of the samples [26,29,30]. For this, a cell suspension was cultured at a concentration of 2.5 to 3.0×104 cells/ml using Roswell Park Memorial institute medium (RPMI 1640), 10 % bovine fetal serum and 1 % solution of penicillinstreptomycin- amphotericin B solution at 37° and 5 % CO2. After adding the samples at a concentration of 1000 μg/ml, the cultured cells were incubated for 24 h and the indicator, which help to evaluate the efficiency in cell metabolism, was added using a prepared tetrazolium compound 3-(4,5-dimethylthiazol-2-yl)- 5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2Htetrazolium (MTS) 20:1 solution (2 mg/ml) and 1-methoxy-phenazine-methosulfate (0.92 mg/ml) in Dulbecco's phosphate-buffered saline. Trials were performed in triplicate and etoposide (10 mg/ml) was used as the positive control.
Subsequently, samples were read at 490 nm in a microplate reader. The mean inhibitory concentration (IC50) representing the concentration of a compound required for 50 % cell inhibition in vitro was calculated with Softmax® PRO software (Molecular Devices Corporation, EU).
Statistical analysis:
Each data point was obtained by making at least 3 independent measurements. Data were analyzed by analysis of variance (p<0.05) and the means separated by one-way ANOVA. To reject null hypothesis F value> F critical and p<α (0.05).
Results and Discussion
Schiff bases were synthesized from substituted 3-aminopyrazoles and the corresponding dialdehyde, according to figure 1, to afford bis(imino)pyridine (2a-e) and bis(imino)benzene (3a-e) compounds, in moderate to good yields. In general, the compounds have high mp and very low solubility in the majority of organic solvents. The latter can be attributed to intra or inter molecular hydrogen bonding with N(1)-H bond [31]. Alkylation at the N(1)-H position (-R1) as in compound 2d and 3d (figure 1) decreased the mp and increased the solubility of the ligands. This methylation appeared to disrupt N-N…H interactions, thus preventing the formation of hydrogen bonds, which might lead to the formation of supramolecular structures [32]. Increasing the steric factor at position -R2 in the heterocyclic ring with aromatic or branch alkyl groups, increased the solubility only slightly (figure 1).
The diagnostic peak for the CH=N vibration appears in the range of 1612-1649 cm−1, in agreement with other related imine heterocyclic compounds [17,19,33,34]. The ν(C=N) corresponding to the breathing or ring stretching modes of pyrazole ring were observed (Table 1) at 1509-1602 cm−1 [13,35-39]. Although some ν(HC=N) of the new compounds coincided with the vibration of the C=N band in amino-pyrazole parent compound; further evidence of the formation of the imine bond [40,41], came from the disappearance of the ν(NH2) bands due to the asymmetric and symmetric stretch of the 3-aminopyrazole starting material and by the presence of N-H peak absorption as a broad band between 3205-3239 cm−1[18]. In compounds 2d and 3d, this latter band was absent by cause of the methyl group substitution at N(1) position. Absorption peaks ν(C-H) were seen in the range of 2902-2980 cm−1 for alkyl groups. No significant pattern between the two families was observed.
| Compound | N-H | Alk | HC=N | C=C, C=N |
|---|---|---|---|---|
| 2a | 3217 | -- | 1649 | 1574 |
| 2b | 3213 | 2920 | 1636 | 1572 |
| 2c | 3205 | 2962 | 1619 | 1567 |
| 2d | -- | 2980 | 1612 | 1581 |
| 2e | 3224 | -- | 1653 | 1570 |
| 3a | 3209 | -- | 1621 | 1602 |
| 3b | 3239 | 2960 | 1618 | 1589 |
| 3c | 3205 | 2961 | 1618 | 1567 |
| 3d | -- | 2902 | 1621 | 1590 |
| 3e | 3206 | -- | 1629 | 1582 |
Table 1: Selected Infrared Frequencies (Cm−1) for Schiff Bases
The UV spectra data for all the pyrazole-derived Schiff bases in DMSO is summarized in Table 2. The compounds in general have two main peaks between 290-343 and 299-383 nm, attributed to π-π* aromatic and n-π* imine transitions, respectively [13,38,42]. In contrast, the λmax of the free 3-amino-1,2-azoles showed one absorption band between 290-304 nm corresponding to π-π* transition of the pyrazole ring [35]. In general, this peak suffered a bathochromic shift upon the formation of the azomethine bond. There was no apparent correlation between substitution in the heterocyclic ring and the λmax of the new compounds, in agreement with the other relative few reports of the electronic absorption of pyrazole-Schiff base ligands and 3-aminopyrazoles [32,38,43].
| Ligand | λ nm, | ε = L cm−1mol−1 |
|---|---|---|
| 2a | 290, 1650 | 299, 1360 |
| 2b | 303, 3029 | 350, 2291 |
| 2c | 266, 4657 | 329, 3423 |
| 2d* | 322, 640 | 376, 523 |
| 2e | 372, 7880 | |
| 3a | 307, 2713 | 346, 1726 |
| 3b | 304, 2768 | 351, 2251 |
| 3c | 327, 8380 | 346, 9930 |
| 3d* | 343, 1690 | 364, 2320 |
| 3e | 383,8443 |
*1×10−4 M CHCl3
Table 2: UV/VIS Spectra of Compounds in DMSO 1×10−4 M
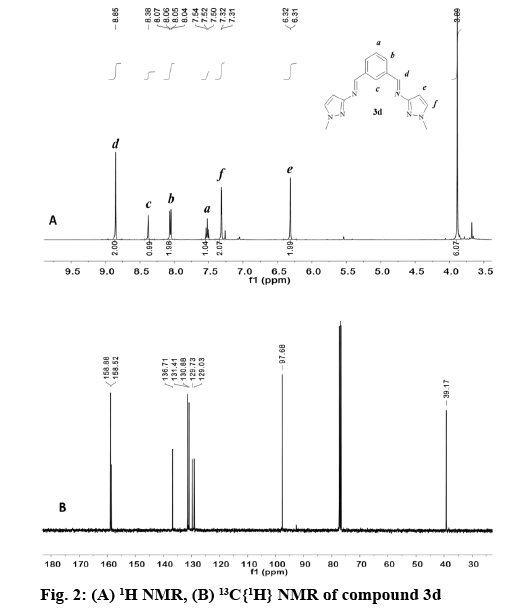
In general, the signal for the iminic proton CH=N in the 1H NMR appeared as a singlet between 8.84 and 9.05 ppm (Table 3) in accordance with other reported bis(imino)pyridine and related ligands [44-46]. The aromatic ring of the bis(imino)benzene ligands (3a-e), was observed as an AB2C system. Proton Hb was split in to a doublet of doublets (3JHb-Ha, 4JHb-Hc) in the range of 7.93-8.07 ppm, while proton Hc was observed as a triplet (4JHb-Hc) at 8.39-8.59 ppm. Proton Ha in compounds 3a-e appears as a triplet in the range of 7.48-7.68 ppm due to the expected ortho coupling with Hb protons (3JHa-Hb=7.7 Hz). In contrast, for bis(imino)pyridine family (2a-e), an AB2 spin system was expected, (figure 2) proton Hb appeared as a doublet at 8.21-8.33 ppm, and Ha is a triplet at 7.60-8.03 ppm. The multiplicity for proton He was determined by the type of substituent at -R2 position of the pyrazole ring, regardless of the aromatic system (figure 1, Table 3). For example, proton He in 2b and 3b appeared as a singlet at 6.27-6.33 ppm (R2=CH3), whereas for 2d and 3d the signal split in to a doublet at 6.32-6.42 ppm (R2=H).
| Compound | CH=N (s, 2H, Hd) | N-H (s, 2H) | -He (s, 2H) | CH=N | C=C-H (Ce) |
|---|---|---|---|---|---|
| 2a | 8.96 | 12.92 | 6.60 (d) | 159.4 | 96.8 |
| 2b | 8.84 | 12.58 | 6.334 | 158.9 | 96.1 |
| 2c | 8.94 | 12.56 | 6.352 | 159.4 | 96.8 |
| 2d* | 8.92 | --- | 6.42 (d) | 158.7 | 97.5 |
| 2e | 8.98 | 13.00 | 6.39 | --- | --- |
| 3a | 8.98 | 12.77 | 6.48 (d) | 158.9 | 96.3 |
| 3b | 8.95 | 12.50 | 6.27 | 159.1 | 97.8 |
| 3c | 8.86 | 12.53 | 6.22 | 157.8 | 91.6 |
| 3d* | 8.86 | --- | 6.32 (d) | 158.9 | 97.7 |
| 3e | 9.05 | 13.30 | 6.97 | 160.8 | 95.0 |
*1H and 13C NMR in CDCl3
Table 3: 1H and 13C{1H} NMR (PPM) Data For Compounds in DMSOD6
The appearance of signals at 2.24, 2.29 and 3.90, 3.93 ppm confirmed the presence of methyl groups for 2b, 3b and 2d, 3d, respectively (figure 2). The latter low field resonance was a result of the EW nature of the pyrrole-like N(1)- in the pyrazole ring. For both families the N-H proton resonance appeared as a broad singlet signal in the range 12.50-13.30 ppm. This fluxional behavior was probably due to a tautomer equilibrium between the N(2)pyridine-like and N(1)-H nitrogen of the 1,2 azole ring [31,32,47]. No tautomerism existed when N(1) was substituted as in compound 2d and 3d (figure 2A).
The 13C NMR spectra of the compounds (2a-d, 3a-e) displayed a signal corresponding to the azomethine carbon HC=N in a narrow range from 157.8- 160.8 ppm (figure 2B). The aromatic carbons in the benzene and pyridine rings were observed between 125-138 ppm, as expected for other heterocyclic Schiff base ligands [45,46,48]. For the pyrazole ring, carbon Ce appears between 91.6-97.8 ppm [35,49] in both families. The methyl group labeled as -R2 in 2b and 3b was observed as a singlet at 11.2 and 11.8 ppm, respectively. In contrast, when the methyl group was attached to N(1) as in 2d and 3d the singlet is shifted downfield to 38.4 and 39.2 ppm.
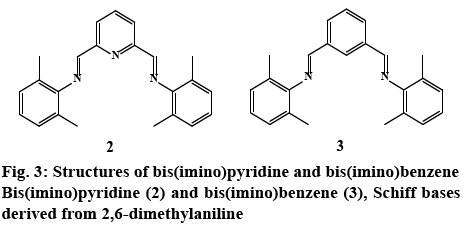
In order to better assess the role of the pyrazole ring on bioactivity, Schiff bases without an aromatic heterocyclic moiety were synthesized 2, 3 (experimental section). The 3-aminopyrazoles were substituted with an aniline derivative, therefore maintaining the aromatic character while eliminating the heterocyclic ring (figure 3). Although these compounds were reported earlier as catalyst for polymerization reactions [50], their cytotoxic properties nonetheless were not evaluated.
The newly synthesized NNN and NN Schiff base ligands were screened for antimicrobial activity. MIC was determined using the micro dilution assay [26] (Table 4). All compounds showed broad spectrum activity against the pathogenic Gram-negative bacteria Escherichia coli, Pseudomonas aeruginosa and Grampositive bacteria Staphylococcus aureus, compared to the standard drug gentamicin, an aminoglycoside mainly effective against Gram-negative bacteria [51].
| Compound | Escherichia coli ATCC 25922 | Pseudomona aeruginosa ATCC 27853 | Staphylococcus aureus ATCC 25923 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| µg/ml | 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | 24 h | 48 h | 72 h |
| 2a | 12.5 | 12.5 | -- | -- | -- | -- | 3.125 | 3.125 | -- |
| 2b | -- | -- | -- | -- | -- | -- | -- | -- | -- |
| 2c | 3.125 | 3.125 | -- | -- | -- | -- | 400 | 400 | -- |
| 2d | 400 | 400 | 400 | 400 | 400 | 400 | 3.125 | 3.125 | 6.25 |
| 2e | 200 | 200 | -- | 12.5 | 12.5 | -- | 400 | 400 | -- |
| 2* | 400 | -- | -- | -- | -- | -- | 400 | 400 | -- |
| 3a | 3.125 | 3.125 | -- | -- | -- | -- | 400 | 400 | -- |
| 3b | 400 | --- | --- | 400 | -- | -- | 100 | 400 | -- |
| 3c | 400 | 200 | -- | 6.25 | 6.25 | -- | -- | -- | -- |
| 3d | 100 | 100 | -- | -- | -- | -- | 400 | 400 | -- |
| 3e | 200 | 200 | -- | 6.25 | 6.25 | -- | -- | -- | -- |
| 3* | -- | -- | -- | 400 | -- | -- | -- | -- | -- |
| Gentamicin | 3.125 | 3.125 | -- | 100 | 100 | -- | 400 | 400 | -- |
-- No inhibition, *control compound without the pyrazole ring. Minimum inhibitory concentration (MIC) expressed in (µg/ml)
Table 4: MIC of Compounds with Different Pathogenic Bacteria at 24, 48 and 72 H
Both bis(imine) pyridine and bis(imine) benzene derived pyrazole ligands presented activity against E. coli, with MIC values in the range of 400-3.125 μg/ml. NNN tridentate ligands, (2c) displayed comparable antibacterial activity with the standard against E. coli at 3.125 μg/ml. When comparing to the control imine 2 (400 μg/ml) all of the compounds except 2b and 2d exhibited higher bioactivity (figure 3). However for 2d effectiveness was maintained for 72 h, in contrast to compound 2 and gentamicin that lost activity after 24 and 48 h, respectively; despite this, a higher concentration than the control drug was needed (400 μg/ml) to maintain bioactivity.
In the case of benzene pyrazole derivatives; 3a had an equivalent activity to gentamicin with a MIC of 3.125 μg/ml. Control ligand 3 showed no activity against E. coli. (Table 4). Therefore, all compounds in this series had better bioactivity than the benzene aniline derivative 3 (figure 3). These results suggest that for this bacterial strain, the incorporation of the heterocyclic fragment enhances the bioactivity of pyrazole bis(imine) compounds, when compared to the ligands derived from 2,6 dimethyl aniline (figure 2). Substitutions on the pyrazole ring also have an effect on bioactivity: compounds 2d, 3d (R1=methyl, R2=H), exhibited higher activity than isomers 2b, 3b (R1=H, R2=methyl). Interestingly, 2b and 3b were the least bioactive compounds in the two families. These results suggest that the inclusion of a methyl group at -R2 in the heterocyclic moiety diminished bioactivity compared with the other pyrazole substitutions (figure 1). With respect to the pyridine or benzene ring in the compounds, there’s no apparent difference between these aromatic systems i.e. 2e and 3e have same MIC values.
Among the screened samples, only 50 % displayed inhibitory activity against P. aeruginosa. In general, NN-bidentate ligands 3b, 3c and 3e were more effective (6.25-400 μg/ml), than the NNN tridentate ligands 2d, 2e (12.5-400 μg/ml). For instance, the activity of 3e (6.25 μg/ml) was greater than 2e (12.5 μg/ml); although there both several orders of magnitude more potent than the commercial drug. A similar behavior was observed in the control ligands; 2 had no activity against P. aeruginosa, while 3 only exhibited activity at the highest concentration (400 μg/ml) for just 24 h. Thus, the difference in cytotoxicity can be attributed to the incorporation of the pyrazole fragment or benzene ring into the Schiff base compounds.
In general, NNN bis (imino)-derived pyrazole compounds exhibited excellent activity against S. aureus (3.125-400 μg/ml), with activities comparable or several times more potent that the displayed by gentamicin (400 μg/ml). The compounds with strongest cytotoxicity were 2a and 2d with MIC values of 3.125 μg/ml; but for the latter, activity was maintained for 72 h in contrast to 2a and the standard drug. Moreover, control 2 exhibited a similar antibacterial activity to gentamicin (400 μg/ml) and appears to be most active toward this Gram-positive bacterium (Table 4). Thus the increase in activity for 2a and 2d could be attributed to the pyrazole ring.
NN-Bidentate-bis(imino) compounds on the other hand, possess higher MIC values in the range of 100- 400 μg/ml; the most active compound 3b (100 μg/ml) was four times more active than the standard drug gentamicin (400 μg/ml) at 24 h. Control ligand 3, was inactive towards Staphylococcus aureus, thus the cytotoxicity exhibited by 3a, 3b, and 3d could be attributed to the pyrazole moiety. From the controls 2 and 3, it could be perceived that the pyridine ring played a role in eliciting activity. Since control compound 2 exhibited the same cytotoxic activity than 2c, 2e, 3a, 3d, therefore, the azomethine bond appeared to be responsible for the observed activity. In general, for this Gram-positive bacterium, bulky substituents (t-Bu, Ph) at -R2 of the pyrazole ring (2c, 2e, 3c, 3e) lacked or had no effect on activity (figure 1). Finally, the absence of specificity between Gram-positive and Gram-negative bacteria might suggest a similar mode of action in the cell.
In this study, in vitro cytotoxicity against HCT-116 colorectal cancer cells was tested for all compounds and reported as IC50, representing the concentration of a compound required for 50% of cell inhibition in μM [26,29,30]. Etoposide was used as the standard drug. Results from Table 5, showed that anticancer activity for compound 2c (0.40 μM) is higher than the standard drug (0.50 μM), and also greater than the controls 2 and 3 at a concentration of 1.24-1.32 μM, respectively; followed by the Schiff base 3b (0.97 μM) with almost double of the concentrations of etoposide. Compounds 2a, 3a and 3d were also effective at higher concentrations of 3.40 μM. Finally, ligands 2b, 2d, 3c, 3e had no effect against this cell line.
| Compound | IC50 ±SDµM |
|---|---|
| 2a | 3.40±0.033 |
| 2b | -- |
| 2c | 0.40±0.004 |
| 2d | -- |
| 2e | 2.63±0.121 |
| 2* | 1.24±0.035 |
| 3a | 3.40±0.203 |
| 3b | 0.97±0.113 |
| 3c | -- |
| 3d | 3.42±0.535 |
| 3e | -- |
| 3* | 1.32±0.142 |
| Etoposide | 0.50±0.048 |
-- No cytotoxic activity, *Compounds without the pyrazole ring, SD is standard deviation, mean inhibitory concentrations (IC50)
Table 5: Mean IC50 against HCT-116 Colorectal Cancer Cells at 24 H
To understand if these results were statistically significantly, a one-way ANOVA was performed, on all the compounds including the controls; the results indicated that there’s a significant difference between the activity exerted by all the Schiff bases in both bis(imino) pyridine (2a, 2c, 2e) and bis(imino) benzene (3a, 3b, 3d) compounds. Further analysis on bis(imino) pyridine compounds revealed that there was a significant difference between all the Schiff bases, including the control 2. This suggested that the difference in cytotoxicity within the group could be ascribed to substituents (-R2, -R1) on the pyrazole ring of the compounds (figure 1).
Results from ANOVA indicated that for bis(imino) benzene compounds only 3b showed a significant difference in activity in this group; and although statistically different with control 3, this analysis suggested that in general, the pyrazole ring was not crucial to elicit bioactivity. On assessing the role of the pyridine/benzene aromatic systems on cytotoxicity, results from Table 5 (2a,3a), suggested that there was no difference in cytotoxicity of these compounds against HCT-116 cells. Given that a similar behavior was exhibited by the controls 2 and 3 (ANOVA), this suggested that pyridine/benzene aromatic systems have no clear effect on cytotoxic activity, so biological activity can be attributed mainly to the HC=N bond or the pyrazole ring. Finally, these result suggested that a t-butyl group in -R2 position of the pyrazole ring (2c) is required to enhanced the anticancer activity compared to the control ligands 2, this behavior is similar to the results for E. coli.
Two new families of pyrazole derived NNN and NN Schiff bases were synthesized and characterized by spectral studies. The biological studies suggest that the compounds exhibit broad-spectrum antimicrobial activity. Bioactivity for these compounds appeared to be a combination of mainly two factors, inclusion and substitutions on the pyrazole ring, and the presence of the azomethine bond. The antimicrobial results showed a dependence on the type of bacteria, which suggested that compounds 2c, 2d, 3c, 3e, warranted further investigation as promising new chemotherapeutics. Furthermore, our preliminary studies showed that anticancer activity within a group depended on the pyrazole fragment, but the control group demonstrated that the bioactivity is also dependent on the azomethine bond, nonetheless the enhancement in biological activity on compounds 2c and 3b demonstrated that substitutions in the pyrazole moiety were also responsible for bioactivity. Work is currently underway to evaluate these compounds against other pathogenic bacteria and yeast.
Acknowledgement
This research was funded by Consejo Nacional de Ciencia y Tecnología (CONACyT), grant number 169578. JGMO thanks CONACyT for graduate fellowship. We thank Instituto Tecnologico de Tijuana (ITT) for NMR and HRMS facilities (Grants INFR- 2011-3-173395 and INFR-2012-01-187686) and to Dr. Daniel Chavez for GC-MS spectra.
Conflict of interest
The authors declared that they have no conflict of interest.
References
- Vijesh AM, Isloor AM, Shetty P, Sundershan S, Fun HK. New pyrazole derivatives containing 1,2,4-triazoles and benzoxazoles as potent antimicrobial and analgesic agents. Eur J Med Chem 2013;62:410-5.
- André T, Boni C, Mounedji-Boudiaf L, Navarro M, Tabernero J, Hickish T, et al. Oxaliplatin, Fluorouracil, and Leucovorin as Adjuvant Treatment for Colon Cancer. N Engl J Med 2004;350:2343-51.
- Fatfat M, Merhi RA, Rahal O, Stoyanovsky DA, Zaki A, Haidar H, et al. Copper chelation selectively kills colon cancer cells through redox cycling and generation of reactive oxygen species. BMC Cancer 2014;14:527.
- Genin MJ, Allwine DA, Anderson DJ, Barbachyn MR, Emmert DE, Garmon SA, et al. Substituent Effects on the Antibacterial Activity of Nitrogen−Carbon-Linked (Azolylphenyl)oxazolidinones with Expanded Activity Against the Fastidious Gram-Negative Organisms Haemophilus influenzae and Moraxella catarrhalis. J Med Chem 2000;43:953-70.
- Abrigach F, Bouchal B, Riant O, Macé Y, Takfaoui A, Radi S, et al. New N,N,N’,N’-tetradentate Pyrazoly Agents: Synthesis and Evaluation of their Antifungal and Antibacterial Activities. Med Chem 2016;12:83-9.
- Kalanithi M, Rajarajan M, Tharmaraj P, Raja SJ. Synthesis, spectroscopic characterization, analgesic, and antimicrobial activities of Co(II), Ni(II), and Cu(II) complexes of 2- N,N-bis-(3,5-dimethyl-pyrazolyl-1-methyl) aminothiazole. Med Chem Res 2015;24:1578-85.
- Bekhit AA, Ashour HM, Abdel Ghany YS, Bekhit Ael-D, Baraka A. Synthesis and biological evaluation of some thiazolyl and thiadiazolyl derivatives of 1H-pyrazole as anti-inflammatory antimicrobial agents. Eur J Med Chem 2008;43:456-63.
- Damljanović I, Vukićević M, Radulović N, Palić R, Ellmerer E, Ratković Z, et al. Synthesis and antimicrobial activity of some new pyrazole derivatives containing a ferrocene unit. Bioorg Med Chem Lett 2009;19:1093-96.
- Hassan YS. Synthesis, Antibacterial and Antifungal Activity of Some New Pyrazoline and Pyrazole Derivatives. Molecules 2013;18:2683-711.
- Güniz Küçükgüzel Ş, Rollas S, Erdeniz H, Kiraz M, Cevdet Ekinci A, Vidin A. Synthesis, characterization and pharmacological properties of some 4-arylhydrazono-2-pyrazoline-5-one derivatives obtained from heterocyclic amines. Eur J Med Chem 2000;35:761-71.
- Sangani CB, Makwana JA, Duan YT, Tarpada UP, Patel YS, Patel KB, et al. Design, synthesis, and antibacterial evaluation of new Schiff's base derivatives bearing nitroimidazole and pyrazole nuclei as potent E coli FabH inhibitors. Res Chem Intermed 2015;41:10137-49.
- Farag AM, Mayhoub AS, Barakat SE, Bayomi AH. Synthesis of new N-phenylpyrazole derivatives with potent antimicrobial activity. Bioorg Med Chem 2008;16(8):4569-78.
- El-Gamel NEA, Farghaly TA. Design and synthesis of novel complexes containing N-phenyl-1H-pyrazole moiety: Ni complex as potential antifungal and antiproliferative compound. Spectrochim Acta A: Mol Biomol Spectrosc 2013;115:469-75.
- Joksović MD, Marković V, Juranić ZD, Stanojković T, Jovanović LS, Damljanović IS, et al. Synthesis, characterization and antitumor activity of novel N-substituted α-amino acids containing ferrocenyl pyrazole-moiety. J Organometg Chem 2009;694:3935-42.
- Sztanke K, Maziarka A, Osinka A, Sztanke M. An insight into synthetic Schiff bases revealing antiproliferative activities in vitro. Bioorganic Med Chem 2013;21:3648-66.
- da Silva CM, da Silva DL, Modolo LV, Alves RB, de Resende MA, Martins CVB, et al. Schiff bases: A short review of their antimicrobial activities. J Adv Res 2011;2:1-8.
- Panneerselvam P, Priya MG, Kumar NR, Saravanan G. Synthesis and pharmacological evaluation of Schiff bases of 4-(2-aminophenyl)-morpholines. Indian J Pharm Sci 2009;71(4):428-32.
- Neelakantan MA, Esakkiammal M, Mariappan SS, Dharmaraja J, Jeyakumar T. Synthesis, Characterization and Biocidal Activities of Some Schiff Base Metal Complexes. Indian J Pharm Sci 2010;72(2):216-22.
- Ren T, Wang J, Li G, Li Y. Synthesis, Characterization and in vitro Antitumor Activity of Novel Schiff Bases Containing Pyrazole Group. Asian J Chem 2014;26:8309-13.
- Gama S, Mendes F, Marques F, Santos IC, Carvalho MF, Correia I, et al. Copper(II) complexes with tridentate pyrazole-based ligands: synthesis, characterization, DNA cleavage activity and cytotoxicity. J Inorg Biochem 2011;105:637-44.
- Abu Bakr S, Abd El-Karim SS, Said MM, Youns MM. Synthesis and anticancer evaluation of novel isoxazole/pyrazole derivatives. Res Chem Intermed 2016;42:1387-99.
- Subhashini NJP, Amanaganti J, Nagarjuna PA. Synthesis, characterization and biological activity of (N1E,N2Z)-N1,N2-Bis((1-Phenyl-3-Aryl-1H-Pyrazol-4-yl)methylene)benzene-1,2-diamines. J Applicable Chem 2014;3:2358-64.
- Iglesias AL, Aguirre G, Somanathan R, Parra-Hake M. New chiral Schiff base–Cu(II) complexes as cyclopropanation catalysts. Polyhedron 2004;23:3051-62.
- Iglesias AL, García JJ. Homogeneous hydrogenation of fluoroaromatic imines with Ni compounds, evidence for η2-CN intermediate in the catalytic cycle. J Mol Catal A Chem 2009;298:51-9.
- Iglesias AL, Muñoz-Hernández M, García JJ. Fluoro aromatic imine nickel(0) complexes: Synthesis and structural studies. J Organomet Chem 2007;692:3498-507
- Villarreal-Gómez LJ, Soria-Mercado IE, Guerra-Rivas G, Ayala-Sánchez NE. Antibacterial and anticancer activity of seaweeds and bacteria associated with their surface. Rev Biol Mar Oceanogr 2010;45:267-75.
- Gibson VC, Redshaw C, Solan GA. Bis(imino)pyridines: Surprisingly Reactive Ligands and a Gateway to New Families of Catalysts. Chem Rev 2007;107(5):1745-76.
- Vance AL, Alcock NW, Heppert JA, Busch DH. An Octahedral Template Based on a New Molecular Turn: Synthesis and Structure of a Model Complex and a Reactive, Diphenolic Ligand and Its Metal Complexes. Inorg Chem 1998;37:6912-20.
- ATCC. Protocol for Thawing, Propagation and Cryopreservation of NCI-PBCF-CCL247 (HCT 116) (ATCC®CCL-247 ™) colorectal carcinoma. Rockville, Maryland: American Type Culture Collection; 2012. Available from: https://physics.cancer.gov/docs/bioresource/colorectal/NCI-PBCF-CCL247_HCT116_SOP-508.pdf.
- ATCC. Culture of Animal Cells: A Manual of Basic Technique. In: Freshney RI, editor. Culture of Animal Cells. 5th ed. New York: Wiley-Liss; 2005.
- Luis G. de Paz J, Elguero J, Foces-Foces CL, Llamas-Saiz A, Aguilar-Parrilla F, Klein O, et al. Theoretical study of the structure and tautomerism of N 1-unsubstituted pyrazoles in the solid state. J Chem Soc Perkin Trans 2 1997;101:10.
- Emelina EE, Petrov AA, Borissova AO, Filyukov DV, Antipin MY. The interplay of hydrogen bonds in the solid state structure of NH-pyrazoles bearing cyano and amino substituents. J Mol Struct 2012;1014:63-9.
- Ainooson MK, Guzei IA, Spencer LC, Darkwa J. Pyrazolylimine iron and cobalt, and pyrazolylamine nickel complexes: Synthesis and evaluation of nickel complexes as ethylene oligomerization catalysts. Polyhedron 2013;53:295-303.
- Abu-Surrah AS, Abu Safieh KA, Ahmad IM, Abdalla MY, Ayoub MT, Qaroush AK, et al. New palladium(II) complexes bearing pyrazole-based Schiff base ligands: Synthesis, characterization and cytotoxicity. Eur J Med Chem 2010;45:471-75.
- Katritzky AR, Ramsden CA, Joule JA, Zhdankin VV. Handbook of Heterocyclic Chemistry. 3rd ed. Amsterdam (NE): Elsevier; 2010.
- Santos IF, Guedes GP, Mercante LA, Bernardino AMR, Vaz MGF. Synthesis, crystal structure and magnetism of three novel copper(II) complexes with pyrazole-based ligands. J Mol Struct 2012;1011:99-104.
- Sobiesiak M, Lorenz IP, Mayer P, Woźniczka M, Kufelnicki A, Krajewska U, et al. Synthesis, X-ray structure and cytotoxic effect of nickel(II) complexes with pyrazole ligands. Eur J Med Chem 2011;46:5917-26.
- Issa RM, Khedr AM, Rizk HF. UV–vis, IR and 1H NMR spectroscopic studies of some Schiff bases derivatives of 4-aminoantipyrine. Spectrochim Acta A Mol Biomol Spectrosc 2005;62:621-29.
- Grazul M, Besic-Gyenge E, Maake C, Ciolkowski M, Czyz M, Sigel RKO, et al. Synthesis, physico-chemical properties and biological analysis of newly obtained copper(II) complexes with pyrazole derivatives. J Inorg Biochem 2014;135:68-76.
- Malladi S, Isloor AM, Isloor S, Akhila DS, Fun HK. Synthesis, characterization and antibacterial activity of some new pyrazole based Schiff bases. Arab J Chem 2013;6:335-40.
- Barra CV, Rocha FV, Netto AVG, Frem RCG, Mauro AE, Carlos IZ, et al. New palladium(II) complexes with pyrazole ligands. J Therm Anal Cal 2011;106:489-94.
- Yusnita J, Puvaneswary S, Mohd Ali H, Robinson WT, Kwai-Lin T. Synthesis, structural characterization and antibacterial activity of 2,6-diacetylpyridine bis(benzenesulfonohydrazide) Schiff bases and their copper(II) complexes. Polyhedron 2009;28:3050-54.
- Eicher T, Hauptmann S, Speicher A. The Chemistry of Heterocycles: Structures, Reactions, Synthesis, and Applications. 3rd ed. Weinheim (Germany): Wiley-VCH; 2012.
- Gong D, Wang B, Cai H, Zhang X, Jiang L. Synthesis, characterization and butadiene polymerization studies of cobalt (II) complexes bearing bisiminopyridine ligand. J Organomet Chem 2011;696:1584-90.
- Zhao M, Yu Z, Yan S, Li Y. Ruthenium (II) complex catalysts bearing a pyridyl-supported pyrazolyl-imine ligand for transfer hydrogenation of ketones. J Organomet Chem 2009;694:3068-75
- Gibson VC, Gregson CKA, Halliwell CM, Long NJ, Oxford PJ, White AJP, et al. The synthesis, coordination chemistry and ethylene polymerisation activity of ferrocenediyl nitrogen-substituted ligands and their metal complexes. J Organomet Chem 2005;690:6271-83.
- Viciano-Chumillas M, Tanase S, de Jongh LJ, Reedijk J. Coordination Versatility of Pyrazole-Based Ligands towards High-Nuclearity Transition-Metal and Rare-Earth Clusters. Eur J Inorg Chem 2010;2010:3403-18.
- Lang JRV, Denner CE, Alt HG. Homogeneous catalytic dimerization of propylene with bis(imino)pyridine vanadium(III) complexes. J Mol Catal A Chem 2010;322:45-9.
- Pinto DCGA, Santos MMC, Silva MSA. Advanced NMR techniques for structural characterization of heterocyclic structures. In: Pinho e Melo TMVD, Rocha Gonsalves M.d’A, editors. Recent Research Developments in Heterocyclic Chemistry. Kerala, India: Research Signpost; 2007. p. 397-475.
- Wu C, Zhoua H, He M, Su Q, Li G, Wu Q, et al. Binuclear aluminum complexes with amine–imine type ligands derived from 1,3-benzenedialdehyde: synthesis, structures and their catalytic properties in ring-opening polymerization. J Coord Chem 2016;69(6):1-8
- Aschenbrenner DS, Venable SJ. Drug Therapy in Nursing. 3rd ed. New York: Lippincott Williams & Wilkins; 2009.