- *Corresponding Author:
- M. Mohamed Mahroop Raja
Department of Microbiology, Jamal Mohamed College (Autonomous), Affiliated to Bharathidasan University, Tiruchirappalli, Tamil Nadu 620020, India
E-mail: mahroop2019@gmail.com
| Date of Received | 22 November 2021 |
| Date of Revision | 04 March 2022 |
| Date of Acceptance | 07 May 2023 |
| Indian J Pharm Sci 2023;85(3):603-612 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Marine diversity is clearly considered an interest and most reliable source for the marine microbial community. Accordingly, the synthesis of L-asparaginase was obtained from the natural source of marine actinomycetes. The marine sediment samples were collected from the Gulf of Mannar coastal region, Kayalpatinam, located at Tuticorin district, Tamilnadu, India. A total of 100 marine actinomycetes strains were isolated, among them, 10 isolates belonged to Streptomyces sp. All the Streptomyces strains were screened for L-asparaginase production. Streptomyces sp KPMS7 was showed large pink coloration on L-asparaginase production medium. Based on morphological, biochemical and 16S rRNA sequence studies, the strain was identified as Streptomyces antibioticus KPMS7 and submitted in GenBank with the accession number MH819507. The strain was further studied for production and characterizations of L-asparaginase enzyme carried out by basal medium supplemented with carbon and nitrogen sources. The maximum growth rate was observed in glucose and yeast extract was effectively used as a substrate for L-asparaginase by submerged fermentation. The maximum enzyme activities were obtained in 172 h, pH 7 and temperature 37° respectively. The enzyme was partially purified by ammonium sulphate precipitation and showed a final specific activity of 71.92 IU/mg with 100 % yield was carried out by crude extract enzyme. In Sodium dodecyl-sulfate polyacrylamide gel electrophoresis analysis revealed that the molecular weight of partially purified L-asparaginase was 116 KDa. The finding concludes that the marine isolate Streptomyces antibioticus KPMS7 pointed out a promising novel source for L-asparaginase synthesis used in anticancer therapy.
Keywords
Sediment, enzymes, 16S rRNA, growth and therapy
Microbial L-asparaginase has been applied as the effective therapeutic agent against lymphoblastic leukemia and many other types of cancer caused in humans[1]. Terrestrial environments have been used as a rich source for enzyme production. In the last decades, screening of L-asparaginase production has been greatly enhanced and well documented in a vast diversity of marine organisms like algae, bacteria, fungi, yeast, and actinomycetes[2]. Bacteria such as E. coli, Vibrio succinogenes, Erwinia carotovora and Bacillus sp[3] and the fungi like Mucor sp., Penicillium sp., and yeast-like Candida utilis have been proved to be potential producers of L-asparaginase[4]. Actinomycetes are serving as a good microbial source to explore the new and effective ways for L-asparaginase enzyme production. Actinomycetes have been recorded as the most potent source of bioactive secondary metabolites.
Actinomycetes are groups of thread-like bacterial and look like fungi with high G+C content which form branching filaments or hyphae and asexual reproduction. The most important actinomycetes are derived from single genus Streptomyces which are broadly distributed in marine and terrestrial habitats[5]. The various enzymes production has been reported from marine Streptomycetes such as protease, lipase, chitinase and alginate lyases[6]. Recently, marine Streptomyces has been tapped for enzymes with improved properties[7]. It also serves as a good source of L-asparaginase is an enzyme that converts L-asparagine to L-aspartic acid and ammonia, and has been used as a chemotherapeutic agent in the treatment of acute lymphoblastic leukemia[8]. The clinical action of this enzyme is attributed to the reduction of L-asparagine because the several types of tumor cell unable to synthesis this amino acid is selectively killed by L-asparagine deprivation. L-asparaginase has gained special attention for its carcinogenic activity produced by various species of Streptomyces such as Streptomyces karnatakensis, Streptomyces griseus, Streptomyces venezuelae, Streptomyces longsporusflavus, Streptomyces gulbargensis, marine Streptomyces sp PDK2 because the diversity of the microbial population is being explored for gathering novel sources of L-asparaginase production with fewer side effects and better treatment[9]. These marine habits prove and confer marine enzymes production and application in the field of manufacture of commercial products and in the health sector[10].
The present investigation deals with the isolation, identification by 16S rRNA sequencing studies and characterization of marine sediment isolate Streptomyces antibioticus KPMS7 for L-asparaginase synthesis.
Materials and Methods
Materials:
Starch casein agar medium, Cyclohexamide, L-asparaginase, Phenol red indicator, glucose, sucrose, starch, lactose, mannitol, and xylose and Ethidium bromide were purchased from Himedia. Remi cooling centrifuge was purchased from Bengaluru, Ultraviolet-Visible (UVVis) spectrophotometer SL 159–ELICO from Hyderabad, Bio-Rad Thermal Cycler from Chennai, Gel electrophoresis was bought from Medox Biotech India Pvt. Ltd., Bench top 3 UV Trans Illuminators was from UK.
Sample collection:
Marine sediments sample was collected from Gulf of Mannar coastal region, Kayalpatinam, located at Tuticorin district, Tamil Nadu, India. The collected samples were processed by serial dilution and plating methods. 1 ml of diluted sample was allowed into the starch casein agar medium supplemented with cyclohexamide 100 μg/ml. Individual actinomycetes colonies were identified and re-grown on the International Streptomyces Project medium-2(ISP-2) at 28° and stored at 4° as stock cultures under refrigerator by using 20 % glycerol.
Morphological identification:
The microbiological examinations of pure cultures of isolates were identified by spore chain, spore formation, colour aerial mycelium and type of cell wall. Biochemical characterization such as catalase, oxidase, starch hydrolysis, urease activity and pigment production was studied for the proper identification of isolated strains.
L-asparaginase production:
All isolated colonies were evaluated for their ability to produce L-asparaginase. The plate assay was performed on a minimal medium supplemented with phenol red[11]. The enzyme asparaginase is a substrate for L-asparaginase production. The final pH of the medium was adjusted to pH 7. The pure culture of the isolates was inoculated and incubated at 28° for 7 d. After incubation, the colonies turn pink zone due to the formation of ammonia were indicated as L-asparaginase production.
Optimization of L-asparaginase production:
The effect of carbon sources was studied by basal medium with 1 % concentration of glucose, sucrose, starch, lactose, mannitol, and xylose for L-asparaginase production supplemented with 0.1 % peptone and yeast extract as nitrogen sources and incubated at 28° with shaking at 120 rpm for 7 d.
The effect of different nitrogen sources of L-asparaginase activity was evaluated by using a 1 % concentration of peptone, yeast extract, proline, and glutamine was added into basal medium supplemented with 0.1 % concentration of soluble starch and xylose as carbon sources. The medium was incubated at 28° with shaking at 120 rpm for 7 d.
Purification and characterization of L-asparaginase:
The purification of the enzyme was carried out by using crude enzyme extract[12]. The enzyme purification was performed by ammonium sulphate precipitation. Finely powdered ammonium sulfate was added to the crude extract. The L-asparaginase activity was associated with the fraction precipitated at 80 % saturation. The precipitated was collected by centrifugation at 9000 rpm for 15 min and dissolved in 1 M Tris-HCl buffer and dialyzed against the same buf fer.
Effect of incubation time:
The active strain was cultivated and inoculated into 50 ml of minimal medium and incubated at 28° with shaking at 120 rpm for 10 d.
Effect of pH:
The active strain was taken out from starch casein medium and inoculated into 50 ml of minimal medium was prepared at different pH levels ranging from 3-10. The setup was incubated at 28° with shaking at 120 rpm for 7 d.
Effect of temperature:
The active strain was selected and inoculated into 50 ml of minimal medium and incubated at various temperatures such as 5°, 15°, 25°, 35° and 45° with shaking at 120 rpm for 7 d.
Enzyme assay:
The enzyme L-asparaginase activity was performed on M-9 broth medium and detected by the Nessler reaction method[13]. 900 μl of freshly prepared 40 mM L-asparaginase in Tris-HCL buffer (pH 8.5) and 100 μl L-asparaginase enzyme filtrate were added and incubated at 37° for 30 min. The reaction was then stopped by the addition of 100 μl of 1.5 M Trichloroacetic acid. The reaction mixture was centrifuged at 10 000 rpm for 10 min. After centrifugation, precipitated protein was removed and the liberated supernatant ammonia was determined using UV-Vis spectrophotometer (Systronics) by adding 200 μl of Nessler reaction into the sample contained 200 μl of supernatant and 1.5 ml of distilled water. After 20 min of incubation, the optical density of the sample was recorded at 450 nm. 1 unit (IU) of L-asparaginase activity was defined as the amount of the enzyme that liberates 1 μM of ammonia per min at 37°, using asparagine as substrate.
Protein determination:
Protein was determined by using the Lowry et al method[14]. A stock solution of standard protein, bovine serum albumin a concentration of 1000 μg/ml was made. Folin- Ciocalteu reagents were added to each test tube. After 30 min of incubation, the absorbance was measured at 660 nm by using a UV-Vis spectrophotometer.
Sodium Dodecyl-Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE):
SDS-PAGE was performed with a 10 % separating gel and 5 % stacking gel containing 0.1 % SDS. After gel electrophoresis, the gel was stained with coomassie brilliant blue and destained with a solution of ethanol, acetic acid, and water in the ratio of 4:1:5.
Amplification of 16S rRNA sequence and phylogenetic tree analysis:
16S rRNA sequencing was performed by extracting the genomic DNA from the Streptomyces cells and amplified by the eubacteria specific primer F243 (5’- GGATGAGCCCGCGGCCTA-3’) and reverse primer R513GC (5’- CGGCCGCGGCTGCTGGCACGTA-3’). The amplification conditions were as follows: 94° for 1 min (denaturation), 55° for 1 min (annealing), 72° for 1.30 min (elongation) at and 72° for 10 min final elongation. 5 μl of the PCR product was loaded on 1.2 % agarose gel in 1X Tris-borate-EDTA buffer and stained with ethidium bromide 0.5 μl/ml. The polymerase chain reaction product was precipitated by Polyethylene glycol (PEG)-NaCl (20 % PEG in 2.5 M NaCl) precipitation at 37° for 30 min. The reaction mixture was centrifuged at 12 000 rpm for 30 min at room temperature. The supernatant was discarded and the pellet was washed twice with 70 % ethanol.
The amplified PCR product was sequenced in Bioserve Biotechnologies, Hyderabad, India. Forward and reverse DNA sequencing reaction of PCR amplicon was carried out using BDT v 3.1 cycle sequencing kit on ABI 3730xl genetic analyzer The reference sequence required for comparison were downloaded from the Genbank using BLASTN sequence-based retrieval system from Genbank (URL http://www.ncbi.nlm. nih.g) using our sequence as the query. Based on the sequenced data, the phylogenetic tree was constructed using the bioinformatics tool MEGA 5.05 for aligning the sequences by the Neighborjoining method. The 16SrRNA sequences for the Streptomyces antibioticus KPMS7 have been deposited in Gen Bank by using sequin (http://www.ncbi.nlm.nib.gov/genebank).
Results and Discussion
Totally 100 marine actinomycetes were isolated and identified as non-Streptomycetes groups such as Micromonospora sp, Actinomadura sp, Nocardia sp, and Micropolyspora sp. Among 100, 10 % of the marine isolates were presumed to be in genus Streptomyces. According to morphological analysis, all the isolates were differed morphologically based on the color of the colony, types of mycelium, spore and pigmentation (Table 1). Most of the strains produced spiral chains of spore are often spirally coiled and a few of them obtained retractile spiral spore on substrate mycelium indicates the genera of Streptomyces sp. The aerial mycelium was unstained by Sudan black and the spore surface was smooth whereas the substrate mycelium was rough and stained by Sudan black. The aerial mycelia were initially white and turned to ash, grey, and dull white or chalky white on starch casein agar.
| S No | Sample code | Nature of spore morphology | Color of aerial mycelium | Type of Cell wall | Name of the identified genus |
|---|---|---|---|---|---|
| 1 | MSR1 | Long chain of spore | Grey | Gram positive | Streptomyces sp |
| 2 | MSR 2 | Short chain of spore | Greenish ash | Gram positive | Streptomyces sp |
| 3 | MSR3 | Rarely branched chain of spore | Dark grey | Gram positive | Streptomyces sp |
| 4 | MSR4 | Short chain of spore | Greenish Ash | Gram positive | Streptomyces sp |
| 5 | MSR5 | Spiral chain of spore | Dark ash | Gram positive | Streptomyces sp |
| 6 | MSR6 | Moderate length of spiral chain of spore | Grey | Gram positive | Streptomyces sp |
| 7 | MSR7 | Branched chain of spore | Sandal white | Gram positive | Streptomyces sp |
| 8 | MSR8 | Spiral spore | Dull grey | Gram positive | Streptomyces sp |
| 9 | MSR9 | Long chain of spore | Chalky White | Gram positive | Streptomyces sp |
| 10 | KPMS7 | Spiral chain of spore | White | Gram positive | Streptomyces sp |
Table 1: Morphological Examination of Isolated Samples
Species among the genera were differentiated based on the biochemical characterization (Table 2). Among the Streptomycetes sp, five isolates were found in catalase- positive (MSR1, MSR2, MSR 5, MSR8 & KPMS7). Three isolates of Streptomyces sp such as MSR3, MSR4 and MSR9 were negatively found in oxidase reaction. All Streptomyces sp were utilized starch except MSR2, MSR4 and MSR9. Similarly, five isolates of Streptomyces sp were urease positive and others were failed to obtain urease reaction. Out of 10 isolates, four Streptomyces sp designated as MSR2, MSR5, MSR6 and MSR8 were found in pigment productions.
| S.No | Isolated sample code | Catalase | Oxidase | Starch | Urease | Pigment Productions | MSR 2 |
|---|---|---|---|---|---|---|---|
| 1 | MSR1 | + | + | + | + | - | MSR 2 |
| 2 | MSR2 | + | + | - | + | + | MSR 2 |
| 3 | MSR3 | - | - | + | + | - | MSR 2 |
| 4 | MSR4 | - | - | - | - | - | MSR 2 |
| 5 | MSR5 | + | + | + | - | + | MSR 2 |
| 6 | MSR6 | - | + | + | + | + | MSR 2 |
| 7 | MSR7 | - | + | + | - | - | MSR 2 |
| 8 | MSR8 | + | + | + | - | + | MSR 2 |
| 9 | MSR9 | - | - | - | + | - | MSR 2 |
| 10 | KPMS7 | + | + | + | - | - | MSR 2 |
Note: (+): Positive; (-): Negative
Table 2: Biochemical Characteristics of Isolated Samples
All the Streptomyces strains were adapted to preliminary screening for L-asparaginase production by plate assay method at 37° for 24-72 h. Among 10 isolates, KPMS7 isolate was produced pink colour zone appeared on the growth which indicates a positive result of L-asparaginase.
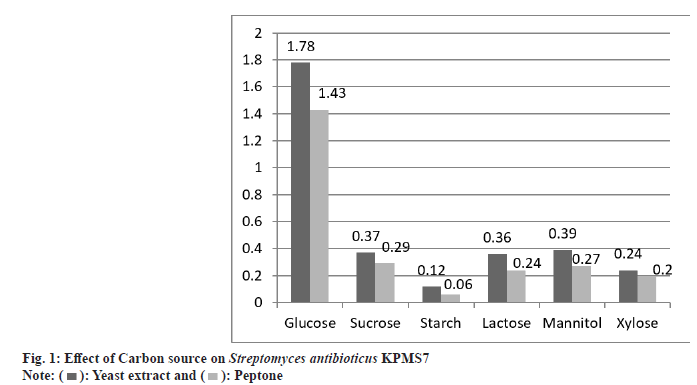
The utilization of different carbon and nitrogen sources was studied on active strain KPMS7. The effect of carbon sources was achieved by 0.1 % concentration nitrogen sources such as peptone and yeast extract. The maximum growth rate was obtained on glucose medium with yeast extract and optimal density value as 1.78 mM. The least growth rate was found in soluble starch as 0.12 mM. In peptone source, the maximum and minimum utilization of carbon sources were found in glucose and soluble starch, and the optimal density was measured as 1.43 mM and 0.06 mM respectively (fig. 1).
Similarly, the effect nitrogen sources were determined by 0.1 % concentration of soluble starch and xylose acted as nitrogen sources. The isolate KPMS7 exhibited maximum growth rate was found on yeast extract supplemented with soluble starch at 1.38 mM optimal density value and minimum growth rate in proline at 0.19 mM (fig. 2). In xylose, the maximum growth rate was 1.13 mM and the least rate as 0.08 mM was observed on yeast extract and proline.
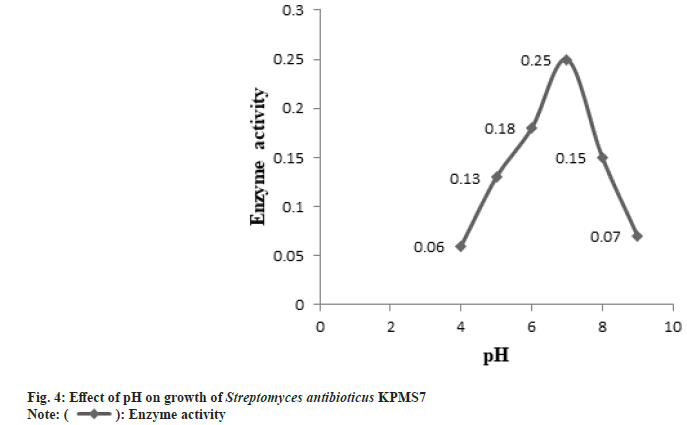
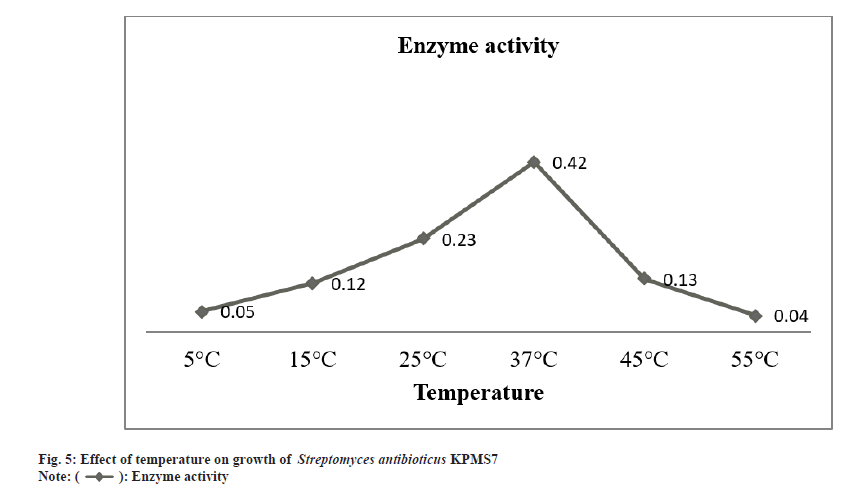
The partial purification of L-asparaginase was carried out by the ammonium sulphate precipitation method. After partial purification, the enzyme was characterized based on biomass production, optimum pH, and temperature. The L-asparaginase enzyme production was started after 24 h of incubation time. The maximal biomass production for enzyme activity was obtained after 172 h (7 d) and then started to decrease the growth rate (fig. 3). The enzyme activity gradually increased and the maximal enzyme production was observed in pH 7 whereas, at higher pH, enzyme production was declined (fig. 4). The enzyme production was found at the optimum temperature of 37° (fig. 5). At higher temperatures, the growth rate was felt down sharply .
The final crude enzyme was partially purified by the ammonium sulfate precipitation method and it was analyzed for protein. The 100 % yield was obtained by crude enzyme extract. The highest enzyme activity was determined as 35.45 IU/ml, total protein was found in 138 mg and specific activity occurred in 71.92 IU/ mg with 1.0 purification fold. The total protein content was found to be 138 mg in marine Streptomyces antibioticus KPMS7. The SDSPAGE analysis showed that the protein band with molecular weight 116 KDa was observed in marine Streptomyces antibioticus KPMS7.
The genomic DNA was extracted from Streptomyces antibioticus KPMS7 amplified for 16S rRNA gene using the primers F243 and R513GC. The size of amplified 16S rRNA gene product of Streptomyces antibioticus KPMS7 was approximately 750 bp. The obtained sequence was compared for homology with other sequences deposited in the BLAST. The Gen Bank accession number of Streptomyces antibioticus KPMS7 was MH819507. The evolutionary analyses were conducted in MEGA5. The phylogenetic tree of the isolate Streptomyces antibioticus KPMS7 was constructed using 16S rRNA gene sequence with that of other Streptomyces species by Neighbor Joining method[15]. The optimal tree with the sum of branch length=0.61904762 was analyzed (fig. 6). The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) are shown next to the branches[16]. The tree was drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the p-distance method and are in the units of the number of base differences per site. The analysis involved 9 nucleotide sequences. Codon positions included were 1st+2nd+3rd+Noncoding. All positions containing gaps and missing data were eliminated. There were a total of 21 positions in the final dataset.
In this present study, actinomycetes were abundantly found in marine sediments. Totally 100 actinomycetes were screened and they are preliminarily characterized morphologically and biochemically. Among 100 actinomycetes, 10 isolates were showed typical shape and spore chain under the microscope. The physiological and biochemical analysis suggested that the strains be classified to Streptomyces group as reported early by many workers[17,18]. The isolated Streptomyces sp was currently updated with a spore-forming gram-positive cell wall[19]. According to the fundamental examinations and 16S rRNA studies, the marine isolated strain was identified as Streptomyces antibioticus KPMS7. The Morphological, biochemical characterizations and sequence analysis were tested for the identification of potent and promising strain Streptomyces is a major research area for many years worldwide[20,21]. Streptomyces sp are characterized by the production of secondary metabolites and moreover, these metabolites included many pigments. Pigmentation by marine Streptomyces sp has been noticed as an important cultural characteristic for defining the organisms[22]. Currently, the genus Stretomyces are accommodates spore forming, Gram-positive filamentous bacteria that developed branching hyphae. Based on these morphological characters, cell wall nature and mycelial formation, the Streptomyces genus were documented [23,24].
All ten isolates were permitted secondary screening for enzyme activity. Among 10 isolates, Streptomyces antibioticus KPMS7 was involved effectively to be produced L-asparaginase by plate method supplemented with phenol red dye. In addition to morphological characteristics showed that the marine Streptomyces is serving as a good source of L-asparaginase enzymeproducing strains by using phenol red dye for screening techniques and converts L-asparagine to L-aspartic acid and ammonia have been reported by other researchers[25,26]. The isolation of 10 actinobacterial strains from marine sources, three isolate S3, S4, and S8 belongs to Streptomyces sp were showed extracellular production of enzyme L-asparaginase[27]. The production of L-asparaginase from marine Streptomyces like Streptomyces aurantiacus[28].
The nutritional characteristics of active strain were studied by using different carbon and nitrogen sources. Among the carbon and nitrogen sources tested, the strain Streptomyces antibioticus KPMS7 was growing well on glucose with yeast extract medium and proved to be the best source of L-asparaginase production under submerged conditions. The maximum rate of enzymatic activity was carried out in Streptomyces sp under submerged condition[29,30]. As previously described, L-asparaginase enzyme production was highly found in the growth medium containing glucose as a carbon substrate by a river isolate of Streptomyces ginsengisoli[31]. Enhanced L-asparaginase synthesis in Streptomyces albidoflavus and Streptomyces glubargensis was observed in yeast extract medium is essential for maximal growth and acted as potent nitrogen source [32,33].
All microorganisms have their own different parameters especially pH and temperature for maximum growth formation that leads to yield maximum enzyme productions. These parameters strongly affect the enzymes production and transport of various components across the cell membrane. The optimal condition is required for maximum enzymes production[34]. In these studies, the enzyme production by Streptomyces antibioticus KPMS7 was started after 24 h of incubation period and reached maximum level was observed in 172 h (7 d). Further studies were revealed that the maximum growth rate for enzyme production was observed in pH 7 and in temperature profile, the highest yield of the enzyme was found at 37°. Most of the marine sources of bacteria and actinomycetes were exhibited maximum L-asparaginase synthesis was found within pH 7-8.5 and the optimum temperature for using marine microorganisms ranging from 28°- 37°[35,36]. Similarly, the maximum L-asparaginase enzyme production from Streptomyces albidoflavus was obtained at pH 7.5 and when cultured at 35° [12].
The purification of an enzyme L-asparaginase synthesized by marine Streptomyces antibioticus KPMS7. The proteins from the crude enzyme extract were precipitated by ammonium sulphate saturation. The crude enzyme extract yielded 100 % of L-asparaginase with 71.92 IU/mg specific activity. Several studies have shown that the crude enzyme extract was secured maximum yield for L-asparaginase synthesis obtained from various marine Streptomyces sp[37-39]. In SDS-PAGE studies, the molecular weight of partially purified L-asparaginase enzyme from Streptomyces antibioticus KPMS7 was found to be 116 kDa. As previous studies reported that the L-asparaginase from Streptomyces sp. PDK2, Streptomyces albidoflavus, and Streptomyces gulbargensis exhibited molecular weights of 140, 140, and 112 kDa, respectively[40-42]. Marine sources have unique features to produce novel bioactive metabolites such as antibiotics, enzymes and antitumor compounds against all infectious diseases.
Molecular analysis was useful for the identification and reliable detection of different Streptomyces as well as monitoring of the species. The genomic DNA was extracted from Streptomyces antibioticus KPMS7 amplified for 16S rRNA gene using the primers F243 and R513GC[43]. The sequences obtained were compared for homology with other sequences deposited in the public databases using the Basic Local Alignment Search Tool. The sequence analysis usually includes alignment of sequences, construction of a phylogenetic tree and testing the reliability of the constructed tree. The relationships of the aligned sequences are usually shown as a tree, in which the branching pattern of the tree displays the evolutionary relationships of the Streptomyces strains. The most commonly applied tree construction method is distance method in which neighbour joining use pair-wise distances calculated from aligned sequences before correcting to evolutionary distances in a substitution model[44]. The shortest distant sequences were clustered together in a tree and its length was optimized to correspond to the distance matrix. The evolutionary analyses were conducted using the software MEGA5 [45].
A marine environment offers plentiful sources for biological reactions. It provides the highest place for various kinds of microorganisms for the production of secondary metabolites. Among marine microbial communities, actinomycetes are predominantly screened for their metabolic significance in the medical and pharmaceutical industries. Among actinomycetes genera, Streptomycetes actively produced one-third of metabolites like antibiotics and Enzymes. From an industrial point of view, Enzymes after antibiotics have a key role in general health. With the increasing advancement in medical science, there would be greater demand for L-asparaginase enzyme from marine sediment sources for clinical trial and food safety control.
This study concludes that Streptomyces sp is the most efficient actinomycetes produced L-asparaginase from marine sediment sources. Marine Streptomyces sp. are well organized L-asparaginase producer is required for treatment of several human dreadful diseases. Currently, L-asparaginase from marine Streptomyces antibioticus KPMS7 (MH819507) sources has been suggested as the promising therapeutic applicant in the clinical research to benefit the general health sector to compete against leukemia and other types of cancer disorder .
Acknowledgements:
We would like to thank the PG and Research Department of Microbiology, Jamal Mo¬hamed College (Autonomous), Affiliated to Bharathidasan University, Tiruchirappalli-620020, Tamil Nadu, India, towards this research work hereby acknowledged.
Conflicts of interest:
The authors declared that there is no conflict of interest.
References
- Labrou NE, Papageorgiou AC, Avramis VI. Structure-function relationships and clinical applications of L-asparaginases. Curr Med Chem 2010;17(20):2183-95.
[Crossref] [Google Scholar] [PubMed]
- Shojaei F, Homaei A, Taherizadeh MR, Kamrani E. Characterization of biosynthesized chitosan nanoparticles from Penaeus vannamei for the immobilization of P. vannamei protease: An eco-friendly nanobiocatalyst. Int J Food Prop 2017;20(sup2):1413-23.
- D’Souza R, Abraham A. Cytotoxic potential of L-asparaginase from Bacillus sp. in vitro. Bact Emp 2019;2(2):49-53.
- Isaac GS, Abu-Tahon MA. Production of extracellular anti-leukemic enzyme L-asparaginase from Fusarium solani AUMC 8615 grown under solid-state fermentation conditions: purification and characterization of the free and immobilized enzyme. Egypt J Bot 2016;56:799-816.
- Peela S, Kurada VB, Terli R. Studies on antagonistic marine actinomycetes from the Bay of Bengal. World J Microbiol Biotechnol 2005;21:583-5.
- E-A El-Naggar N. Extracellular production of the oncolytic enzyme, L-asparaginase, by newly isolated Streptomyces sp. strain NEAE-95 as potential microbial cell factories: Optimization of culture conditions using response surface methodology. Curr Pharm Biotechnol 2015;16(2):162-78.
[Crossref] [Google Scholar] [PubMed]
- Grayson I. Chemocatalysis or biocatalysis? Continuous or Batch? Teckno Scienze Srl 2016;34(5):27.
- Pui CH, Robison LL, Look AT. Acute lymphoblastic leukaemia. Lancet 2008;371(9617):1030-43.
[Crossref] [Google Scholar] [PubMed]
- El-Naggar NE, Moawad H, El-Shweihy NM, El-Ewasy SM. Optimization of culture conditions for production of the anti-leukemic glutaminase free L-asparaginase by newly isolated Streptomyces olivaceus NEAE-119 using response surface methodology. BioMed Res Int 2015;2015:627031.
[Crossref] [Google Scholar] [PubMed]
- Trincone A. Enzymatic processes in marine biotechnology. Mar Drugs 2017;15(4):93.
[Crossref] [Google Scholar] [PubMed]
- Gulati R, Saxena RK, Gupta R. A rapid plate assay for screening L?asparaginase producing micro?organisms. Lett Appl Microbiol 1997;24(1):23-6.
[Crossref] [Google Scholar] [PubMed]
- Narayana KJ, Kumar KG, Vijayalakshmi M. L-asparaginase production by Streptomyces albidoflavus. Ind J Microbiol 2008;48:331-6.
[Crossref] [Google Scholar] [PubMed]
- Wriston JC, Yellin TO. L-asparaginase: A review. Adv Enzymol Relat Areas Mol Biol 1973;39:185-248.
[Crossref] [Google Scholar] [PubMed]
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the Folin phenol reagent. J Biol Chem 1951;193:265-75.
[Google Scholar] [PubMed]
- Saitou N, Nei M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol Biol Evol 1987;4(4):406-25.
[Crossref] [Google Scholar] [PubMed]
- Felsenstein J. Confidence limits on phylogenies: An approach using the bootstrap. Evolution 1985;39(4):783-91.
- Hassan AA, El-Barawy AM, El Mokhtar MN. Evaluation of biological compounds of Streptomyces species for control of some fungal diseases. J Am Sci. 2011;7(4):752-60.
- Laidi RF, Kansoh AL, Elshafei AM, Cheikh B. Taxonomy, identification and biological activities of a novel isolate of Streptomyces tendae. Arab J Biotechnol 2006;9(3):427-36.
- Waksman SA. The Actinomycetes Classification, identification and description of genera and species. Baltimore: Lippicncott Williams and Wilkins 1961; 2:377-75.
- Shaik M, Sankar GG, Iswarya M, Rajitha P. Isolation and characterization of bioactive metabolites producing marine Streptomyces parvulus strain sankarensis-A10. J Genetic Engi Biotechnol 2017;15(1):87-94.
[Crossref] [Google Scholar] [PubMed]
- Abbas IH. A biological and biochemical studies of actinomycetes isolated from Kuwait saline soil-Kuwait. J Appl Sci Res 2006;2(10):809-15.
- Abraham J, Chauhan R. Profiling of red pigment produced by Streptomyces sp. JAR6 and its bioactivity. 3 Biotech 2018;8(1):22.
[Crossref] [Google Scholar] [PubMed]
- Waksman SA. The Actinomycetes. Vol. II. Classification, identification and descriptions of genera and species. Baltimore: The Williams and Wilkins Co1961;1-363.
- Anderson AS, Wellington EM. The taxonomy of Streptomyces and related genera. Int J Syst Evol Microbiol 2001;51(3):797-814.
[Crossref] [Google Scholar] [PubMed]
- Arango C, Acosta-Gonzalez A, Parra-Giraldo CM, Sánchez-Quitian ZA, Kerr R, Díaz LE. Characterization of actinobacterial communities from Arauca river sediments (Colombia) reveals antimicrobial potential presented in low abundant isolates. Open Microbiol J 2018;12:181.
[Crossref] [Google Scholar] [PubMed]
- Sarquis MI, Oliveira EM, Santos AS, Costa GL. Production of L-asparaginase by filamentous fungi. Mem Inst Oswaldo Cruz 2004;99:489-92.
[Crossref] [Google Scholar] [PubMed]
- Basha NS, Rekha R, Komala M, Ruby S. Production of extracellular anti-leukaemic enzyme lasparaginase from marine actinomycetes by solidstate and submerged fermentation: Purification and characterisation. Tropic J Pharm Res 2009;8(4):353-60.
- Gupta N, Mishra S, Basak UC. Occurrence of Streptomyces aurantiacus in mangroves of Bhitarkanika. Malaysian J Microbiol 2007;3:7-14.
- Khamna S, Yokota A, Lumyong S. L-asparaginase production by actinomycetes isolated from some Thai medicinal plant rhizosphere soils. Int J Int Biol 2009;6(1):22-6.
- Sahu MK, Sivakumar K, Poorani E, Thangaradjou T, Kannan L. Studies on L-asparaginase enzyme of actinomycetes isolated from estuarine fishes. J Environ Biol 2007;28(2):465-74.
[Google Scholar] [PubMed]
- Deshpande N, Choubey P, Agashe M. Studies on optimization of growth parameters for L-asparaginase production by Streptomyces ginsengisoli. Sci World J 2014;2014:895167.
[Crossref] [Google Scholar] [PubMed]
- Amena S, Vishalakshi N, Prabhakar M, Dayanand A, Lingappa K. Production, purification and characterization of L-asparaginase from Streptomyces gulbargensis. Braz J Microbiol 2010;41:173-8. [Crossref] [Google Scholar] [PubMed]
- Verma N, Kumar K, Kaur G, Anand S. L-asparaginase: A promising chemotherapeutic agent. Crit Rev Biotechnol 2007;27(1):45-62.
[Crossref] [Google Scholar] [PubMed]
- Rahimzadeh M, Poodat M, Javadpour S, Qeshmi FI, Shamsipour F. Purification, characterization and comparison between two new L-asparaginases from Bacillus PG03 and Bacillus PG04. Open Biochem 2016;10:35-45.
[Crossref] [Google Scholar] [PubMed]
- Sudha K., Rani J, Sivasankari TR. Extracellular L-asparaginase production by halotolerant of strain Enterobacter hormacchie isolated from marine fishes. Int J Adv Res 2017;5(1):2619-25.
- Nageswara S, Guntuku G, Tadimalla P. Production of L-asparaginase by solid state fermentation using marine fungus. BioMed Res 2014:1-9.
- Meena B, Anburajan L, Dheenan PS, Begum M, Vinithkumar NV, Dharani G, et al. Novel glutaminase free L-asparaginase from Nocardiopsis alba NIOT-VKMA08: Production, optimization, functional and molecular characterization. Bioprocess Biosyst Eng 2015;38:373-88.
[Crossref] [Google Scholar] [PubMed]
- Sivasankar P, Sugesh S, Vijayanand P, Sivakumar K, Vijayalakshmi S, Balasubramanian T, et al. Efficient production of L-asparaginase by marine Streptomyces sp. isolated from Bay of Bengal, India. Afr J Microbiol Res 2013;7(31):4015-21.
- Dharmaraj S. Study of L-asparaginase production by Streptomyces noursei MTCC 10469, isolated from marine sponge Callyspongia diffusa. Iran J Biotechnol 2011;9(2):102-8.
- Dhevagi P, Poorani E. Isolation of L-asparaginase producing actinomycetes from marine sediments. Int J Curr Res Aca Rev 2016;4:88-97.
- Dharmaraj S, Dhevendaran K. Evaluation of Streptomyces as a probiotic feed for the growth of ornamental fish Xiphophorus helleri. Food Technol Biotechnol 2010;48(4):497-504.
- Dharmaraj S, Ashokkumar B, Dhevendaran K. Fermentative production of carotenoids from marine actinomycetes. Iran J Microbiol 2009;1(4):36-41.
- Ravikumar S, Fredimoses M, Gokulakrishnan R. Biodiversity of actinomycetes in Manakkudi mangrove ecosystem, Southwest coast of India. Annal Biol Res 2011;2(1):76-82.
- Nei M, Kumar S. Molecular Evolution and Phylogenetics. New York: Oxford University Press; 2000.
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: Molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol 2011;28(10):2731-9.
[Crossref] [Google Scholar] [PubMed]
 ): Yeast extract and (
): Yeast extract and ( ): Peptone
): Peptone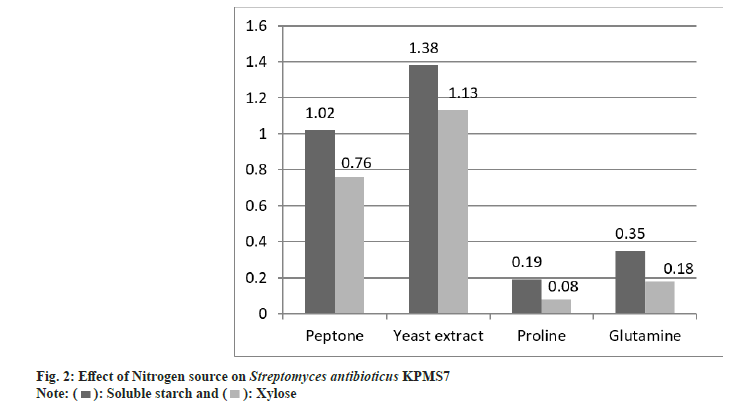
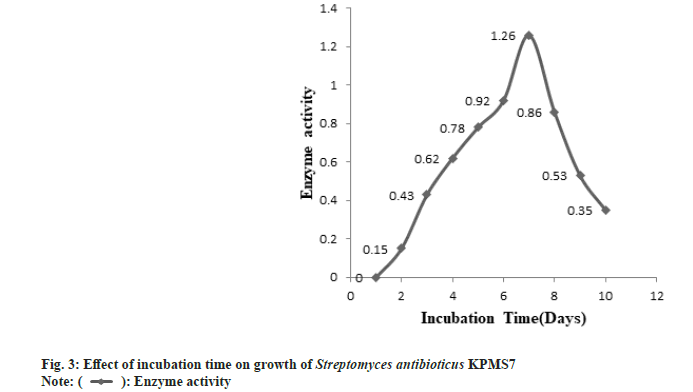
 ): Enzyme activity
): Enzyme activity