- *Corresponding Author:
- GA. Garoosi
Department of Biotechnology,
Qazvin 34149 16818,
Iran
E-mail: agaroosi90@yahoo.com
| Date of Received | 05 July 2019 |
| Date of Revision | 03 October 2021 |
| Date of Acceptance | 05 July 2021 |
| Indian J Pharm Sci 2021;83(4):663-670 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Stevia rebaudiana (Bert.) produces steviol glycoside sweeteners that are sweeter than sucrose. Stevioside and rebaudioside A are important steviol glycosides used in food production. This study was conducted to investigate the effect of different concentrations of cadmium chloride (0, 20, 40, 60 and 80 mg/L) and silver nitrate (0, 15, 30, 45 and 60 mg/L) and different sampling times on stevioside and rebaudioside A production and Ent-kaurenoic acid 13-hydroxylase, uridine diphosphate-glycosyltransferase 74G1 and uridine diphosphate-glycosyltransferase 76G1 genes expression in in vitro conditions. The results showed that the application of cadmium nitrate and silver nitrate decreased stevioside and rebaudioside A production. Among various concentrations of cadmium chloride, the highest concentrations of stevioside and rebaudioside A were respectively 39.76 mg/g and 2.27 mg/L dry weight 96 and 72 h after elicitation with 20 mg/L cadmium chloride. Among different concentrations of silver nitrate, the highest amounts of stevioside and rebaudioside A were respectively 33.33 mg/g dry weight was 2.19 mg/L obtained at control condition after 96 h and elicitation with 60 mg/L for 24 h. In elicitation with cadmium chloride the expression all three genes were increased but silver nitrate elicitation increased uridine diphosphate-glycosyltransferase 76G1 gene expression and decreased the Ent-kaurenoic acid 13-hydroxylase and uridine diphosphateglycosyltransferase 74G1 genes expression. The correlation analysis showed that Ent-kaurenoic acid 13-hydroxylase gene expression was not correlated with uridine diphosphate-glycosyltransferase 74G1 and uridine diphosphate-glycosyltransferase 76G1 genes expression, while uridine diphosphateglycosyltransferase 74G1 was inversely correlated with uridine diphosphate-glycosyltransferase 76G1. Also, positive correlations were observed between uridine diphosphate-glycosyltransferase 74G1 gene expression with stevioside synthesis and uridine diphosphate-glycosyltransferase 76G1 gene expression with rebaudioside A.
Keywords
Cadmium chloride, gene expression, silver nitrate, Stevia rebaudiana (Bert.), rebaudioside A, stevioside
Stevia rebaudiana Bert. (S. rebaudiana) is a medicinal plant with different valuable compounds such as steviol glycosides (SGs) [1]. S. rebaudiana has 34 types of SGs such a stevioside, rebaudioside A, rebaudioside C and dulcoside A [2,3]. The sweetness of stevioside, rebaudioside A, rebaudioside C and dulcoside A is respectively about 110-270, 150-320, 40-60 and 30 times higher than that of sucrose [4]. These compounds have different medicinal applications for diabetes, dental maladies, obesity, hypertension and cancer treatments [5]. Stevioside is one of the most important active compounds applied in many food products in different countries [6]. The studies showed that different enzymes are involved in SGs production such as KA13H, UGT74G1 and UGT76G1 [7,8].
Different factors such as genotype, propagation methods, environmental conditions and agronomic practices affect the amount of SGs production [9]. Plant cell and tissue culture technique is an important strategy for production of highly valuable compounds [10]. Although using this system allows producing more predictable and stable secondary compounds, but the amount of these compounds is very low [11]. To enhance the secondary metabolites production in in vitro condition, different techniques such as elicitation are developed [12]. Researchers have shown that the production and accumulation of secondary metabolites are a part of defense responses against pathogens, physical and chemical stresses [13].
Elicitors are compounds that trigger the formation of secondary metabolites and stimulate their accumulation [14,15]. Elicitors are classified according to their origin as biotic and abiotic (such as cadmium chloride and silver nitrate) compounds [2]. The studies showed that cadmium chloride increased secondary metabolites production in Vitis vinifera cv. (V. vinifera cv) [16], Chataranthus roseus [17], Artemisia annua L. [18], Glycine max L. (G. maxL.) [19] and Salvia miltiorrhiza (S. miltiorrhiza) [20] and silver nitrate have been found to increase secondary metabolites production in Malva sylvestris L. [21], Genista tinctoria L. [22], Agastache rugosa (A. rugosa) [23] and Silybum marianum [24]. Therefore, in this study, we report for the first time the effect of cadmium chloride, silver nitrate and exposure times of elicitors on stevioside and rebaudioside A production and on Ent-kaurenoic acid 13-hydroxylase (KA13H), uridine diphosphate-glycosyltransferase 74G1 (UGT74G1) and uridine diphosphate-glycosyltransferase 76G1 (UGT76G1) genes expression in S. rebaudiana (Bert.) in vitro culture.
Materials and Methods
Plant material:
The Stevia plant was obtained from the College of Agriculture and Natural Resource, University of Tehran, Karaj, Iran. Stem cutting with 10-15 cm size was prepared and washed for 0.5 h with tap water. The cutting surface was sterilized with 70 % (v/v) ethanol for 1 min, 5.1 % (w/v) sodium hypochlorite solution for 15 min and rinsed 4 times in sterile distilled water for 6 min. For plant propagation, node explants with axillary buds (10-20 mm) size were prepared and transferred to Murashige and Skoog medium (MS medium) [25] supplemented with 0.5 mg/l Blood-agar Plate (BAP) and 0.1 mg/l Naphthaleneacetic acid (NAA), maintained in a growth chamber at 25±2° with 16 h photoperiod and sub-cultured every 21 d. For the equal growth of the plants, the seedlings with 6-8 leaflets and without lateral buds and roots were selected and transferred to MS medium for 15 d.
Elicitor treatment:
In this study, we used silver nitrate (AgNO3) (PanreacQuimicasa, Spain) and cadmium chloride (CdCl2.H2O) (Riedel-De, Germany) as elicitors. Stock solutions were prepared individually and sterilized with 0.2 μm syringe filter. To investigate the effect of cadmium chloride and silver nitrate, cadmium chloride at concentrations of 0, 20, 40, 60 and 80 mg/L and silver nitrate at concentrations of 0, 15, 30, 45 and 60 mg/L were added to MS medium after autoclaving. Subsequently, seedlings were transferred to elicitation MS media including 6.5 mg/L plant agar. Sampling was performed at 24, 48, 72 and 96 h after elicitation. After sampling, some of the samples were frozen in liquid nitrogen and maintained at -80° for gene expression analysis.
Extraction of stevioside and rebaudioside A:
The stevioside and rebaudioside A extraction from shoots of elicited and control seedlings were prepared as described by Kolb et al. [26] with a few modifications. The samples were oven dried for 24 h at 50°. The dried samples were powdered in the porcelain dish. Then, 2 ml of 70 % ethanol was added to 20 mg powdered sample and mixed. Next, the samples were placed for 0.5 h at 70° in a water bath (Techno, England) and shaken every 5 min. Finally, the mixture was centrifuged at 14 000 rpm for 10 min and the supernatant was transferred to a new tube for high-performance liquid chromatography (HPLC) analysis and maintained at -20°.
HPLC analysis of stevioside and rebaudioside A:
HPLC analysis was performed on a Knauer HPLC system (Ultraviolet (UV) detector, Germany). A volume of 20 μl of samples was injected into a C-18 reverse-phase Tosoh column (TSKgel-ODS C-18, 5 μm, 4.6×250 mm). The mobile phase for stevioside and rebaudioside A elution was 32 % water and 68 % methanol. The effluent was monitored at 210 nm and curves of stevioside and rebaudioside A standards (Sigma-Aldrich Chemical Co, USA) were used to calculate the stevioside and rebaudioside A concentration in the samples [27].
Extraction of total Ribonucleic acid (RNA):
RNA was extracted from the treatments with the highest and lowest contents of stevioside related samples with an RNX-Plus kit (CinnaGen, Iran) based on the manufacturer’s instructions. To evaluate the quality of RNA, the extracted RNA samples were loaded on 1.5 % agarose gel. In this work, NanoDrop 2000c spectrophotometer (Thermo Scientific, USA) was used to evaluate the quantity of RNA.
Synthesis of total complementary Deoxyribonucleic acid (cDNA):
For the synthesis of a single strand of total cDNA, RNA stock was prepared at 1 μg/μl concentration. First, 3 μg of RNA was mixed with 1 μg/μl Oligo (dT)18 primer (CinnaGen, Iran) and 9 μl deionized water in the 0.5 ml microtube, maintained at 70° for 5 min and immediately transferred to the ice. Then, 2 μl 10X reverse transcription buffer (CinnaGen, Iran) with 4 μl deoxynucleotide triphosphates, mix 10 mm (CinnaGen, Iran) was added to microtube and maintained at 37° for 5 min. Subsequently, 1 μl of Reverse Transcriptase enzyme (200 u/μl, CinnaGen, Iran) was added and the reaction mixture was incubated at 42° for 1 h. The reaction was stopped by heating the mixture at 70° for 10 min. The synthesized template cDNA was maintained at -20° [28].
Primer design:
The primers of KA13H, UGT74G1 and UGT76G1 genes studied in the present work were designed by Mandal et al. [5]. The actin gene was used as a house keeping gene, which was specific for Stevia and its primer designed by Oligo v7.56 software (Table 1). The primers were tested by Primer Blast of NCBI (https://www.ncbi.nlm.nih.gov) and synthesized by BioNEER Corporation (South Korea).
| Gene (Accession number) | Primer sequence (forward primer, F and reverse primer, R) (5'-3') | Amplicon length (bp) |
|---|---|---|
| KA13H (DQ398871.3) | F: CCTATAGAGAGGCCCTTGTGG | 102 |
| R: TAGCCTCGTCCCTTTGTGTC | ||
| UGT74G1 (AY345982.1) | F: GGTAGCCTGGTGAAACATGG | 115 |
| R: CTGGGAGCTTTCCCTCTTCT | ||
| UGT76G1 (AY345974.1) | F: GACGCGAACTGGAACTGTTG | 121 |
| R: AGCCGTCGGAGGTTAAGACT | ||
| Actin (AF548026.1) | F: GCCCCTAGCAGCATGAAGAT | 162 |
| R: GCACTTCCTGTGGACAATGG |
Table 1: List of Gene Specific Primers Used in Pcr and Qrt-Pcr Analysis
To verify the authenticity of primers on cDNA, the polymerase chain reaction (PCR) was performed by a combination of 7 μl deionized water, 1 μl of each forward and reverse primers, 1 μl cDNA and 10 μl Master PCR. Thermal cycles of PCR reaction included 1 cycle at 94° for 5 min, 35 cycles at 94° for 30 s, 58° for 45 s, 72° for 30 s and 1 cycle at 72° for 10 min.
Statistical analysis:
All experiments were performed based on a completely randomized design in triplicate. Data analyses were performed using IBM statistical package for the social sciences (SPSS) statistics for Windows, Version 23.0 (Armonk, NY, USA). Mean comparisons were carried out using Duncan’s multiple range test at a probability level of 0.05. T-test was applied to compare the expression of the gene in the treatments with the highest and lowest contents of stevioside.
Results and Discussion
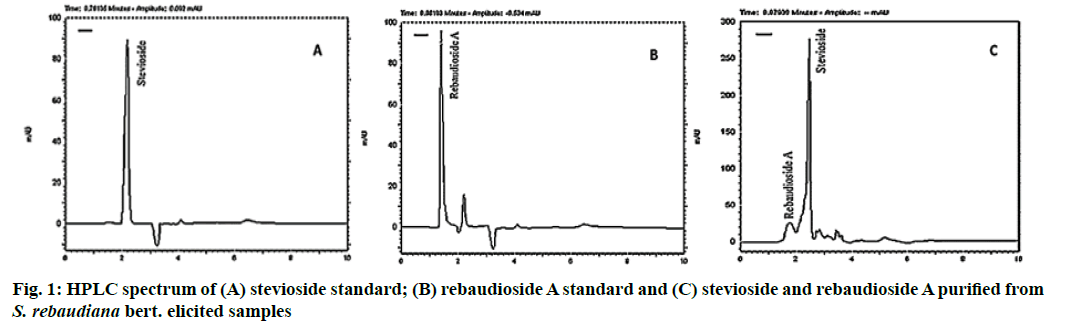
The stevioside and rebaudioside A extracted from samples were detected using the HPLC method with the C-18 column. Fig. 1 shows the HPLC spectrum of stevioside and rebaudioside A in samples. Elicitation of Stevia seedlings with different concentration of cadmium chloride had a significant effect of stevioside and rebaudioside A production (p<0.01). The stevioside production was decreased by increasing cadmium chloride concentration to 40 mg/L but then increased by increasing cadmium chloride concentration from 40 mg/L to 80 mg/L. Generally, application of different concentrations of cadmium chloride reduces stevioside production than the control. Therefore, by using 20 mg/L (31.42 mg/g dry weight (DW)), 40 mg/L (25.30 mg/g DW), 60 mg/L (27.12 mg/g DW) and 80 mg/L (30.63 mg/g DW) cadmium chloride the stevioside production decreased 0.85 %, 20.14 %, 14.42 % and 3.34 % than the control respectively (Table 2). Also, elicitation with different concentrations of cadmium chloride had a negative effect on rebaudioside A production by increasing cadmium chloride concentrations. On the other hand, the using 20 mg/L (1.03 mg/g DW), 40 mg/L (1.12 mg/g DW), 60 mg/L (0.21 mg/g DW) and 80 mg/L (0.34 mg/g DW) of cadmium chloride decreased rebaudioside A production 34.81 %, 29.75 %, 87.34 % and 78.48 % compared to the control respectively (Table 2). Among different concentrations of cadmium chloride, the highest amount of stevioside and rebaudioside A production were respectively 31.96 and 1.58 mg/g DW obtained in the control condition.
| Cadmium chloride | Silver nitrate | ||||||
|---|---|---|---|---|---|---|---|
| Concentrations (mg/L) | Sampling times(H) | Stevioside(mg/g DW) | Rebaudioside A(mg/g DW) | Concentrations(mg/l) | Sampling times(H) | Stevioside (mg/g DW) | Rebaudioside A(mg/g DW) |
| Control | 24 | 30.97±0.55bcd | 1.82±0.39ab | Control | 24 | 30.97±0.55abc | 1.82±0.39ab |
| 48 | 29.84±0.88bcd | 1.68±0.56ab | 48 | 29.84±0.88abc | 1.68±0.56ab | ||
| 72 | 32.64±0.6abc | 1.49±0.35abc | 72 | 32.64±0.60ab | 1.80±0.16ab | ||
| 96 | 33.33±0.96abc | 1.33±0.20bcd | 96 | 33.33±0.96a | 1.33±0.20a-e | ||
| 20 | 24 | 23.52±0.77de | 0.78±0.10cde | 15 | 24 | 29.71±1.51abc | 0.71±0.00c-g |
| 48 | 32.22±1.85abc | 0.71±0.29cde | 48 | 29.40±3.38abc | 2.06±0.66ab | ||
| 72 | 30.18±2.7bcd | 2.27±0.39a | 72 | 26.00±3.40a-d | 1.15±0.31b-f | ||
| 96 | 39.76±3.13a | 0.37±0.13e | 96 | 25.58±0.66b-e | 1.82±0.38ab | ||
| 40 | 24 | 26.46±2.83cde | 1.44±0.30bc | 30 | 24 | 21.51±1.66d-g | 1.51±0.00abc |
| 48 | 26.4±1.78cde | 0.54±0.24de | 48 | 21.88±3.63d-g | 0.11±0.00g | ||
| 72 | 20.17±1.26e | 2.00±0.19ab | 72 | 19.36±2.31d-g | 1.42±0.05a-d | ||
| 96 | 28.19±1.29cde | 0.48±0.26e | 96 | 19.73±1.91d-g | 0.17±0.04g | ||
| 60 | 24 | 25.48±5.51cde | 0.37±0.05e | 45 | 24 | 23.65±1.12c-f | 0.16±0.02g |
| 48 | 26.67±4.37cde | 0.08±0.06e | 48 | 18.11±0.66efg | 0.14±0.00g | ||
| 72 | 29.04±2.24bcd | 0.07±0.00e | 72 | 16.72±1.77fg | 0.19±0.00g | ||
| 96 | 27.29±3.04cde | 0.31±0.13e | 96 | 15.22±0.56g | 0.26±0.03fg | ||
| 80 | 24 | 25.84±2.36cde | 0.71±0.18cde | 60 | 24 | 23.93±4.84c-f | 2.19±0.42a |
| 48 | 27.91±1.15cde | 0.00±0.00e | 48 | 26.51±0.86a-d | 0.56±0.15d-g | ||
| 72 | 32.3±2.44abc | 0.44±0.14e | 72 | 20.79±2.70d-g | 0.48±0.27efg | ||
| 96 | 36.46±1.87ab | 0.21±0.09e | 96 | 20.58±3.24d-g | 0.32±0.16fg | ||
Table 2: The Effect of Interactions Between Different Concentrations of Cadmium Chloride and Silver Nitrate with Sampling Times on Stevioside and Rebaudioside a Production in S. rebaudiana BERT. Seedlings
The results showed that the sampling time has a significant effect on stevioside and rebaudioside A production (p<0.01) and the stevioside production had increased over time. Among different sampling times, the highest amount of stevioside production was 33 mg/g DW at 96 h after treatment, which was 24.76 %, 15.34 % and 14.30 % higher than that produced 24 h (26.45 mg/g DW), 48 h (28.61 mg/g DW) and 72 h (28.87 mg/g DW) after treatment respectively (Table 2). Also, among different sampling times, the highest amount of rebaudioside A was 1.25 mg/g DW obtained 72 h after elicitation with cadmium chloride, which was respectively 22.54 %, 108.33 % and 131.48 % higher than that produced 24 h (1.03 mg/g DW), 48 h (0.60 mg/g DW) and 96 h 0.54 mg/g DW) after the elicitation (Table 2).
In the interaction of elicitor concentrations and sampling times, the highest amount of stevioside production was 39.76 mg/g DW observed at 96 h after elicitation with 20 mg/L cadmium chloride and the highest amount of rebaudioside A production was 2.27 mg/g DW obtained 72 h after elicitation with 20 mg/L cadmium chloride (Table 2). Cetin et al. [16] reported that different concentrations of cadmium chloride have a significant effect on secondary metabolites production in cell suspension cultures of V. vinifera cv. The results of this study indicated that the highest value of total phenolic (168.82 mg/100 g), total flavanol (15.94 mg/100 g), total flavonol (14.73 mg/100 g) and trans-resveratrol (490.76 μg/100 g) were obtained 2 d after treatment with 1.0 mm cadmium chloride. Also, it has been shown that cadmium chloride affects secondary compounds production in field conditions. For example, applying different concentrations of cadmium chloride (0.25, 0.5, 1.0 and 2 mg/L) leads to an increase in proline, protein, amino acid and daidzein in G. max L. and liposoluble and hydrosoluble compounds in the shoots of S. miltiorrhiza [19,20].
The results showed that elicitation with different concentrations of silver nitrate had a significant effect (p<0.01) on stevioside and rebaudioside A production, but different sampling times did not affect stevioside and rebaudioside A production. Also, elicitation with silver nitrate had a negative effect on stevioside production. In the control condition, the stevioside production was 27.67 mg/L, which was higher than elicitation conditions. By using silver nitrate the stevioside content decreased 12.68 %, 34.93 %, 41.87 % and 27.58 % at 15 mg/L (27.67 mg/g DW), 30 mg/L (20.62 mg/g DW), 45 mg/L (18.42 mg/g DW) and 60 mg/L (22.95 mg/g DW) compared to the control, respectively (Table 2). Using different concentrations of silver nitrate also resulted in a decrease of the rebaudioside A production. By application of 15 mg/L (1.43 mg/g DW), 30 mg/L (0.80 mg/g DW), 45 mg/L (0.19 mg/g DW) and 60 mg/L (0.89 mg/g DW) silver nitrate the rebaudioside A production decreased 13.3 %, 51.51 %, 89.10 % and 46.67 % compared to the control (1.43 mg/g DW), respectively (Table 2).
Stevioside production, unlike rebaudioside A production, was not affected by different sampling times. The highest stevioside production was 25.95 mg/g DW at 24 h after elicitation but was not statistically significant different from other sampling times (Table 2). The highest rebaudioside A production was 1.28 mg/g DW obtained 24 h after elicitation, which was 41.11 %, 27 % and 62.82 % higher than of 48 h (0.91 mg/g DW), 72 h (1.01 mg/g DW) and 96 h (0.78 mg/g DW) of sampling times, respectively (Table 2).
By investigating the different concentrations of silver nitrate and different sampling times, the highest amount of stevioside was 33.33 mg/g DW observed at control condition 96 h after elicitation, while the highest amount of rebaudioside A was 2.19 mg/g DW obtained from 60 mg/L silver nitrate after 24 h of elicitation (Table 2). Jayalakshmi et al. [21] reported that spraying silver nitrate on Malva sylvestris L. leaves increases anthocyanin production. Also, Vildova et al. [24] showed that treating of Silybum marianum cell suspension culture increases taxifolin production. In that research, the highest amount of taxifolin was 2.2 mg/g in treatment with 5.887×104 mol/l of silver nitrate. Deepthi and Satheeshkumar et al. [30] reported that silver nitrate significantly increases biomass and camptothecin (CPT) yield of Ophiorrhiza mungos L, but its higher concentrations result in a decrease in cell growth and CPT level.
After analysis of stevioside and rebaudioside A production by HPLC system, the samples with highest and lowest amount of stevioside at different concentrations of cadmium chloride and silver nitrate were selected for RNA extraction (Table 2).
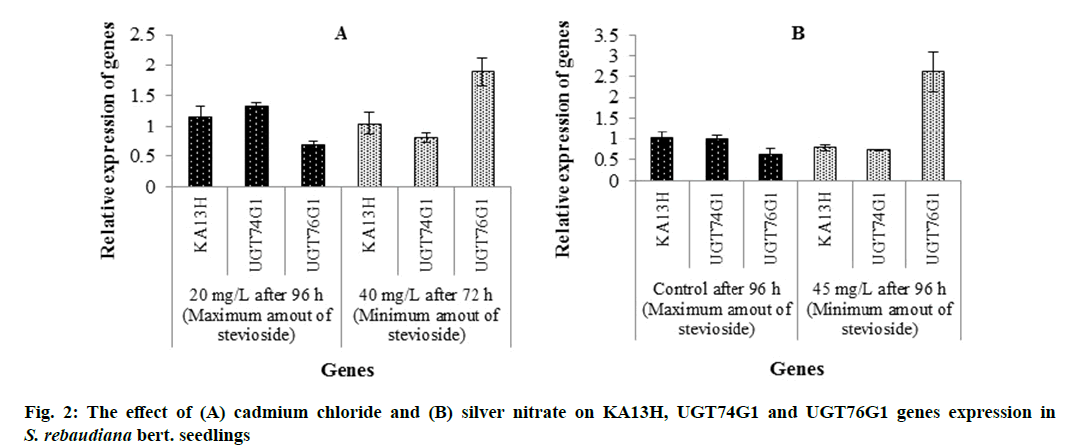
The results showed that by 96 h elicitation with 20 mg/L cadmium chloride the KA13H gene expression increases 1.10-fold compared to the elicitation with 40 mg/L after 72 h, but which is not statistically significant. Also, the expression of UGT74G1 gene at 96 h after elicitation with 20 mg/L cadmium chloride was 1.65-fold higher than 72 h after elicitation with 40 mg/L cadmium chloride. However, the expression of UGT76G1 gene was decreased 2.78-fold 96 h after elicitation with 20 mg/L cadmium chloride compared to the 76 h after elicitation with 40 mg/L cadmium chloride. Therefore, under cadmium chloride elicitation by increasing the KA13H and UGT74G1 genes expression and decreasing the UGT76G1 gene expression, the stevioside production was increased (fig. 2A). The effect of cadmium chloride on genes expression was investigated in different studies. For example, it was found that treatment of Arabidopsis thaliana with cadmium chloride increased OsMSR3 gene expression [31]. Charfeddine et al. [32] studied the effect of cadmium chloride on genes expression of wild and transgene-species of Solanum tuberosum and reported that application of cadmium chloride in transgenic plants improved plant growth, proline production and antioxidant production by increasing the stress dehydration responsive element binding (StDREB) gene expression.
The results indicated that in the control condition after 96 h the KA13H gene expression increases 1.29-fold compared to the 96 h after elicitation with 45 mg/L silver nitrate. The expression of UGT74G1 gene at control condition after 96 h was 1.39-fold higher than that at 96 h after elicitation with 45 mg/L silver nitrate. In comparison, the expression of UGT76G1 gene is decreased in control condition after 96 h compared to that at 96 h after elicitation with 45 mg/L silver nitrate. Therefore, by elicitation with silver nitrate the KA13H and UGT74G1 genes expression were decreased, UGT76G1 gene expression was increased and stevioside production was decreased (fig. 2B). However, it seems that cadmium chloride and silver nitrate have different effects on the expression of these genes. On the other hand, it was expected that with increasing UGT76G1 gene expression, the amount of rebaudioside A to be increased, but on the contrary, it was decreased (Table 2). These findings suggest that a deeper and more accurate study of the effect of elicitors and elicitation time on gene expression and metabolite production should be performed. In different studies, the effect of silver nitrate on genes expression was evaluated. Park et al. [23] reported that treatment of A. rugosa by silver nitrate increases phenylalanine ammonia-lyase (PAL) expression.
The correlation analysis showed that in elicitation with different concentrations of cadmium chloride, the KA13H gene expression was not correlated with UGT74G1 and UGT76G1 genes expression and stevioside and rebaudioside A production. The UGT74G1 gene expression had a negative correlation with UGT76G1 and rebaudioside A production while it had a positive correlation with stevioside production. In addition, the UGT76G1 gene expression had a positive correlation with rebaudioside A production and negative correlation with stevioside production. The stevioside production had a negative correlation with rebaudioside A production (Table 3A). Also, in elicitation with different concentrations of silver nitrate, the KA13H gene expression was not correlated with UGT74G1 and UGT76G1 genes expression and stevioside and rebaudioside A production. Another result of this work was that UGT74G1 gene expression had a negative correlation with UGT76G1 and had a positive correlation with stevioside and rebaudioside A production. The UGT76G1 gene expression had a negative correlation with stevioside and rebaudioside A production. The stevioside production had a positive correlation with rebaudioside A production (Table 3B).
| A | |||||
|---|---|---|---|---|---|
| KA13H | UGT74G1 | UGT76G1 | Stevioside | Rebaudioside A | |
| KA13H | 1 | ||||
| UGT74G1 | -0.144 | 1 | |||
| UGT76G1 | 0.061 | -0.940** | 1 | ||
| Stevioside | 0.357 | 0.827* | -0.907* | 1 | |
| Rebaudioside A | -0.043 | -0.925** | 0.987** | -0.936** | 1 |
| B | |||||
| KA13H | UGT74G1 | UGT76G1 | Stevioside | Rebaudioside A | |
| KA13H | 1 | ||||
| UGT74G1 | 0.744 | 1 | |||
| UGT76G1 | -0.492 | -0.850* | 1 | ||
| Stevioside | 0.684 | 0.944** | -0.902* | 1 | |
| Rebaudioside A | 0.63 | 0.856* | -0.855* | 0.950** | 1 |
*p<0.05 and **p<0.01
Table 3: Correlation of KA13H, UGT74G1 and UGT76G1 Genes and Stevioside and Rebaudioside A Production in S. rebaudiana BERT. Seedlings
The results of this study showed that the stevioside and rebaudioside A production are significantly affected by different concentrations of cadmium chloride, silver nitrate and sampling times. The highest amount of stevioside and rebaudioside A were 39.76 mg/g DW and 2.19 mg/g DW obtained 96 h after elicitation with 20 mg/L cadmium chloride and 24 h after elicitation with 60 mg/L silver nitrate respectively. The qRT-PCR analysis revealed that by increasing stevioside production under cadmium chloride and silver nitrate elicitation, the KA13H and UGT74G1 genes expression was increased while the UGT76G1 gene expression was decreased. Therefore, by increasing of KA13H and UGT74G1 genes expression, the GT76G1 gene expression was decreased.
Acknowledgments:
This work was extracted from M. Sc thesis of the first author submitted to Department of Biotechnology at Imam Khomeini International University (IKIU), Qazvin, Iran and especial thanks go to faculty members for their professional supporting helps. Authors thank from IKIU for financial support and especial thanks go to plant tissue culture and molecular biology technician Labs (Dr. Reza Fajaminezhad and Dr. Reza Heidari).
Conflicts of Interest:
There are no conflicts of interest.
References
- Khalil SA, Ahmad N, Zamir R. Gamma radiation induced variation in growth characteristics and production of bioactive compounds during callogenesis in Stevia rebaudiana (Bert.). New Negat Plant Sci 2015;1:1-5.
- Bayraktar M, Naziri E, Akgun IH, Karabey F, Ilhan E, Akyol B, et al. Elicitor induced stevioside production, in vitro shoot growth, and biomass accumulation in micropropagated Stevia rebaudiana. Plant Cell Tiss Org 2016;127(2):289-300.
- Ceunen S, Geuns JMC. Influence of photoperiodism on the spatio-temporal accumulation of steviol glycosides in Stevia rebaudiana (Bertoni). Plant Sci 2013;198:72-82.
- Gupta P, Sharma S, Saxena S. Biomass yield and steviol glycoside production in callus and suspension culture of Stevia rebaudiana treated with proline and polyethylene glycol. Appl Biochem Biotech 2015;176(3):863-874.
- Mandal S, Upadhyay S, Singh VP, Kapoor R. Enhanced production of steviol glycosides in mycorrhizal plants: a concerted effect of arbuscular mycorrhizal symbiosis on transcription of biosynthetic genes. Plant Physiol Biochem 2015;89:100-6.
- Dey A, Kundu S, Bandyopadhyay A, Bhattacharjee A. Efficient micropropagation and chlorocholine chloride induced stevioside production of Stevia rebaudiana Bertoni. C R Biol 2013;336(1):17-28.
- Humphrey TV, Richman AS, Menassa R, Brandle JE. Spatial organisation of four enzymes from Stevia rebaudiana that are involved in steviol glycoside synthesis. Plant Mol Biol 2006;61(1-2):47-62.
- Yoneda Y, Nakashima H, Miyasaka J, Ohdoi K, Shimizu H. Impact of blue, red and far-red light treatments on gene expression and steviol glycoside accumulation in Stevia rebaudiana. Phytochem 2017;137:57-65.
- Brandle JE, Rosa N. Heritability for yield, leaf: stem ratio and stevioside content estimated from a landrace cultivar of Stevia rebaudiana. Canad J Plant Sci 1992;72(4):1263-6.
- Farjaminezhad R, Garoosi GA. Establishment of green analytical method for ultrasound-assisted extraction of azadirachtin, mevalonic acid and squalene from cell suspension culture of Azadirachta indica using response surface methodology. Ind Crop Prod 2020;144:111946.
- Farjaminezhad R, Garoosi GA. New biological trends on cell and callus growth and azadirachtin production in Azadirachta indica. 3 Biotech 2019;9(8):1-7.
- Sivanandhan G, Dev GK, Jeyaraj M, Rajesh M, Arjunan A, Muthuselvam M, et al. Increased production of withanolide A, withanone and withaferin A in hairy root cultures of Withania somnifera (L.) Dunal elicited with methyl jasmonate and salicylic acid. Plant Cell Tiss Org 2013;114(1):121-9.
- Zare N, Farjaminezhad R, Asghari-Zakaria R, Farjaminezhad M. Enhanced thebaine production in Papaver bracteatum cell suspension culture by combination of elicitation and precursor feeding. Nat Prod Res 2014;28(10):711-7.
- Akula R, Ravishankar GA. Influence of abiotic stress signals on secondary metabolites in plants. Plant Signal Behav 2011;6(11):1720-31.
- Murthy HN, Lee EJ, Paek KY. Production of secondary metabolites from cell and organ cultures: strategies and approaches for biomass improvement and metabolite accumulation. Plant Cell Tiss Org Culture 2014;118(1):1-6.
- Cetin ES, Babalik Z, Hallac-Turk F, Gokturk-Baydar N. The effects of cadmium chloride on secondary metabolite production in Vitis vinifera cv. cell suspension cultures. Biol Res 2014;47(1):47.
- Zheng Z, Wu M. Cadmium treatment enhances the production of alkaloid secondary metabolites in Catharanthus roseus. Plant Sci 2004;166(2):507-14.
- Zhou L, Yang G, Sun H, Tang J, Yang J, Wang Y, et al. Effects of different doses of cadmium on secondary metabolites and gene expression in Artemisia annua L. Front Med 2017;11(1):137-46.
- Srivastava R, Khan R, Nasim SA, Manzoor N. Cadmium treatment alters phytochemical and biochemical activity in Glycine max L. Int J Botany 2011;7(4):305-9.
- Li X, Wang S, Guo L, Huang L. Effect of cadmium in the soil on growth, secondary metabolites and metal uptake in Salvia miltiorrhiza. Toxi Environ Chem 2013;95(9):1525-38.
- Jayalakshmi NR, Saraswathi KJ, Vijaya B, Prasad DP, Suresh R. Morphological variations with enhanced accumulation of anthocyanins in Malva sylvestris L. with accumulation of silver nitrate treatment. J Plant Sci 2012;7(1):23.
- Kubeš J, T?mová L, Martin J, Vildová A, Hendrychová H, Sojková K. The production of isoflavonoids in Genista tinctoria L. cell suspension culture after abiotic stressors treatment. Nat Prod Res 2014;28(24):2253-63.
- Park WT, Arasu MV, Al-Dhabi NA, Yeo SK, Jeon J, Park JS, et al. Yeast extract and silver nitrate induce the expression of phenylpropanoid biosynthetic genes and induce the accumulation of rosmarinic acid in Agastache rugosa cell culture. Molecules 2016;21(4):426.
- Vildová A, Hendrychova H, Kubeš J, T?mová L. Influence of AgNO3 treatment on the flavonolignan production in cell suspension culture of Silybum marianum (L.) Gaertn. Int J Agri Biosyst Eng 2016;8(8):959-62.
- Murashige T, Skoog F. A revised medium for rapid growth and bio assays with tobacco tissue cultures. Physiol Plantarum 1962;15(3):473-97.
- Kolb N, Herrera JL, Ferreyra DJ, Uliana RF. Analysis of sweet diterpene glycosides from Stevia rebaudiana: improved HPLC method. J Agri Food Chem 2001;49(10):4538-41.
- Serfaty M, Ibdah M, Fischer R, Chaimovitsh D, Saranga Y, Dudai N. Dynamics of yield components and stevioside production in Stevia rebaudiana grown under different planting times, plant stands and harvest regime. Ind Crop Prod 2013;50:731-6.
- Japelaghi RH, Haddad R, Garoosi GA. Isolation, identification and sequence analysis of a thioredoxin h gene, a member of subgroup III of h-type Trxs from grape (Vitis vinifera L. cv. Askari). Mol Biol Rep 2012;39(4):3683-93.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔct method. Methods 2001;25(4):402-8.
- Deepthi S, Satheeshkumar K. Enhanced camptothecin production induced by elicitors in the cell suspension cultures of Ophiorrhiza mungos Linn. Plant Cell Tiss Org 2016;124(3):483-93.
- Cui Y, Xu G, Wang M, Yu Y, Li M, da Rocha PS, et al. Expression of OsMSR3 in Arabidopsis enhances tolerance to cadmium stress. Plant Cell Tiss Org 2013;113(2):331-40.
- Charfeddine M, Charfeddine S, Bouaziz D, Messaoud RB, Bouzid RG. The effect of cadmium on transgenic potato (Solanum tuberosum) plants overexpressing the StDREB transcription factors. Plant Cell Tiss Org 2017;128(3):521-41.