- *Corresponding Author:
- Vandana Pandey
Department of Chemistry
Kurukshetra University
Kurukshetra, Haryana 136119, India
E-mail: vandana_p71@rediffmail.com
| Date of Received | 21 November 2019 |
| Date of Revision | 10 February 2021 |
| Date of Acceptance | 19 May 2021 |
| Indian J Pharm Sci 2021;83(3):504-514 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
In the present work, two dimensional quantitative structure activity relationship, molecular docking and absorption, distribution, metabolism, excretion and toxicity analyses were performed to pyrrolo[3,4-c] quinoline-1,3-diones derivatives, previously reported as caspase-3 inhibitors. A total of one hundred fifteen compounds were used to build linear multiple linear regression (multiple linear regression) and non-linear (artificial neural networks) quantitative structure activity relationship models, using genetic algorithm as a feature selection method. Both models were thoroughly validated following Organization for economic cooperation and development principles by internal and external validation as well as the domain of application (antiphase domain). Both Genetic algorithm-multiple linear regression (Rtrain=0.88, Rtest=0.94, mapetest=5.3 and rmsetest=0.41) and Genetic algorithm-artificial neural network (Rtrain=0.9, Rtest=0.93, mapetest=4.5 and rmsetest=0.4) models are statistically robust with high external predictive ability. Molecular docking simulations were performed on selected inhibitors revealed that binding energy values are in accordance with inhibitory activity values against caspase-3, which is modulated by hydrogen bondings, Pi stacking and hydrophobic interactions. The docking studies suggest that the inhibitors bind with an allosteric site of the enzyme formed by ARG207B, SER251B, PHE250 and PHE256 of the B chain. Besides, in silico, absorption, distribution, metabolism, excretion and toxicity profiles of selected inhibitors were checked to evaluate the key pharmacokinetic, physiochemical and druglikeness features.
Keywords
Apoptosis, descriptors, genetic algorithm, artificial neural network
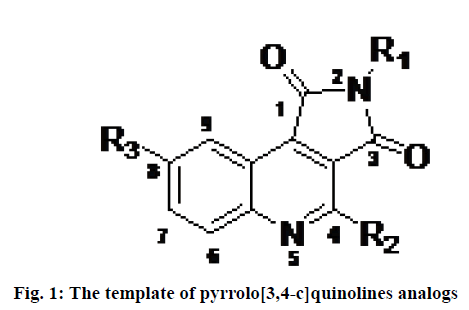
Programmed cell death also known as apoptosis is a normal physiological process, essential for the development and health of organisms. A family of cysteine proteases known as caspase (cysteine-dependent aspartate protease) plays important role in inducing apoptosis[1]. Caspases are strict endonuclease[2]. Dysregulation of apoptosis is believed to play a role in various chronic diseases including neurodegenerative disorders, autoimmune disorders, stroke and myocardial infarction and several forms of cancers[3]. Therefore the development of a drug that can regulate the process of apoptosis has been a challenging task for researchers and pharmaceutical companies to treat some pathological conditions caused by abnormal apoptosis. Among 14 members of the caspase family, caspase-3 (apopain) is a key executioner enzyme involved in apoptosis. It is responsible for the physiological and morphological changes that occur in apoptosis and is expressed in almost all tissues at a relatively high level[4]. Therefore, it is considered an interesting therapeutic target for the treatment of diseases caused by undesirable apoptosis. Numerous peptide and nonpeptide inhibitors have been reported in the literature[5]. In this context, different classes of the reversible and irreversible non-peptide small molecule inhibitors have been described including dithiocarbamate, N-nitrosoanilines, isatin sulfonamide derivatives, anilinoquinazolines, 2-(2,4-dichlorophenoxy)-N- (2-mercaptoethyl)-acetamide, 5-fluoro-1H-indole- 2-carboxylic acid (2-mercaptoethyl)-amide 1 and pyrrolo[3,4-c]quinolines-1,3-diones[6-10]. Among them, pyrrolo[3,4-c]quinoline-1,3-diones derivatives (fig. 1) are considered as one of the less explored heterocyclic structures with promising caspase-3 inhibition activities to prevent abnormal apoptosis.
In silico methods are becoming imperative tools in pharmaceutical biology especially in drug designing and drug refinement. In chemometric research, quantitative structure activity relationships (QSAR) studies in combination with molecular docking and Absorption, distribution, metabolism, excretion and toxicity (ADMET) analysis offers the advantage of being more simple, environment friendly and cost-effective than experimental approaches in novel drug design and sustainable pharmacy. Ligand-based (QSAR) and receptor-base (molecular docking) prediction methods are complementary to each other[11]. QSAR models are mathematical equations, constructing the linear or non-linear relationship between biological activity and chemical structure presented in the form of theoretical descriptors[12]. There are several approaches in QSAR modeling. Linear modeling approaches include multiple linear regression (MLR), partial least square (PLS), etc. are developed to extract the maximum information from complex data matrices based on their linear behavior. In contrast, artificial neural networks (ANNs) have been used for exploring nonlinear modeling and optimization when underlying mechanisms are very complex[13]. Molecular docking, as an optimization problem, has played an important role in the understanding of drug/receptor interactions[14]. In this in silico approach, compounds are usually ranked through scoring functions, mainly categorized into force field-based, empirical or knowledge-based methods. Studies have demonstrated the applicability of molecular docking methods to in silico campaigns, including those targeting caspase-3[15,16]. Virtual ADMT/ tox profiling is also an important aspect of the drug design process prior to in vivo studies which explain the pharmacokinetics aspects of a drug molecule[17].
Considering all these realities, two dimensional QSAR (2D QSAR), molecular docking and ADMET estimations have been performed on a progression of substituted, pyrrolo[3,4-c]quinolines-1,3-diones as inhibitors of caspase-3 in the present investigation. The primary goal of the study was to utilize different computational techniques for the assessment of the key structural features required of pyrrolo[3,4-c]quinolines- 1,3-diones derivatives (fig. 1) as effective caspase-3 inhibitors. Sharma et al. have performed a molecular modeling study on the small dataset (25 compounds) of pyrrolo[3,4-c]quinolines derivatives[18]. Here, a dataset containing 115 inhibitors was selected to develop robust, reliable linear and non-linear QSAR models following Organization for Economic Co-operation and Development (OCED) standards. Besides, molecular docking studies and ADME/tox profiles of the inhibitors were explored to provide further insights for the design and development of inhibitors of caspase-3.
Materials and Methods
Dataset and software:
For the present molecular modeling study, a set of one hundred fifteen pyrrolo[3,4-c]quinolines-1,3-diones derivatives with a wide activity range was retrieved from the literature[10,19-21]. This data set represents inhibition in terms of Half-maximal inhibitory concentration (IC50) values for each molecule. The biological activity data were then converted to pIC50 values and were used as the dependent variable. The structures of all the compounds along with their actual biological activities are presented in supplementary Table 1. All calculations presented in this work were carried out on a personal computer with Windows 7 operating system. Online facilities eDragon and genetic algorithm 1.4 tool were used for descriptor generation and best subset selection respectively[22,23]. ANN calculations were performed with Matlab software. Avogadro, iGEMDOCK, Pyrx softwares were used for optimization and docking study respectively[24-26].
| Comp. | pIC50a | MLRb | ANNc | NMD | Comp. | pIC50a | MLRb | ANNc | NMD |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 4.2 | 4.0134 | 4.1182 | 0.229 | 61 | 7.4 | 7.6609 | 7.303 | 0.263 |
| 2* | 4.43 | 4.8120 | 4.7168 | 0.230 | 62 | 7.8 | 7.8696 | 7.1906 | 0.127 |
| 3 | 6.68 | 6.9373 | 6.363 | 0.007 | 63 | 7.45 | 7.6556 | 7.6879 | 0.005 |
| 4 | 5.2 | 4.8474 | 4.827 | 0.239 | 64 | 7.77 | 7.4492 | 7.4739 | 0.005 |
| 5 | 5.8 | 5.5465 | 5.4182 | 0.233 | 65 | 7.88 | 7.5597 | 7.7131 | 0.018 |
| 6 | 7.36 | 7.2911 | 7.2309 | 0.002 | 66 | 7.85 | 7.4477 | 7.5175 | 0.006 |
| 7* | 5.33 | 5.8753 | 5.4531 | 0.192 | 67 | 8.1 | 8.0307 | 8.3658 | 0.004 |
| 8 | 5.6 | 6.2940 | 5.632 | 0.182 | 68 | 8.04 | 8.1147 | 8.1335 | 0.008 |
| 9 | 6.34 | 6.2673 | 6.4584 | 0.186 | 69 | 8.22 | 7.7640 | 7.8691 | 0.000 |
| 10 | 7.8 | 7.7081 | 7.2486 | 0.017 | 70 | 8.22 | 8.5105 | 8.4566 | 0.057 |
| 11 | 4.63 | 5.7111 | 5.3788 | 0.205 | 71 | 8.3 | 8.6394 | 8.446 | 0.060 |
| 12 | 5.26 | 5.5865 | 5.1161 | 0.197 | 72 | 8.3 | 8.3760 | 8.3713 | 0.066 |
| 13 | 5.96 | 5.3788 | 5.9492 | 0.206 | 73 | 8.52 | 8.6421 | 8.5043 | 0.057 |
| 14 | 7.43 | 7.0041 | 7.6167 | 0.024 | 74* | 8.39 | 8.7554 | 8.516 | 0.056 |
| 15 | 5.1 | 5.5301 | 5.2876 | 0.228 | 75 | 7.92 | 7.8977 | 8.0749 | 0.004 |
| 16 | 5.59 | 5.6832 | 5.4911 | 0.228 | 76 | 7.74 | 7.7181 | 7.7953 | 0.007 |
| 17* | 7.82 | 7.5274 | 7.5425 | 0.006 | 77 | 7.85 | 7.5593 | 7.6245 | 0.003 |
| 18 | 6.44 | 6.9703 | 5.7962 | 0.245 | 78* | 7.88 | 7.8123 | 7.8977 | 0.010 |
| 19 | 7.04 | 7.3211 | 6.9813 | 0.085 | 79 | 8.52 | 8.0960 | 8.2285 | 0.014 |
| 20 | 7.8 | 7.2273 | 7.862 | 0.228 | 80* | 7.08 | 7.7928 | 7.9492 | 0.005 |
| 21 | 7.48 | 7.3032 | 7.3404 | 0.059 | 81 | 7.82 | 7.5101 | 7.9431 | 0.036 |
| 22 | 7.7 | 8.0910 | 7.8191 | 0.068 | 82 | 6.4 | 6.8127 | 6.2939 | 0.045 |
| 23 | 7.68 | 7.7481 | 7.6241 | 0.072 | 83 | 7.74 | 7.8080 | 7.825 | 0.011 |
| 24* | 7.64 | 7.5245 | 7.2602 | 0.140 | 84 | 5.52 | 7.0327 | 6.6622 | 0.011 |
| 25* | 7.26 | 7.6405 | 6.6378 | 0.226 | 85* | 7.4 | 7.0101 | 6.7968 | 0.015 |
| 26* | 8.4 | 9.0319 | 8.6123 | 0.060 | 86 | 7.57 | 7.3480 | 7.5389 | 0.006 |
| 27* | 7.27 | 7.3562 | 7.4054 | 0.002 | 87 | 7.44 | 7.0023 | 6.9782 | 0.007 |
| 28 | 7.64 | 7.1876 | 7.2511 | 0.140 | 88* | 7.48 | 7.7198 | 8.1925 | 0.091 |
| 29* | 7.39 | 7.9458 | 7.576 | 0.011 | 89* | 4.96 | 5.4058 | 5.1024 | 0.070 |
| 30 | 5.26 | 7.0742 | 6.5041 | 0.007 | 90 | 6.53 | 5.8390 | 5.2456 | 0.069 |
| 31 | 7.7 | 7.4732 | 7.1713 | 0.005 | 91 | 6.59 | 6.3183 | 6.5763 | 0.094 |
| 32 | 7.5 | 7.4211 | 7.5392 | 0.028 | 92 | 5.9 | 6.2269 | 5.8901 | 0.090 |
| 33 | 7.01 | 6.8764 | 6.7799 | 0.196 | 93 | 4.81 | 5.2477 | 4.8084 | 0.072 |
| 34* | 7.25 | 6.9016 | 7.2179 | 0.195 | 94 | 5.28 | 5.2370 | 5.1895 | 0.160 |
| 35 | 7.24 | 6.4213 | 6.0653 | 0.196 | 95 | 4.55 | 6.4107 | 5.7141 | 0.067 |
| 36 | 7.26 | 6.9051 | 7.1518 | 0.197 | 96* | 6.45 | 7.0406 | 6.9235 | 0.106 |
| 37 | 7.27 | 6.9819 | 7.5226 | 0.020 | 97 | 6.44 | 7.1000 | 6.5842 | 0.129 |
| 38 | 7.62 | 7.5450 | 7.767 | 0.198 | 98 | 7.23 | 7.4718 | 7.1927 | 0.015 |
| 39* | 6.82 | 7.6084 | 7.629 | 0.465 | 99 | 7.48 | 6.7914 | 7.2347 | 0.008 |
| 40 | 6.71 | 7.4556 | 6.6959 | 0.602 | 100 | 7.48 | 7.5859 | 7.7289 | 0.011 |
| 41 | 7.79 | 7.2196 | 7.0143 | 0.006 | 101 | 4.16 | 4.2338 | 4.2707 | 1.000 |
| 42 | 7.52 | 7.3268 | 7.3313 | 0.012 | 102 | 4.39 | 4.5204 | 4.2391 | 0.860 |
| 43 | 7.67 | 7.0167 | 7.5067 | 0.023 | 103* | 5.67 | 6.0758 | 5.8514 | 0.094 |
| 44 | 7.07 | 6.9526 | 7.4821 | 0.034 | 104 | 6.06 | 7.0006 | 6.2552 | 0.126 |
| 45 | 7.96 | 6.9398 | 7.5212 | 0.021 | 105 | 6.27 | 6.4734 | 6.6021 | 0.197 |
| 46 | 7.96 | 7.0104 | 7.4315 | 0.096 | 106 | 6.42 | 6.6659 | 7.0486 | 0.097 |
| 47 | 6.18 | 6.3656 | 6.1521 | 0.096 | 107 | 6.49 | 6.2621 | 6.6143 | 0.092 |
| 48 | 7.13 | 6.9072 | 7.1333 | 0.090 | 108 | 6.51 | 6.6374 | 6.8851 | 0.093 |
| 49* | 7.43 | 7.3299 | 7.8379 | 0.027 | 109 | 6.86 | 5.9949 | 6.3898 | 0.090 |
| 50 | 7.72 | 7.3137 | 7.1861 | 0.005 | 110 | 6.51 | 6.2502 | 6.2676 | 0.010 |
| 51* | 7.82 | 7.9757 | 8.0591 | 0.008 | 111* | 6.51 | 6.6062 | 6.6333 | 0.006 |
| 52 | 7.62 | 8.0930 | 7.8641 | 0.121 | 112 | 6.57 | 6.5181 | 6.4397 | 0.024 |
| 53 | 5.64 | 7.0673 | 6.7915 | 0.006 | 113 | 6.59 | 6.8742 | 6.7374 | 0.005 |
| 54 | 7.62 | 7.1885 | 7.0165 | 0.006 | 114* | 6.61 | 6.3143 | 6.4481 | 0.022 |
| 55 | 8.1 | 7.7631 | 7.8323 | 0.008 | 115 | 6.66 | 6.5078 | 6.5764 | 0.004 |
| 56 | 7.65 | 7.4151 | 7.5447 | 0.005 | |||||
| 57 | 7.69 | 7.2280 | 7.4175 | 0.010 | |||||
| 58 | 8 | 7.9794 | 8.1278 | 0.005 | |||||
| 59* | 8.04 | 7.9503 | 8.0953 | 0.005 | |||||
| 60 | 7.65 | 7.1595 | 6.9641 | 0.003 |
Table 1: Observed and Calculated Data of Caspase-3 Inhibition Activity Of PYRROLO[3,4-c]Quinolines
Calculation and selection of molecular descriptors:
The three dimensional (3D) structures of 115 compounds were drawn and the starting geometries of the molecular structures were optimized using the built-in Avogadro minimization algorithm based on the Merck Molecular Force Field 94 (MMFF94) force field using the Steepest Descent Algorithm with 500 steps of minimization. Standard database format (SDF) files of all these compounds were generated anduploaded to Dragon software for descriptor generation. A large pool of descriptors was generated for each molecule including molecular properties, constitutional descriptors, topological descriptors, connectivity indices, information indices, topological charge indices, geometrical descriptors, weighted holistic invariant molecular (WHIM), 3D-Morse, Getaway and Resource description framework (RDF) descriptors. For the development of a robust QSAR model, a key step is the selection of the optimal subset of variables. Objective and subjective feature selection methods were applied to get the appropriate subset for linear and non-linear QSAR mapping. Genetic algorithm (GA) applied for subjective feature selection after preprocessing of the data set. It was used to search feature space and to select descriptors relevant to the inhibitory values. The important parameters that contributed to the GA performance are listed as follows: crossover probability=1, mutation probability=0.5, the initial number of equation generated=100, total number of iteration=100.
Selection of training and test set:
The optimal division of the data set is an important and critical step in QSAR modeling. For meaningful prediction, the entire dataset was divided into two subsets: training and test set. The division was done by the Euclidean distance-based method so that both sets cover the entire chemical space of the whole dataset. Finally, 92 and 23 compounds from the whole dataset were used as the training set and test set, respectively.
Linear and non-linear modeling:
Linear and non-linear mapping of the refined data set was carried out by MLR and ANN method respectively. The advantage of MLR is its simple form and easily interpretable mathematical expression. For improvement of the performance of the generated model and to explore the non-linear relationship between selected independent variables and inhibitory activity, a fully connected feed-forward artificial neural network was implemented. Mainly two major components are essential to the effectiveness of a neural network at solving a particular problem: its architecture and the algorithm by which it is trained. A three-layered feed-forward ANN trained with the Levenberg-Marquardt algorithm was used for non-linear mapping.
Model validation:
Model validation is an important feature in QSAR modeling to confirm the reliability, robustness, and quality of the developed QSAR model. This is done to test the internal stability and predictive ability of the selected models. The developed QSAR models in this study were validated by internal as well as external validation. The methods of least-squares fit (R2), leave one out cross-validation (Q2LOO), adjusted R2 (R2adj), root-mean-squared error (RMSE) and scrambling (Y-randomization) were used for internal validation of the models. For estimating external predictability of proposed models, R2 (test), mean absolute percent error (mapetest), root mean squared error (rmsetest) as well as parameters recommended by Golbraikh and Tropsha were estimated[27]. Defining the Applicability Domain (AD) is an important aspect according to OECD. In the present study, AD was verified by calculating normalized mean Euclidean distance value for each compound as well as using the Leverage approach[28]. Williams’s plots were generated for the detection of response outlier and structurally influential chemicals in each developed model.
Molecular docking:
The crystal structure of caspase-3 (PDB entry: 1gfw) in complex with the ligand (MSI) were downloaded from the RCSB Protein Data Bank[29]. The selected protein has an X-ray resolution factor of 2.8 Å. Structure based virtual screening and post-screening analysis was carried by using iGEMDOCK tool. Standard docking protocol was followed by setting a population size of 200 with 70 generations and 2 solutions. The docking procedure was validated by removing crystallographic bound ligand (MSI) from the binding sites of 1GFW and redocking it. Then, the selected energy minimized inhibitors were docked into the receptor. Protein compound interaction profiles were generated and analyzed by post-analysis tools. To verify the results obtained by iGEMDOCK, again molecular docking was performed by PyRx. For docking simulations, a grid box covering all the binding sites present in the 1GFW was constructed.
ADME/Tox properties:
Physicochemical properties, pharmacokinetics, druglikeness and medicinal chemistry of the selected inhibitors were also computed. The simplified molecular-input line-entry system (SMILES) of the set of compounds with the lowest binding energy were submitted to Swiss ADME online server for the study of their pharmacokinetics and drug-likeness properties[30]. Selected ligands were also analyzed for bioavailability property using Boiled egg analysis[31].
Results and Discussion
The dataset with 1666 descriptors for each molecule was generated online using eDragon descriptor calculation facility. The descriptor pool was screened first to decrease the redundancy existing in the descriptor data matrix. The objective feature selection procedure was performed in two steps. At first, descriptors with constant and near-constant values for all molecules were removed (1324 descriptors). Then highly intercorrelated descriptors were removed using Volume- Weighted Average Price (VWAP) algorithm proposed by Ballabio et al. with variance and correlation coefficient cut-off values 0.01 and 0.7, respectively[32]. Applying these two steps, the number of descriptors was considerably reduced from 1666 to 422. The entire dataset was then utilized for the subjective feature selection procedure. Genetic algorithm was implemented for selecting relevant descriptors. In the present case, a string composed of 422 genes representing the presence and absence of a descriptor is coded by 1 or 0 respectively. The chromosomes with less number of selected descriptors, a high value of fitness function and a low value of Lack of fit (LOF) were marked as informative chromosomes. Finally, nine descriptors demonstrating high accordance with inhibitory activity were selected for linear and nonlinear regression. A correlation matrix was obtained among all nine descriptors selected finally because the regression equation is useless if descriptors are highly correlated. It can be seen from the correlation matrix (supplementary Table 2), there is no significant correlation among the selected descriptors. The splitting of the dataset into training and test sets was performed rationally using Euclidean distance based technique. As a result, 80 % of the data set (92 compounds) was used as the training set and the remaining 20 % (23 compounds) was used as the test set. The best multivariate linear model representing the linear relationship between selected nine descriptors for pyrrolo[3,4-c]quinoline-1,3-diones derivatives and corresponding inhibitory activity is shown in the following eqn: pIC50=2.41547(±1.49026)- 2.34372(±0.52346) MATS8v-1.35568(±0.44891) GATS6e-1.13556(±0.22567) Lop+0.11274(±0.10776) MAXDN+0.96173 (±0.38519) EEig02 d-2.45236 (±0.41102) Mor28u+0.66974(±0.15395) -028+1.59022 (±0.49535) Mor28m-0.26551(±0.04402) nCs (1), Here N=92, R2: 0.75327, R2 Adj: 0.7261, Q2 (Loo) :0.69003, MAPE(train)=6.45.
The QSAR model presented by equation (1) was internally cross-validated by leave one out (LOO) method. The value of Q2 (Loo) (0.69, >0.5) for the training set can serve as an indicator of a high predictive ability of the proposed model. Y randomization technique was also performed by randomly shuffling the dependent variable while keeping the independent variable as it is. Low values of average R2 (0.10) and average Q2 (-0.14) resulted after the generation of fifty random models, which confirmed that the developed QSAR model is reliable. From all the statistical parameters, it can be seen that the proposed linear model is stable, robust and predictive, consequently was used for the prediction of activities of the test set data. The prediction results obtained from the GA-MLR approach for the entire dataset are given in Table 1.
The contribution of the selected descriptors present in the equation can be interpreted as fol1ows: The descriptors Moran autocorrelation of lag 8 weighted by vander Waals volume (MATs8v) and Geary autocorrelation of lag 6 weighted by Sanderson electronegativity (GATS6e) belong to 2D autocorrelations. Their Negative Contribution towards inhibitory activity indicates an inverse relationship with vander Waals volume and Sanderson electronegativity, respectively. The negative relationship is consistent with the observation that inhibitors with bulky, non-polar and lipophilic substituents at position 2 are less active. It may be due to the unavailability of the inhibitor to accommodate in the sterically hindered binding site of caspase-3. The descriptor Lop (or Loc, lopping centric index,) corresponds to topological indices. This radial centric information indices display negative contribution in the eq1. The descriptor that affects the activity with a low positive coefficient is maximal electrotopological negative variation (MAXDN). It is related to the nucleophilicity of the inhibitor. It may influence the electrostatic interactions between the inhibitor and the receptor site. Eigenvalue 02 from Edge adjacency matrix weighted by dipole moment (EEig02d), Edge adjacency indices displays a positive coefficient in the eq.1, indicating the pIC50 value directly relates to this descriptor. Mor28u (signal 28/unweighted) and Mor28m (signal 28/weighted by mass) are two 3D-MoRSE descriptors present in equation (1). Mor28u has the highest contribution towards pIC50 value. In the present case, inhibitors with a high Mor28u value show poor inhibitory activity. C-028 descriptor belongs to Atom-centered fragments. It provides information about the number of predefined structural features in the molecule which is in the present case is R--CR- -X. i,e central carbon atom (C) that has two carbon neighbors (R2) and one heteroatom neighbors (X). The positive sign indicating that pIC50 is directly correlated to this descriptor. The last descriptor in the eq. 1 is nCs (number of total secondary C (sp3), functional group counts), which also negatively correlated to the inhibitory activity. It is worth mentioning that the binding interactions of the inhibitor to the target site depend upon the shape, size, polarizability, etc. The descriptors selected by GA account for these features.
All nine selected descriptors from the genetic algorithm method were used as input for three-layer backpropagation ANN models to explore the non-linear relationship between descriptors and reported pIC50 values. The number of nodes in the input layer was nine, as the input vectors were set of nine descriptors selected by the GA. In order to optimize the number of nodes in the hidden layer, the concept of ρ as proposed by Andrea and Kalayeh was used[33]. The output layer was the pIC50 value. A feed-forward neural network trained with Levenberg-Marquardt (L-M) algorithm was used for non-linear mapping. L-M algorithm allows the network to learn more crafty features of complex mapping. The transfer function in the first layer was tan-sigmoid and the output layer transfer function was linear. MSE value for the test set was calculated by changing the number of nodes in the hidden layer. Several training sessions were conducted with the different number of hidden nodes ranging from 4 to 11. Finally, a network of 9-5-1 was selected and calculated pIC50 values for the training and test set are present in Table 1. Statistical parameters for the training set are as follows: R2=0.9, RMSE=0.47, MAPE(train)=4.45.
The prediction performances of the GA-MLR and GA-ANN models were evaluated using some common external validation parameters such as R2 pred, RMSE(Pred), MAPE(pred). The Golbraikh-Tropsha criteria have also been applied to test sets, to provide confidence in the validation methodology (Table 2). The calculated values of the above mentioned parameters are in good agreement with the proposed criteria. The statistical quality of both the QSAR models is comparable showing significant validation results. The acceptable R(pred) values of (0.94, 0.93) indicate a good prediction capability of the models. It is clear from Table 2, that both GA-MLR and GA-ANN models are comparable, however, the GA-ANN model is superior due to the low MAPE value for the prediction.
| Internal validation | External validation | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| R(train) | rmse | mape | R | rmse | mape | r02 | r'02 | k | k' | R02 | R'02 | r02-r'02 | |
| GA-MLR | 0.88 | 0.507 | 6.45 | 0.94 | 0.41 | 5.3 | 0.88 | 0.86 | 0.97 | 1.026 | 0.96 | 0.97 | 0.02 |
| GA-ANN | 0.9 | 0.47 | 4.45 | 0.93 | 0.4 | 4.5 | 0.87 | 0.87 | 0.98 | 1.01 | 0.98 | 0.99 | 0.002 |
Table 2: Comparision of Validation Parameters of GA-MLR and GA-ANN Models
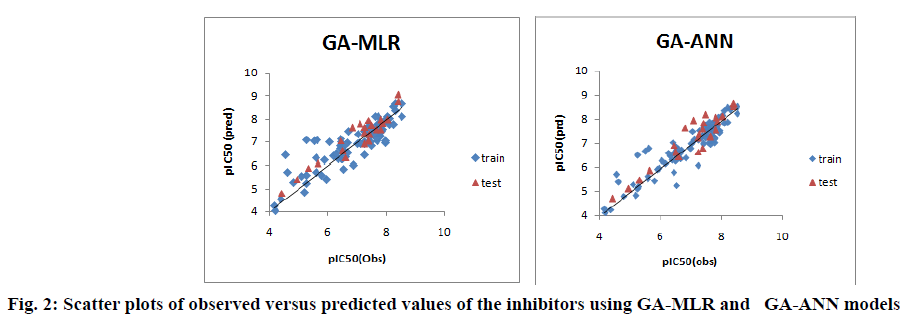
Furthermore, the proximity between the observed and predicted pIC50 values for both linear and nonlinear models of pyrrolo[3,4-c]quinolines1,3-dione derivatives is graphically represented through the scatter plot, shown in fig. 2.
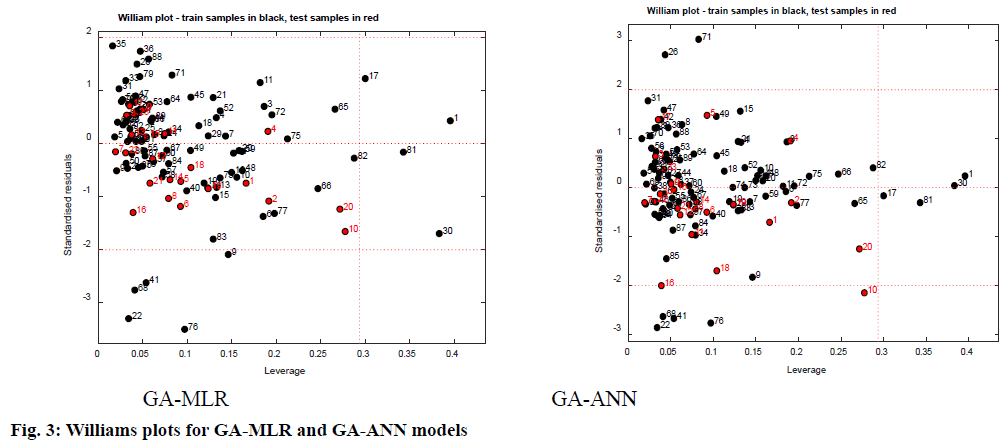
AD represents the chemical space defined by the structural information extracted from the chemical used as training set compounds in the QSAR modeling. Both QSAR models were verified for the applicability of the domain to generate reliably predicted values of pIC50 for the inhibitors. The applicability domain of the model was analyzed by the Euclidean distance method as well as using Williams’s plots (fig. 3). The outcomes from applicability domain analysis for the GA-MLR model, by Euclidean distance method are quite satisfactory within the normal distribution range and normalized mean distance values are reported in Table 1. The Williams plot was used to visualize influential chemicals, i.e., chemicals with leverage greater than the critical hat value, h*( h*=3(P+1)/n (where P=number of model descriptors and n=number of chemicals in the training set), as well as outlier chemicals, i.e., chemicals with standardized residual greater than three standard deviations (SD) units (3σ). The Williams plots for GA-MLR and GA-ANN models (fig. 3) show the presence of four training samples (1,17, 30 and 81) having a great influence on the models (i.e., greater than the warning leverage h*). From the Williams plot for the GA-MLR method, it is observed that, though the compounds 22, 41, 68 and 76 lie within the AD of the model, they are outside the 3σ limit. For the GA-ANN method, it is evident that no outlier is found, since all the compounds for both the training and test set were within the applicability domain of the square area, indicating reliable predictive power of the proposed model.
The inhibition activity of the molecules in the dataset strongly relies upon substitution at the 2, 4 and 8 positions on the core scaffold. Results suggest that activity strongly depends on the nature of the substituent at position 2. The presence of 2-hetroaryl substituent increases the potency of the inhibitor. The significantly more potent compounds in the dataset have pyrazol-4-yl(26, 73,74), 1-phenyl-pyrazole-5- yl(71,72) and 4-pyridyl(79) substituent at position-2. The polyfunctional and hydrophilic substituent at this position increases the activity may be due to the possibility of more non-covalent interaction with the binding pocket.
The activity of these ligands is enhanced by the presence of nucleophilic substituent at 4- position. It makes imide carbonyl more electrophilic in nature which is one of the main contributors to inhibition activity. Alkyl, aryl and heteroaryl groups (1-84, 86-88, 99, 100) at this position enhance the activity, whereas, compounds with electronegative substituents (89-97) at the 4- position are less active. The electron-withdrawing capacity of 8-substituent also has a very significant and direct relationship with the activity of these inhibitors. The bulky group with electron-withdrawing capacity is favorable at position 8. Compounds with 1,3,5-trimethyl-1H-pyrazol-4-yl at 2-position, methyl group at 4-position and morphonyl sulphonyl moiety at 8-position are the lead compounds.
Docking studies were employed to position the inhibitors into the capase-3 binding pocket to determine the optimum binding conformation and to elucidate the interaction with amino acid residues present within the pocket of the receptor. As reversible and noncompetitive nature of the pyrrolo[3,4-c]quinolines-1,3- dione derivatives were reported. It was clear that the inhibitors would dock to the allosteric site other than the active site. Docking software iGemdock was used to dock the selected inhibitors with the target enzyme. It is an integrated virtual screening (VS) environment for docking, screening, post-analysis and visualization of pharmacological interactions. It provides interactive interfaces to prepare both the binding site of the target enzyme and the screening compound library. In the present case, ten inhibitors having pIC50 value of more than 8.1 were considered as promising candidates for docking study. Each energy minimized compound from the library was docked into the binding site. Subsequently, iGemdock generated protein-compound interaction profiles for each compound. Table 3 illustrates the result of the compounds based on the most favorable binding energy.
| Pyrx score | Igemdock score | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ligand | pIC50 | Energy | VDW | HBond | Elec | MW | #Rotatable bonds | #H-bond acceptors | #H-bond donors | TPSA | WLOGP | MLOGP | ESOL Log S | |
| 26 | 8.4 | -9.6 | -108.93 | -93.98 | -14.94 | 0 | 469.51 | 3 | 8 | 0 | 123.08 | 2.03 | 1.13 | -3.53 |
| 55 | 8.1 | -8.9 | -117.99 | -97.72 | -20.27 | 0 | 453.47 | 3 | 8 | 1 | 125.49 | 2.39 | 1.32 | -3.79 |
| 67 | 8.1 | -8.8 | -120.93 | -101.57 | -19.36 | 0 | 485.47 | 5 | 10 | 1 | 160.24 | 1.19 | 0.14 | -3.09 |
| 69 | 8.22 | -8.8 | -110.59 | -93.25 | -17.33 | 0 | 455.49 | 4 | 8 | 0 | 123.08 | 1.9 | 0.91 | -3.28 |
| 70 | 8.22 | -9 | -103.48 | -75.29 | -28.19 | 0 | 455.49 | 3 | 8 | 0 | 123.08 | 1.73 | 0.91 | -3.41 |
| 71 | 8.3 | -9.9 | -108.99 | -80.91 | -28.08 | 0 | 517.56 | 4 | 8 | 0 | 123.08 | 3.18 | 2.03 | -4.84 |
| 72 | 8.3 | -9.5 | -120.52 | -83.48 | -33.01 | -4.02 | 561.57 | 5 | 10 | 1 | 160.38 | 2.88 | 1.46 | -4.72 |
| 73 | 8.52 | -9.8 | -105.32 | -83.53 | -21.79 | 0 | 455.49 | 3 | 8 | 1 | 133.94 | 2.02 | 0.91 | -3.44 |
| 74 | 8.39 | -9.7 | -127.87 | -97.42 | -30.44 | 0 | 497.52 | 4 | 9 | 0 | 140.15 | 2.16 | 1.33 | -3.54 |
| 79 | 8.52 | -9.2 | -106.08 | -90.66 | -15.41 | 0 | 438.46 | 3 | 8 | 0 | 118.15 | 2.08 | 0.84 | -3.27 |
Table 3: Docking Score and Important ADME/T Parameters of Selected Inhibitors
Results obtained by iGEMDOCK were again validated by using AutoDock vina in PyRx. Both tools identified the same binding site in 1gfw. Outcomes obtained from PyRx supported the igemdock results with a very good binding efficiency between the receptor and ligands. The results are in reasonable correlation with the corresponding pIC50 values of the inhibitors. The docking scores of the ten molecules selected as top hits are presented in Table 3. The superimposition of the conformations of selected inhibitors at the binding pocket is presented in the supplementary file (fig. S1).
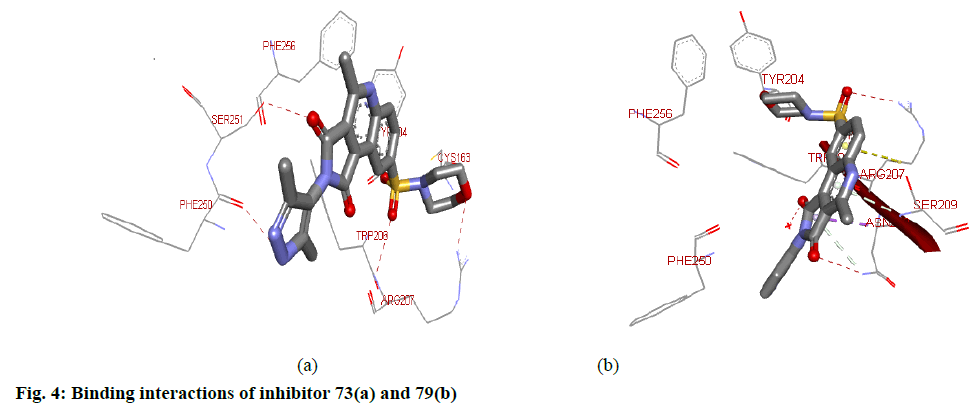
For target protein, total binding energy values (igemdock score) for all the compounds range from −103.48 to −127.87 kcal/mol as reported in Table 3. Whereas, the Binding affinity of the top ten hits has binding values from -8.8 to -9.9 (pyrx score). Residues present in the Chain B of the target protein were involved in the interaction with the inhibitors. Docking interactions between the top two (73 aand79) hits based on the highest pIC50 values and calculated binding affinity are shown in fig. 4. Hydrogen bond interactions were found between 73 and 1gfw binding pocket. Here, ARG207B, PHE250B and SER251B participated in H-bond interactions. Nitrogen atoms of the ARG207B are involved in two H-bond interactions, one with O atom of SO2 group and the other with O atom of morphonyl moiety. The oxygen atom of PHE250B forms H-bond interaction with N atom of the pyrazole-4-yl group. O atom of SER251B makes hydrogen bond interaction with O atom of one of the imide carbonyl groups. The presence of the other imide carbonyl may be considered redundant. Inhibitor 73 also formed hydrophobic interactions with TRP206B and PHE256B of the target site. Besides, TYR204B and PHE256B were involved in pi stacking with the aromatic rings of the 73. Inhibitor 79 made H-bond interaction with ARG207B, ASN208B and SER209B of the target protein. Here, N atoms of ARG207B involves in interaction with O atoms the SO2 group. ASN208B forms H-bond interaction with one of the O atom of imide carbonyl. The oxygen atom of the SER209B forms an H-bond interaction with the nitrogen atom of quinoline moiety. Hydrophobic interactions were also observed with PHE250B and TRP206B present at the binding pocket.
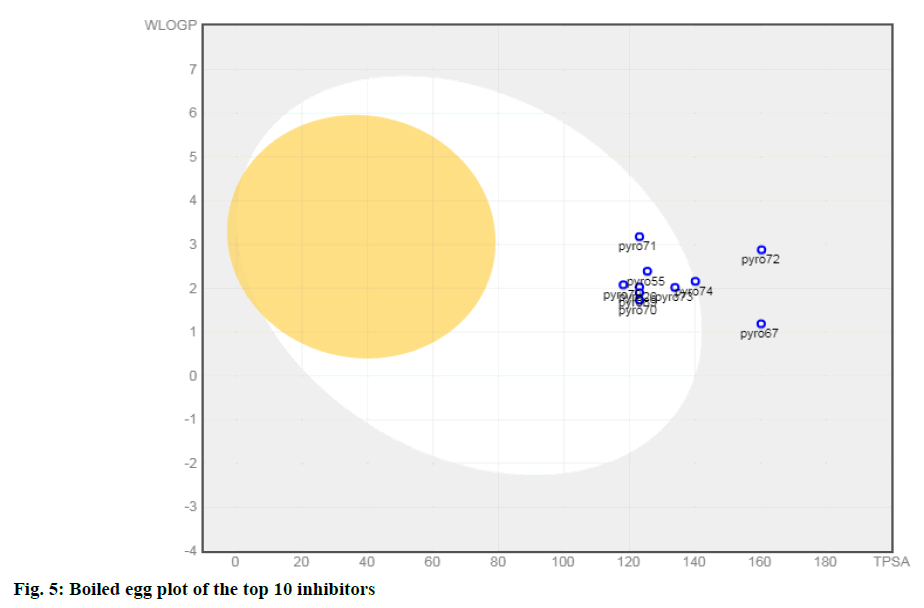
In silico ADMET analysis is proved to be a good tool in drug discovery. The physicochemical properties of a drug have a compelling impact on the metabolic fate and pharmacokinetics. Some of the important ADME/ Tox parameters of the top ten molecules selected based on pIC50 value are shown in Table 3. The molecular weight of selected inhibitors lies in the range of 438.46 and 497.52 (except 71 and 72) and thus follows one of the criteria of the Lipinski rule of five. These inhibitors possess less than ten rotatable bonds; therefore, satisfy the criteria for oral bioavailability. One of the important parameters to understand the passive molecular transport of a drug candidate is the Topological polar surface area (TPSA). It is clear from Table 3; molecules have TPSA value ranging from 118.15 to 160.38. Thus, inhibitors 67 and 72 do not satisfy the criteria defined for good intestinal absorption (TPSA<140 Å2). The values of the water partition coefficient (W log P) are in the range of 1.19 to 3.18 (<5) predicting a low level of toxicity and non-specific binding. Also, all compounds had log S values between -3.09 and -3.79 (except 71and 72), indicating that all are soluble in water. Brain or Intestinal Estimate D permeation method (BOILED-Egg) is an intuitive graphical method to accurately predict the passive human gastrointestinal absorption (HIA) and brain permeability (BBB). This classification model relies on the descriptors: WLOGP and TPSA values. From the Boiled egg analysis of the top 10 molecules (fig. 5), it has been observed that all 10 compounds are found to be the substrate of the P-glycoprotein (PGP+) indicated by the blue dots.
Eight compounds (except 67 and72) are present in the white yolk attributed to being passively absorbed by the gastrointestinal tract (GIT). It is worthwhile to mention that compound 74 has eleven N and O atoms (C23H23N5O6S) (Lipinski’s rule violation, no of N or O>10) also it is present at the boundary of the boiled egg plot. The remaining seven compounds were in the range to satisfy Lipinski’s rule of five to be recognized as drug like potential.
Conclusion
A combined computational approach was applied to give insight into the structural features and mechanism of inhibition for a series of pyrrolo[3,4-c]quinolines- 1,3-diones derivatives as caspase-3 inhibitors. Two hybrid regression methods namely, GA-MLR and GAANN were investigated for building QSAR models for the prediction of inhibition activity. The fitting ability, reliability, stability and predictive potential of the developed models were evaluated by various validation parameters (internal and external) following OECDs principles. Results obtained from molecular docking simulations were in accordance with the inhibition data, in which selected inhibitors were shown to bind to the allosteric binding site of the enzyme. These inhibitors were also subjected to in silico assessment of ADME/ tox properties. Taken together, this study demonstrates that selected six (26, 55, 69, 70, 73 and 79) inhibitors are found to be suitable as potential candidate molecules as a caspase-3 inhibitor and can further be tested under the in vivo and in vitro condition for prediction of new drugs.
Supplementary Information (SI):
Structural information of the 115 pyrrolo[3,4-c] quinolines-1,3-diones derivatives (Table S1), the correlation matrix of the selected descriptors (Table S2), superimposed docked pose of selected compounds (fig .S1) are available as supplementary.
Conflict of interests:
The authors declared no conflicts of interest.
References
- Stennicke HR, Ryan CA, Salvesen GS. Reprieval from execution: the molecular basis of caspase inhibition. Trends Biochem Sci 2002;27(2):94-101.
- Stennicke HR, Salvesen GS. Catalytic properties of the caspases. Cell Death Differ 1999;6(11):1054-9.
- Web SJ, Harrison DJ, Willey A H. In Apoptosis: Pharmacological Implication sand Therapeutic Opportunities, S.H. Kaufmann (Ed.), San Diego, Academic Press; 1997.
- Nuttall ME, Lee D, McLaughlin B, Erhardt JA. Selective inhibitors of apoptotic caspases: implications for novel therapeutic strategies. Drug Discov Today 2001;6(2):85-91.
- Schirmeister T, Kaeppler U. Non-peptidic inhibitors of cysteine proteases. Mini Rev Med Chem 2003;3(4):361-73.
- Nobel CS, Kimland M, Nicholson DW, Orrenius S, Slater AF. Disulfiram is a potent inhibitor of proteases of the caspase family. Chem Res Toxicol 1997;10(12):1319-24.
- Guo Z, Xian M, Zhang W, McGill A, Wang PG. N-nitrosoanilines: a new class of caspase-3 inhibitors. Bioorg Med Chem 2001;9(1):99-106.
- Lee D, Long SA, Murray JH, Adams JL, Nuttall ME, Nadeau DP, et al. Potent and selective nonpeptide inhibitors of caspases 3 and 7. J Med Chem 2001;44(12):2015-26.
- Scott CW, Sobotka-Briner C, Wilkins DE, Jacobs RT, Folmer JJ, Frazee WJ, et al. Novel small molecule inhibitors of caspase-3 block cellular and biochemical features of apoptosis. J Pharmacol Exp Ther 2003;304(1):433-40.
- Kravchenko DV, Kysil VV, Ilyn AP, Tkachenko SE, Maliarchouk S, Okun IM, et al. 1,3-Dioxo-4-methyl-2,3-dihydro-1H-pyrrolo[3,4-c] quinolines as potent caspase-3 inhibitors. Bioorg Med Chem Lett 2005;15(7):1841-5.
- Masand VH, Mahajan DT, Hadda TB, Jawarkar RD, Chavan H, Bandgar BP, et al. Molecular docking and quantitative structure–activity relationship (QSAR) analyses of indolylarylsulfones as HIV-1 non-nucleoside reverse transcriptase inhibitors. Med Chem Res 2014;23(1):417-25.
- Free SM, Wilson JW. A mathematical contribution to structure-activity studies. J Med Chem 1964;7(4):395-9.
- Zupan J, Gasteiger J. Neural networks in chemistry. Wiley-VCH, Weinheim; 1999.
- Kitchen DB, Decornez H, Furr JR, Bajorath J. Docking and scoring in virtual screening for drug discovery: methods and applications. Nat Rev Drug Discov 2004;3(11):935-49.
- Lakshmi PJ, Kumar BS, Nayana RS, Mohan MS, Bolligarla R, Das SK, et al. Design, synthesis, and discovery of novel non-peptide inhibitor of Caspase-3 using ligand based and structure based virtual screening approach. Bioorg Med Chem 2009;17(16):6040-7.
- Sharma S, Basu A, Agrawal RK. Pharmacophore modeling and docking studies on some nonpeptide-based caspase-3 inhibitors. Biomed Res Int 2013;2013.
- Czarnik AW, Mei HY. How, Why to Apply the Latest Technology, In Comprehensive Medicinal Chemistry II, edited by J.B. Taylor and David J. Triggle; 2007.
- Sharma S, Ravichandran V, Jain PK, Mourya VK, Agrawal RK. Prediction of caspase-3 inhibitory activity of 1, 3-dioxo-4-methyl-2, 3-dihydro-1h-pyrrolo[3,4-c] quinolines: QSAR study. J Enzyme Inhib Med Chem 2008;23(3):424-31.
- Kravchenko DV, Kysil VM, Tkachenko SE, Maliarchouk S, Okun IM, Ivachtchenko AV. Pyrrolo[3,4-c]quinoline-1,3-diones as potent caspase-3 inhibitors. Synthesis and SAR of 2-substituted 4-methyl-8-(morpholine-4-sulfonyl)-pyrrolo[3,4-c]quinoline-1,3-diones. European journal of medicinal chemistry 2005;40(12):1377-83.
- Kravchenko DV, Kuzovkova YA, Kysil VM, Tkachenko SE, Maliarchouk S, Okun IM, et al. Synthesis and Structure− Activity Relationship of 4-Substituted 2-(2-Acetyloxyethyl)-8-(morpholine-4-sulfonyl) pyrrolo[3,4-c]quinoline-1,3diones as Potent Caspase-3 Inhibitors. J Med Chem 2005;48(11):3680-3.
- Okun I, Malarchuk S, Dubrovskaya E, Khvat A, Tkachenko S, Kysil V, et al. Screening for caspase-3 inhibitors: a new class of potent small-molecule inhibitors of caspase-3. J Biomol Screen 2006;11(3):277-85.
- Tetko IV, Gasteiger J, Todeschini R, Mauri A, Livingstone D, Ertl P, et al. Virtual computational chemistry laboratory–design and description. J Comput Aided Mol Des 2005;19(6):453-63.
- Roy K, Mitra I. On various metrics used for validation of predictive QSAR models with applications in virtual screening and focused library design. Comb Chem High Throughput Screen 2011;14(6):450-74.
- Hanwell MD, Curtis DE, Lonie DC, Vandermeersch T, Zurek E, Hutchison GR. Avogadro: an advanced semantic chemical editor, visualization and analysis platform. J Cheminformatics 2012;4(1):1-7.
- Yang JM, Chen CC. GEMDOCK: a generic evolutionary method for molecular docking. Proteins 2004;55(2):288-304.
- Dallakyan S, Olson AJ. Small-molecule library screening by docking with PyRx. Chem Biol 2015;1263:243-50.
- Golbraikh A, Tropsha A.Beware of q2!J Mol Graphics Model 2002;20(4):269-76.
- Sahigara F, Mansouri K, Ballabio D, Mauri A, Consonni V, Todeschini R. Comparison of different approaches to define the applicability domain of QSAR models. Molecules 2012;17(5):4791-810.
- Lee D, Long SA, Adams JL, Chan G, Vaidya KS, Francis TA, et al. Potent and selective nonpeptide inhibitors of caspases 3 and 7 inhibit apoptosis and maintain cell functionality. J Biol Chem 2000;275(21):16007-14.
- Zoete V, Daina A, Bovigny C, Michielin O. Swiss Similarity: A Web Tool for Low to Ultra High Throughput Ligand-Based Virtual Screening. J Chem Inf Model2016;56:1399-404.
- Daina A, Zoete V. A boiled‐egg to predict gastrointestinal absorption and brain penetration of small molecules. Chem Med Chem 2016;11(11):1117-21.
- Ballabio D, Consonni V, Mauri A, Claeys-Bruno M, Sergent M, Todeschini R. A novel variable reduction method adapted from space-filling designs. Chemom Intell Lab Syst 2014;136:147-54.
- Andrea TA, Kalayeh H. Applications of neural networks in quantitative structure-activity relationships of dihydrofolate reductase inhibitors. J Med Chem 1991;34(9):2824-36.