- *Corresponding Author:
- Wantong Chen
School of Biology and Biological Engineering, South China University of Technology, Guangzhou, 510006, China
E-mail: wantong.chen.gd@qq.com
| This article was originally published in a special issue, “Drug Discovery and Repositioning Studies in Biopharmaceutical Sciences” |
| Indian J Pharm Sci 2024:86(4) Spl Issue “57-66” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Citrobacter freundii is a kind of pathogen widely existing in environment and human gut, and its drug resistance has been reported in recent years. The objective of this study was to isolate a phage targeting Citrobacter freundii and explore its morphological, biological and genomic characteristics to help the control and antibacterial of Citrobacter freundii. The Citrobacter freundii strain was co-cultured with sewage samples, followed by doublelayer plate assays to isolate a phage specific to Citrobacter freundii. Subsequently, the biological and genomic characteristics of the isolated phage were studied. As a result, a phage with strong stability of Citrobacter freundii was isolated from sewage and named CPB1126. The transmission electron microscopy revealed that CPB1126 presents as a Myoviridae-like morphology, characterized by a polyhedron-shaped head measuring 112±6.2 nm in length and 81±5.4 nm in width, along with a tail measuring of 103±3.7 nm in length. The optimal number of infection is 0.1. The one-step growth curve showed that the incubation period lasts 20 min and the outbreak period lasts 20 min. In addition to its isolated host, a Citrobacter werkmanii ATCC51114 strain was also sensitive to CPB1126. CPB1126 exhibited strong stability. It is resistant to chloroform and maintains high activity at pH 3-10. It also maintains high activity at temperature from -20° to 55°. The antibacterial experiments showed that excellent antibacterial effects could be achieved between 0.1 and 10 multiplicity of infection. The total length of the genome is 172 165 base pairs with 302 coding sequences. The functional annotation ratio is 56.3 %, it is classified as the Caudoviricetes order, Straboviridae family, Tevenvirinae subfamily, Moonvirus genus. CPB1126 is the 4th reported isolated phage of genus of Moonvirus, and its tail protein sequence is <85 % similar to that of reported phages of the same genus. In conclusion, this study reports the isolation of a highly virulent Citrobacter phage exhibiting robust lytic capabilities and strong stability, which would contribute to the accumulation of phage isolates resources and provides great assets for future phage usage especially for infections.
Keywords
Phage, Citrobacter, antibacterial, morphological, genomic, pathogen
Citrobacter freundii (C. freundii), a Gram-negative bacterium belonging to the Enterobacteriaceae family, is an opportunistic pathogen commonly residing in the intestinal tracts of both humans and animals[1]. It has been associated with a spectrum of infections ranging from intestinal to wound and bloodstream infections[1-6]. The pathogenic potential of C. freundii is not confined to terrestrial hosts; it also poses significant threats to aquatic species, leading to substantial economic losses in the aquaculture sector[7-10]. There is more of evidence that highlights the multidrug resistance of C. freundii, a consequence of the rampant use of antibiotics, which complicates treatment strategies in both clinical and aquaculture settings[11-13]. In response to the escalating issue of antibiotic resistance, bacteriophage therapy has emerged as a promising alternative for combating drug-resistant infections[14-19]. Despite its potential, the arsenal of phages that target C. freundii is currently limited. In an effort to augment our phage repository and enhance our anti-infective capabilities, this study aims to isolate and characterize novel bacteriophages targeting C. freundii. We will investigate their morphological, biological, and genomic profiles and assess their antibacterial efficacy to mitigate the challenges posed by C. freundii infections.
Materials and Methods
Bacterial strains:
C. freundii strain RTGS0227, Enterococcus faecalis strains JLDX0005, JDLX0006 and XJY0012, Bacillus cereus strain BC2402 and Cronobacter sakazakii strain CS333 were sourced from laboratory storage, Citrobacter werkmanii (C. werkmanii) ATCC51114 was derived from Shanghai Microbiological Culture Collection (SHMCC) Co., Ltd.
Phage isolation and purification:
We collected sewage water from local hospitals, centrifuged at 8000 rpm for 10 min and sterilized the supernatant with 0.45 μm filters (PALL, 4614). 40 ml filtered sample was mixed with 10 ml quintuplestrength Brain Heart Infusion (BHI) (BD, 237500) broth supplemented with 1 mM Calcium chloride (CaCl2) and Magnesium chloride (MgCl2). Late log-phase bacterial strain was 1:100 inoculated and cultured at 37° overnight. After co-culturing, we centrifuged the culture at 5000 Relative Centrifugal Force (RCF) for 15 min and sterilized the supernatant using a 0.45 μm filter. Next, we added a precipitant solution to the filtered culture (final concentration of 1 M Sodium chloride (NaCl) and 10 % (w/v) Polyethylene Glycol 8000 (PEG-8000)), mix and settle at 4° overnight. The settled mixture was then centrifuged at 4°, 12 000 RCF for 20 min, and the supernatant was discarded. Finally, we resuspended the pellet in 1 ml mixture of NaCl, Magnesium sulphate (MgSO4) and gelatin (SM buffer) to obtain the concentrated phage solution.
To identify isolated phages, we performed spot assays using the concentrated phage solution. 100 μl was mixed with 3 ml BHI semi-solid agar, 10 μl of the concentrated phage solution was dropped on the plate and cultured in 37° overnight. Once there were plaques showed up, we picked the plaques into 1 ml SM buffer. Then we dropped 10 μl of the supernatant of the phage solutions in the SM buffer on a new semi-solid agar plate with host strains and streaked to serial dilute with sterilized paper strips 3-4 times, incubated at 37° overnight. When a single plaque appeared, we picked the single plaque into 1 ml SM buffer and repeat the purification for 3-5 rounds to obtain a pure phage solution.
Phage enrichment and titrating:
1 % inoculate RTGS0227 bacterial suspension and a phage plaque into 50 ml BHI culture medium, add 1 mmol/l CaCl2 and MgCl2, incubate at 37° in a shaker for 6 h. Centrifuge and filter as described earlier. After that, the supernatant of lysate was diluted in gradients and the concentration of phage-rich liquid is determined by spot assays on the double-layer plate.
Transmission Electron Microscopy (TEM):
10 ml of 20 % sucrose solution was carefully injected beneath the lysate. Then we centrifuged it at 23 500 rpm for 3 h at 4°. We discard the supernatant and resuspended the pellet with 1 ml of SM buffer. Subsequently, we performed dialysis using a 100 kD MWCO dialysis in 1 l of SM buffer for 4 h. We replaced the buffer with another 1 l of fresh SM buffer and continued dialysis overnight. Finally, we sent the purified samples to Beijing Geo analysis Technology Co., Ltd., for TEM picture-taking.
Killing curve assay:
We mixed the phage with the log-phase host bacteria at Multiplicity of Infection (MOI) of 0.001, 0.01, 0.1, 1 and 10. The initial Optical Density (OD) 600 nm was adjusted to approximately 0.1. Subsequently, 200 μl of this mixture was added to each well of a 96- well plate and incubated at 37° with orbital shaking. Measurements of OD 600 nm were taken every 10 min. As a control, bacterial hosts were mixed with BHI-YH broth to serve as the free-phage control, while only BHI-YH liquid acted as the blank control. Each group was repeated three times.
pH, thermal and chloroform stability:
For pH stability, 100 μl of phage lysate was mixed with 900 μl of Double Distilled Water (ddH2O) at the pH ranging from 1-12 for 1 h. For thermal stability, phage lysate was place at the temperature of -20°, 4°, 37°, 45°, 55°, 65°, 75° and 85° for 1 h. For chloroform stability, phage lysate was combined with chloroform at the volume ratio of 0 %, 10 %, 20 %, 30 %, 40 % and 50 % for 1 h. After these treatments the phage titer was tested by spot assays.
Host range assays of phage:
Using the spot assays on double-layer plates, CPB1126 was tested its ability to infect host strains RTGS0227, Enterococcus faecalis JLDX0005, JLDX0006, XJY0012, Bacillus cereus strain BC2402, and Cronobacter sakazakii strain CS333 and C. werkmanii ATCC51114. In short, 100 μl of log-phase bacterial culture was combine with 3 ml semi-solid BHI agar. The mixture was then poured onto a BHI plate and allowed to dry completely. Next, we dripped 5 μl of a 108 PFU/ml phage solution onto the plate in triplicate. The plate was subsequently cultured at 37° overnight.
Optimal MOI of phage:
Equal volume of host culture (107 CFU/ml) was mix with phage lysate at different concentration (104, 105, 106, 107 and 108 PFU/ml (MOI of 0.001, 0.01, 0.1, 1 and 10), then cultured at 37° for 3 h. Then the phage titer was tested with spot assay. The optimal MOI was established as the highest titer.
One-step growth curve of phage:
Log-phase of bacterial strain RTGS0227 was diluted to 107 CFU/ml and mixed with 106 PFU/ml, incubated for 5 min at room temperature for absorption. Then the culture was centrifuged at 12 000 RCF for 1 min, the pellet was resuspended, following incubation in the shaker at 37° for 100 min. Samples were taken every 10 min. Then the samples were centrifuged at 12 000 RCF for 1 min and filtered with 0.45 μm filters. Phage titer of each sample was tested by spot assays.
Phage genome extraction:
Deoxyribonuclease (DNase) I (SIGMA, DN25) and Ribonucleases (RNase) A (INVITROGEN, 12091- 021) were added at a final concentration of 10 μg/ ml each into 50 ml of phage lysate, the mixture incubated at 37° for 1 h. Another round of DNase I and RNase A were added and incubated at 37° for 30 min. The nucleases were then inactivated at 65° for 20 min. Next, a precipitant solution was added to the lysate, resulting in a final concentration of 1 M NaCl and 10 % (w/v) PEG-8000, then settled at 4° overnight. Subsequently, the mixture was centrifuged at 4°, 12 000 RCF for 20 min and the pellet was resuspended with 200 μl of SM buffer. Then, 5 μl of proteinase K (NEB, P8107S) supplemented with 0.5 % Sodium Dodecyl Sulfate (SDS) were added to the resuspension, incubated at 56° for 1 h after gently mixing. Then another round of proteinase K was added, and incubated for an additional hour, following the inactivation at 65° for 20 min. The phage Deoxyribonucleic Acid (DNA) was extracted using the GeneClean® turbo kit, following the protocol provided by the kit.
Phage genome assembly and annotation:
For genome assembly, the single-end library of the phage genome DNA was prepared using the MGlEasy universal DNA library prep kit, and then sequenced on the DNBSEQ-E5 platform (MGI, BGIShenzhen, China). Raw data were filtered with fast to remove poor-quality reads and then assembled using SPAdes[20].
For functional gene annotation, gene prediction was performed using prodigal[21]. BLASTp searches against the National Center for Biotechnology Information (NCBI) database[22] and hmmscan searches against the UniProt/Swiss-Prot database were conducted. Final function annotation results were obtained through manual investigation of the amino acid homology search results. The genome map was conducted using Proksee[23].
The virulent factor was searched against databases such as the Virulence Factors Database (VFDB, http://www.mgc.ac.cn/VFs/). The antibiotic gene was identified by searching against the Comprehensive Antibiotic Resistance Database (CARD)[24] and ResFinder Database[25]. The transfer Ribonucleic Acid (tRNA) genes were identified with tRNAscan- SE[26].
Phylogenetic tree construction of the phage:
We searched ‘C. freundi phage’ and downloaded the non-redundant nucleotides from NCBI. Selected their large terminate sequences and performed the multiple sequences alignment with the online software miff[27], then constructed the phylogenetic tree with fast tree and visualized it with MEGA 11[28,29].
Linear comparison of the phages:
To perform the linear comparison of the phage genomes, the Gene bank file of the phage genomes were uploaded to the website https://gbkviz.streamlit. app/, select the ‘Genome Comparison Type’ as ‘Nucleotide One-to-One’, set the ‘Min Identity (%)’ to 85 %.
Results and Discussion
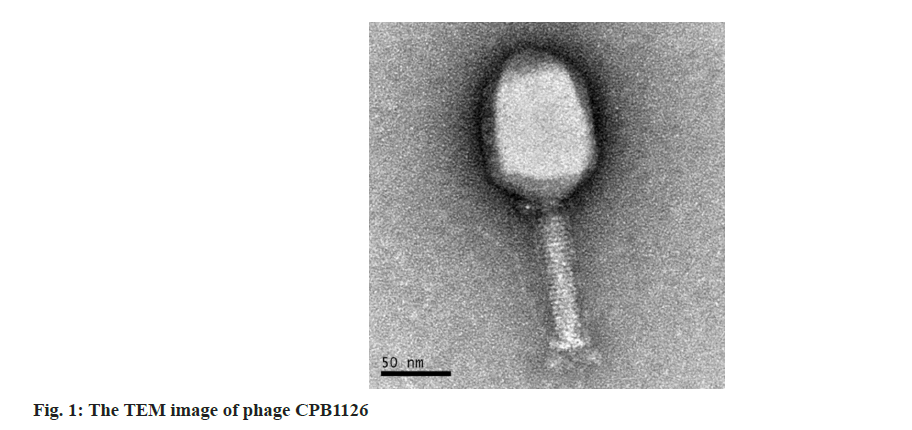
Having performed the phage isolation and purification steps, we obtained a strain of C. freundii phage, and we named it as CPB1126, storage it in Global Phage Hub (GPH). The plaques was clear, has a neat edge and its diameter was about 1 mm. The TEM graphs revealed that it has the morphology of Myoviridae-like phage, characterized with a polyhedron-shaped head measuring 112±6.2 nm (mean±Standard Deviation (SD)) in length and 81±5.4 nm in width, along with tail measuring 103±3.7 nm in length (fig. 1).
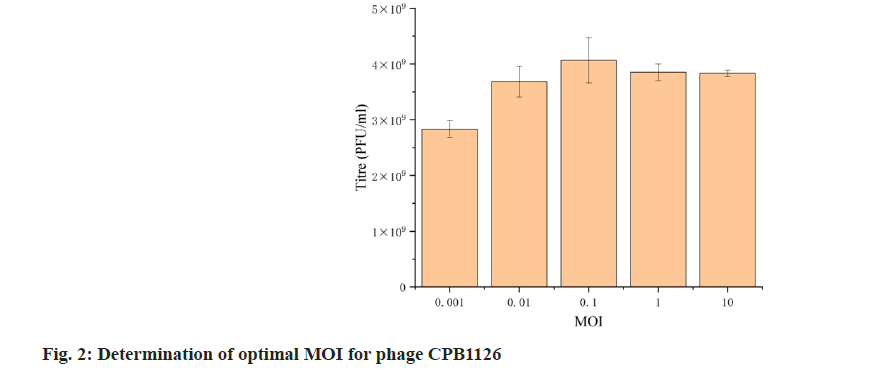
We then tested the optimal MOI of CPB1126. The resulting titers of the phages at MOI 0.001, 0.01, 0.1, 1 and 10 was 2.8×109, 3.68×109, 4.07×109, 3.85×109 and 3.83×10 PFU/ml (fig. 2). Therefore the optimal MOI of CPB1126 targeting C. freundii strain RTGS0227 is 0.1.
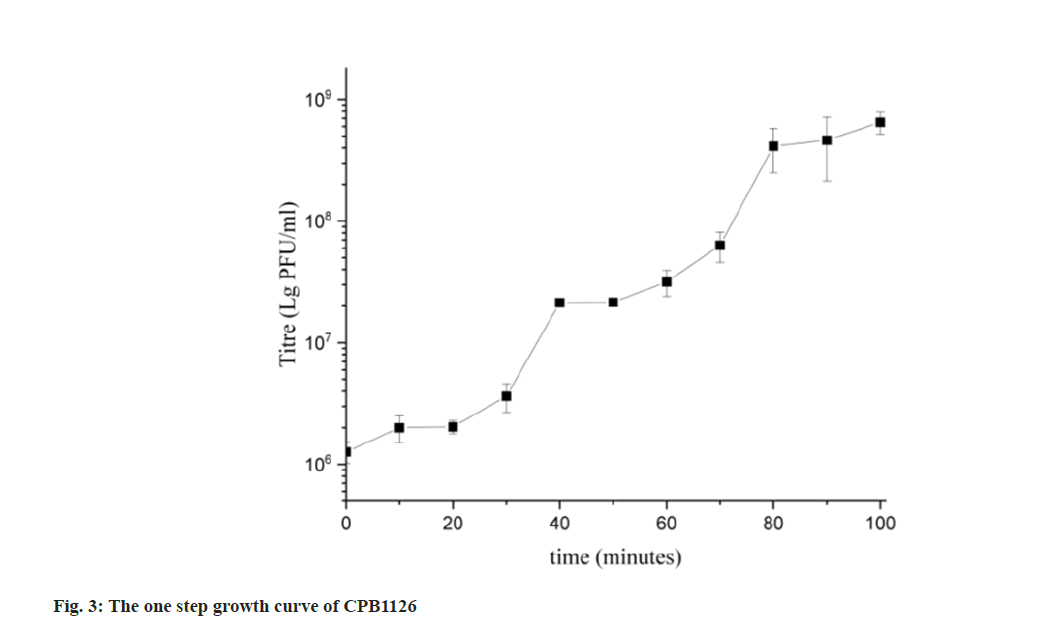
The one-step growth curve revealed that CPB1126 finished two rounds of explosive growth within 100 min infecting RTGS0227, as shown in fig. 3. The latent period and the resting period were both about 20 min. The burst size of the first rising during (20- 40) min was 21.33, and the burst rise of the second rising during (60-80) min was 19.36. Therefore, the burst size of CPB1126 was about 20 phages per infected bacterial cell.
The ability of CPB1126 to infect several common resistant bacteria was tested using spot assays, as shown in Table 1. The results showed that CPB1126 could infect C. freundii strain RTGS0227 and C. werkmanii ATCC51114, indicating CPB1126 is a cross-species infecting Citrobacter phage, and suggesting its potential candidacy for specific biological control applications. For further bacteriaphage interaction investigations, we need to accumulate a broader range of bacterial strains and phage resources for comprehensive studies.
| Species | Strain ID | Infection |
|---|---|---|
| C. freumdii | RTGS0227 | Y |
| Citrobacter werkmanii | ATCC51114 | Y |
| Enterococcus faecalis | JLDX0005 | N |
| Enterococcus faecalis | JLDX0006 | N |
| Enterococcus faecalis | XJY0012 | N |
| Bacillus cereus | BC2402 | N |
| Cronobacter sakazakii | CS3338 | N |
Table 1: The Host Range of Phage Cpb1126
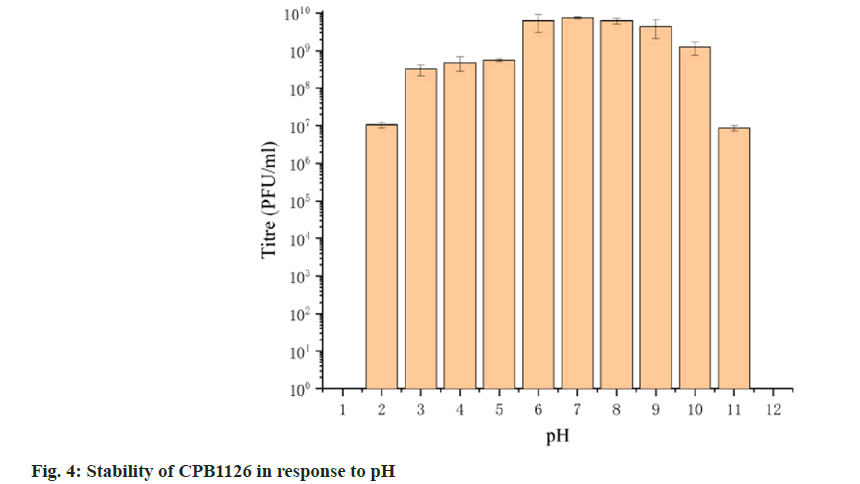
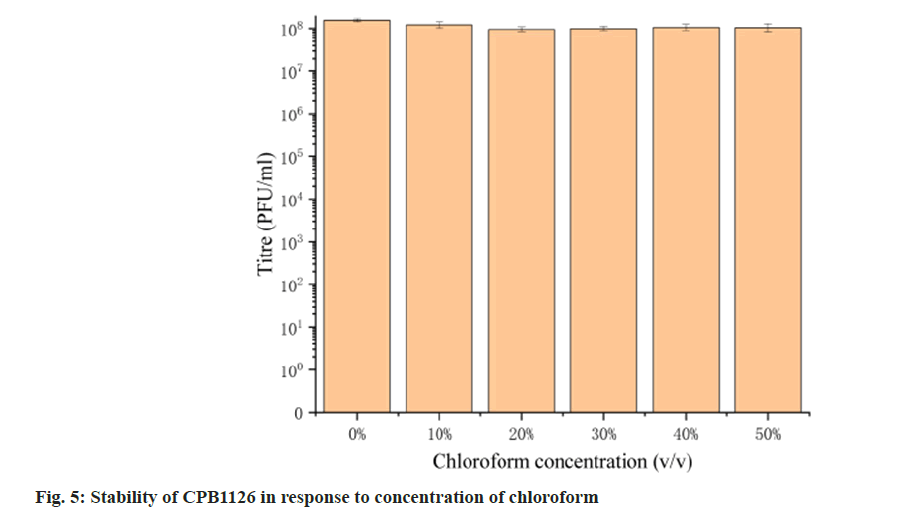
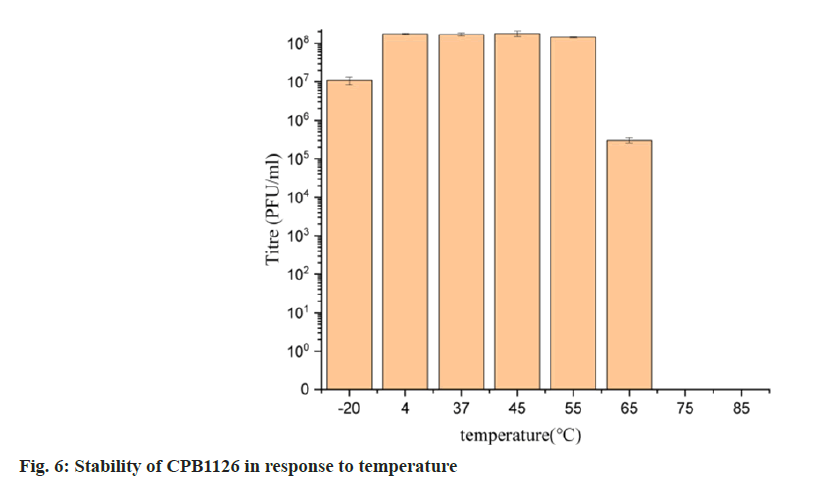
To assess its stability, we examined the activity of CPB1126 following treatment with varying pH levels, temperatures and concentrations of chloroform. The results of pH stability tests revealed that CPB1126 have a broad pH tolerance range from pH (2-11) (fig. 4). The titer was not changed from pH (6-9) and maintains good stability from pH (3-10). When the pH falls below 2 or exceeds 11, the phage loses its activity. CPB1126 has an extremely high tolerance to chloroform, as it does not affect the activity of CPB1126, suggesting that CPB1126 does not have a lipid-containing envelope structure (fig. 5). CPB1126 also has a high tolerance to cold and can maintain its activity at -20°. However, its tolerance to heat is low, with activity partially lost at 65° and completely lost at 85° (fig. 6). Combining these results, we suggest that CPB1126 possesses the potential to be adaptable for conventional transportation and be suitable for environmental and aquaculture disinfection.
The genome of phage CPB1126 was 172 165 bp in length with the GC content of 38.64 %. It carried ten tRNA genes and no integrase was carried. Neither virulent factor nor antibiotic genes was found in CPB1126, suggesting its safety during its applications. There were 302 Coding Sequences (CDSs) in the genome of CPB1126, of which 132 were hypothesis or unknown function proteins, and 170 were annotated as proteins with known functions, resulting in a function annotation ratio of 56.3 %. The genome of CPB1126 was submitted to National Microbiology Data Center (NMDC) with the accession number of NMDCX0000235 (https:// nmdc.cn/resource/genomics/attachment/detail/NMDCX0000235), and the genome map as shown in fig. 7.
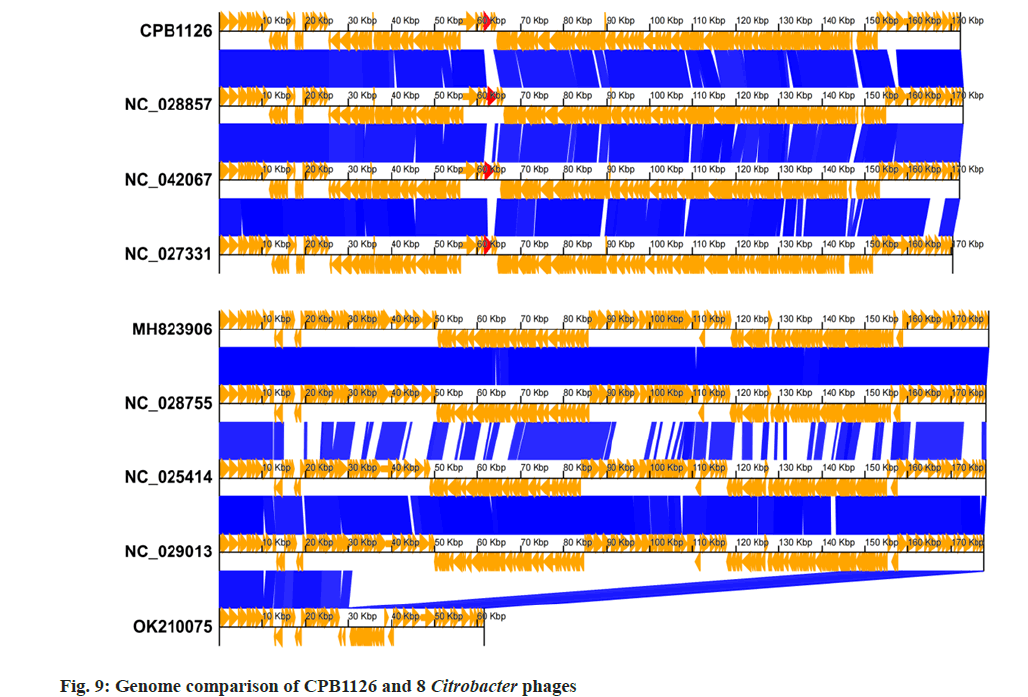
To investigate the relationship between CPB1126 and other C. freundi phages, we obtained 34 nonredundant genomic sequences of C. freundi phages from NCBI for constructing a phylogenetic tree. After performing function annotation, it was determined that only 20 strains harbor the large terminase gene. Subsequently, a phylogenetic tree was constructed using the large terminase sequences from these 20 downloaded strains and CPB1126, as shown in fig. 8. The phylogenetic tree showed that CPB1126 was located in the same branch with the Citrobacter phages NC_028857, NC_042067, and NC_027331.
According to NCBI, the three strains of Citrobacter phages belong to the order of Caudoviricetes, family of Straboviridae, sub-family of Tevenvirinae, and genus of Moonvirus. Therefore the tree suggest that CPB1126 belong to the genus of Moonvirus.
We then performed the genomic linear comparison of CPB1126 and the other 8 strains that located adjacent to CPB1126 on the phylogenetic tree, as shown in fig. 9. The result showed that CPB1126 exhibits a comparable genomic structure to NC_028857, NC_042067, and NC_027331, with the majority of CDSs sharing identities exceeding 85 %. Meanwhile, phage MH823906, NC_028755, NC_025414, and NC_029013, which located on distinct branches of the phylogenetic tree exhibited different genomic structures. Although CPB1126 shared some similar genomic structures with NC_028857, NC_042067, and NC_027331, we noticed that these four strain have great dissimilarities on the CDSs annotated as ‘long tail fiber distal subunit’, suggesting they probably absorb to different receptors on the host bacterial cell wall, and highlighting the novelty of CPB1126. Moreover, the differences on their tails could potentially make them serve as a reserve material for forming of a broad-spectrum bacteriophage cocktail.
The orange arrows indicates the CDSs and their directions. The blue color blocks indicate that the identity between the upper and lower sequences is >85 %. The red arrows are the CDSs annotated as long tail fiber distal subunit mentioned above.
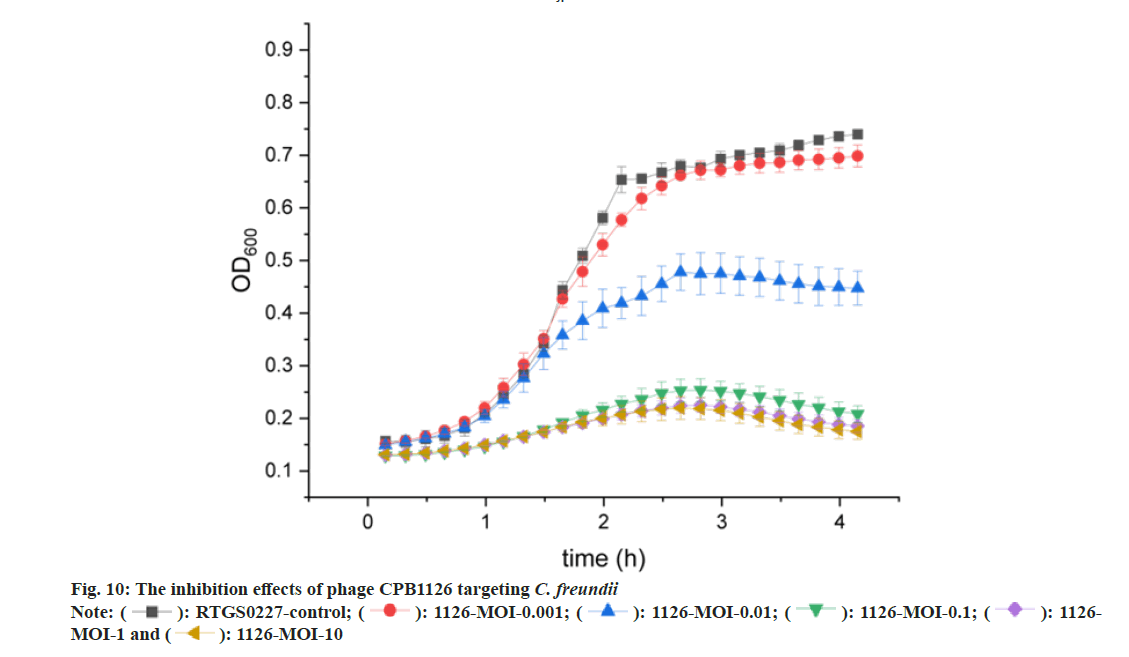
The inhibition effects were shown by the killing curve assays (fig. 10). The results showed that CPB1126 could not effectively kill C. freundii strain RTGS0227 at MOI 0.001. When MOI was 0.01, both the phage-treated bacteria and the phage-free bacteria first rapidly grow together for about 1.5 h, and then the phage-treated bacteria eventually are inhibited by the phages. When MOI was 0.1, 1 or 10, the bacteria no longer rapid grow phase, and the absorbance slightly increased and then decreases again. This result indicates that CPB1126 can already show great inhibition ability at an MOI of 0.1 or higher, and provides great assets for future infection treatments.
In this study, we isolated a stable strain of C. freundii phage named CPB1126 from hospital sewage. It can perform clear and tiny plaques on both C. freundii strain and can C. werkmanii strain. CPB1126 is very stable, remaining good activity at the pH range of 3-10, or at temperatures ranging from 4° to 55° for 1 h, and even maintain partial activity under -20° and 65°. It is also tolerant to high concentration of chloroform as well. Hence, it possesses the potential to be adaptable for conventional transportation and be suitable for environmental and aquaculture disinfection.
The antibacterial ability of CPB1126 is strong. The phage killing curve showed that CPB1126 showed strong bacterial inhibit effects under the range of MOI from 0.1 to 10. MOI 0.1 is effective already thereby a low concentration of phages is need for usage. The genome of CPB1126 is about 172 kb, encoding 302 CDSs but 132 CDS of them are unknown proteins. The genome of CPB1126 does not carry any virulent factors, antibiotic genes, and integrases, thus rendering it a safe lytic phage. There are only 34 nonredundant genomes of C. freundii phages on NCBI. With the phylogenetic tree, CPB1126 is identified as the family of Straboviridae, the sub-family of Tevenvirinae, and the genus of Moonvirus. Through genomic linear comparison, we observed significant dissimilarities in the distal subunit protein of the long tail fiber among these four strains, implying potential variations in receptor attachment and highlighting the novelty of CPB1126. Moreover, the differences on their tails could potentially make them serve as a reserve material for forming of a broad-spectrum bacteriophage cocktail.
Acknowledgements:
The authors acknowledge the GPB for preserving the phages and bacterial strains.
Conflict of interests:
The authors declared no conflict of interests.
References
- Liu L, Chen D, Liu L, Lan R, Hao S, Jin W, et al. Genetic diversity, multidrug resistance and virulence of Citrobacter freundii from diarrheal patients and healthy individuals. Front Cell Infect Microbiol 2018;8:233.
[Crossref] [Google Scholar] [PubMed]
- Liu L, Qin L, Hao S, Lan R, Xu B, Guo Y, et al. Lineage, antimicrobial resistance and virulence of Citrobacter spp. Pathogens 2020;9(3):195.
[Crossref] [Google Scholar] [PubMed]
- Sommer J, Reiter H, Sattler J, Cacace E, Eisfeld J, Gatermann S, et al. Emergence of OXA-48-like producing Citrobacter species, Germany, 2011 to 2022. Eurosurveillance 2024;29(15):2300528.
[Crossref] [Google Scholar] [PubMed]
- Anderson MT, Mitchell LA, Zhao L, Mobley HL. Citrobacter freundii fitness during bloodstream infection. Sci Rep 2018;8(1):11792.
- Uberoi A, McCready-Vangi A, Grice EA. The wound microbiota: Microbial mechanisms of impaired wound healing and infection. Nat Rev Microbiol 2024:1-5.
- Zhanel GG, Pozdirca M, Golden AR, Lawrence CK, Zelenitsky S, Berry L, et al. Sulopenem: An intravenous and oral penem for the treatment of urinary tract infections due to multidrug-resistant bacteria. Drugs 2022;82(5):533-57.
[Crossref] [Google Scholar] [PubMed]
- Gong C, Guo M, Lou J, Zhang L, An Z, Vakharia VN, et al. Identification and characterization of a highly virulent Citrobacter freundii isolate and its activation on immune responses in largemouth bass (Micropterus salmoides). Fish Shellfish Immunol 2023;143:109224.
- An T, Lu X, Han Y, Guo C, Guo J, Zhu G, et al. Exploring the bacterial diversity and composition with special emphasis on pathogens in ship ballast water and sediments using full-length 16S rRNA gene sequencing. Mar Pollut Bull 2023;194:115336.
[Crossref] [Google Scholar] [PubMed]
- Lomiya ME, Raguvaran R, Mondal D, Dosar S, Nair SS, Jitha KR, et al. Mitigating antimicrobial resistance, an approach to stewardship in canine urinary tract infection. Vet Res Commun 2024:1-1.
[Crossref] [Google Scholar] [PubMed]
- Cardoso MD, Maciel OL, de Souza AL, Roges EM, Gonçalves VD, Siciliano S, et al. Smelly shark, smelly ray: What is infecting you? J Appl Microbiol 2024;135(4):lxae068.
[Crossref] [Google Scholar] [PubMed]
- Liu L, Zhang L, Zhou H, Yuan M, Hu D, Wang Y, et al. Antimicrobial resistance and molecular characterization of Citrobacter spp. causing extraintestinal infections. Front Cell Infect Microbiol 2021;11:737636.
[Crossref] [Google Scholar] [PubMed]
- Boutin S, Welker S, Gerigk M, Miethke T, Klaus HE, Nurjadi D. Molecular surveillance of carbapenem-resistant Enterobacterales in two nearby tertiary hospitals to identify regional spread of high-risk clones in Germany, 2019-2020. J Hosp Infect 2024;S0195-6701(24):156-7.
[Crossref] [Google Scholar] [PubMed]
- Zhang F, Li Z, Liu X, Hu Y, Zhao J, Zhang Y, et al. Carbapenem-resistant Citrobacter freundii harboring bla KPC-2 and bla NDM-1: A study on their transferability and potential dissemination via generating a transferrable hybrid plasmid mediated by IS 6100. Front Microbiol 2023;14:1239538.
[Crossref] [Google Scholar] [PubMed]
- Strathdee SA, Hatfull GF, Mutalik VK, Schooley RT. Phage therapy: From biological mechanisms to future directions. Cell 2023;186(1):17-31.
[Crossref] [Google Scholar] [PubMed]
- Bhargava K, Nath G, Bhargava A, Aseri GK, Jain N. Phage therapeutics: From promises to practices and prospectives. Appl Microbiol Biotechnol 2021;105:9047-67.
[Crossref] [Google Scholar] [PubMed]
- Chen P, Liu Z, Tan X, Wang H, Liang Y, Kong Y, et al. Bacteriophage therapy for empyema caused by carbapenem-resistant Pseudomonas aeruginosa. Biosci Trends 2022;16(2):158-62.
[Crossref] [Google Scholar] [PubMed]
- Li J, Yan B, He B, Li L, Zhou X, Wu N, et al. Development of phage resistance in multidrug-resistant Klebsiella pneumoniae is associated with reduced virulence: A case report of a personalised phage therapy. Clin Microbiol Infect 2023;29(12):1601-7.
[Crossref] [Google Scholar] [PubMed]
- Federici S, Kredo-Russo S, Valdés-Mas R, Kviatcovsky D, Weinstock E, Matiuhin Y, et al. Targeted suppression of human IBD-associated gut microbiota commensals by phage consortia for treatment of intestinal inflammation. Cell 2022;185(16):2879-98.
[Crossref] [Google Scholar] [PubMed]
- Galtier M, Sordi LD, Sivignon A, de Vallée A, Maura D, Neut C, et al. Bacteriophages targeting adherent invasive Escherichia coli strains as a promising new treatment for Crohn’s disease. J Crohn's Colitis 2017;11(7):840-7.
[Crossref] [Google Scholar] [PubMed]
- Prjibelski A, Antipov D, Meleshko D, Lapidus A, Korobeynikov A. Using SPAdes de novo assembler. Curr Protoc Bioinformatics 2020;70(1):e102.
[Crossref] [Google Scholar] [PubMed]
- Hyatt D, Chen GL, LoCascio PF, Land ML, Larimer FW, Hauser LJ. Prodigal: Prokaryotic gene recognition and translation initiation site identification. BMC Bioinformatics 2010;11:119.
[Crossref] [Google Scholar] [PubMed]
- Tesson F, Hervé A, Mordret E, Touchon M, D’humières C, Cury J, et al. Systematic and quantitative view of the antiviral arsenal of prokaryotes. Nat Commun 2022;13(1):2561.
[Crossref] [Google Scholar] [PubMed]
- Stothard P, Grant JR, van Domselaar G. Visualizing and comparing circular genomes using the CGView family of tools. Brief Bioinformatics 2019;20(4):1576-82.
[Crossref] [Google Scholar] [PubMed]
- Alcock BP, Huynh W, Chalil R, Smith KW, Raphenya AR, Wlodarski MA, et al. CARD 2023: Expanded curation, support for machine learning and resistome prediction at the comprehensive antibiotic resistance database. Nucl Acids Res 2023;51(D1):D690-9.
[Crossref] [Google Scholar] [PubMed]
- Florensa AF, Kaas RS, Clausen PT, Aytan-Aktug D, Aarestrup FM. ResFinder-An open online resource for identification of antimicrobial resistance genes in next-generation sequencing data and prediction of phenotypes from genotypes. Microb Genom 2022;8(1):000748.
[Crossref] [Google Scholar] [PubMed]
- Chan PP, Lin BY, Mak AJ, Lowe TM. tRNAscan-SE 2.0: Improved detection and functional classification of transfer RNA genes. Nucl Acids Res 2021;49(16):9077-96.
[Crossref] [Google Scholar] [PubMed]
- Katoh K, Rozewicki J, Yamada KD. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief Bioinform 2019;20(4):1160-6.
[Crossref] [Google Scholar] [PubMed]
- Price MN, Dehal PS, Arkin AP. FastTree: Computing large minimum evolution trees with profiles instead of a distance matrix. Mol Biol Evol 2009;26(7):1641-50.
[Crossref] [Google Scholar] [PubMed]
- Tamura K, Stecher G, Kumar S. MEGA11: Molecular evolutionary genetics analysis version 11. Mol Biol Evol 2021;38(7):3022-7.
[Crossref] [Google Scholar] [PubMed]
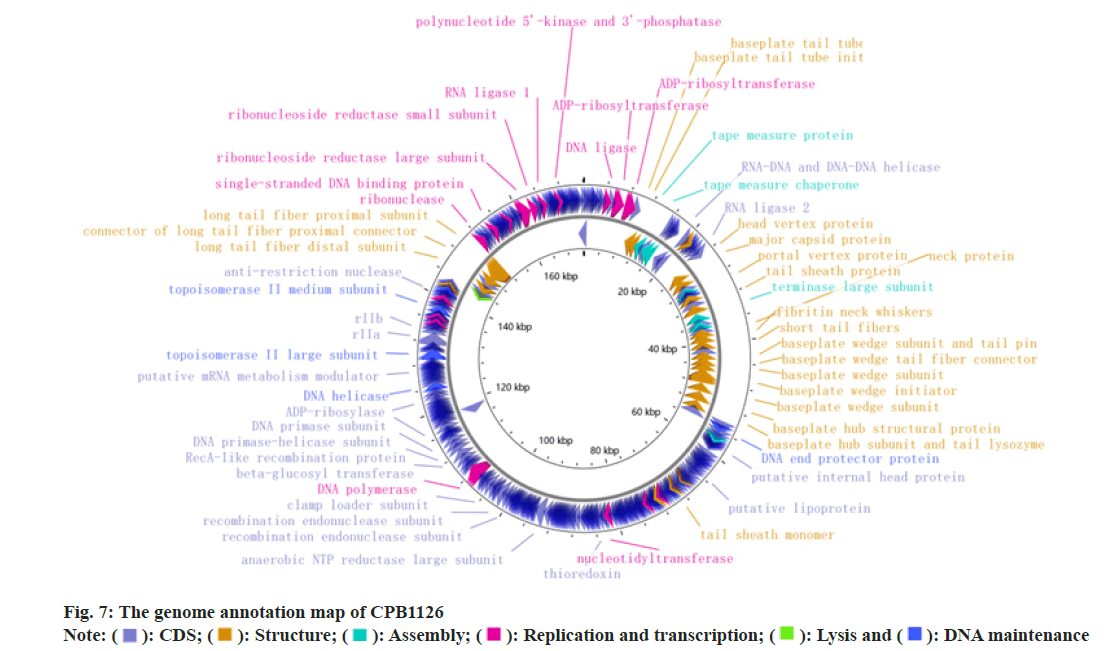
 ): CDS; (
): CDS; ( ): Structure; (
): Structure; ( ): Assembly; (
): Assembly; ( ): Replication and transcription; (
): Replication and transcription; ( ): Lysis and (
): Lysis and ( ): DNA maintenance
): DNA maintenance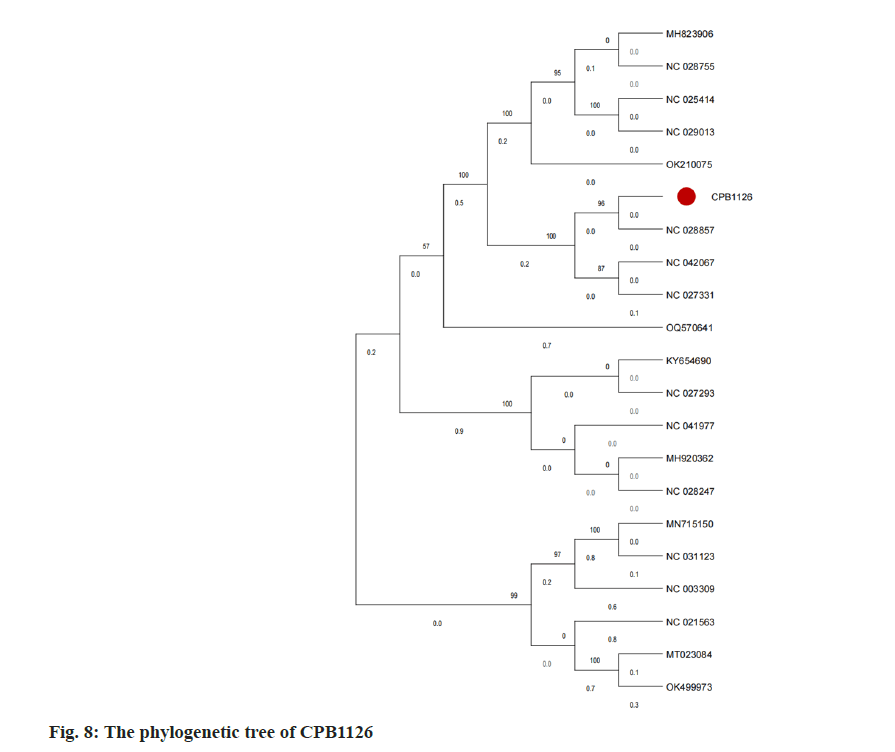
 ): RTGS0227-control; (
): RTGS0227-control; ( ): 1126-MOI-0.001; (
): 1126-MOI-0.001; ( ): 1126-MOI-0.01; (
): 1126-MOI-0.01; ( ): 1126-MOI-0.1; (
): 1126-MOI-0.1; ( ): 1126-
MOI-1 and (
): 1126-
MOI-1 and ( ): 1126-MOI-10
): 1126-MOI-10



