- *Corresponding Author:
- P. Katiyar
Department of Microbiology
Gurukula Kangri Vishwavidyalaya
Haridwar, Uttarakhand 249404, India
E-mail: prashant22021994@gmail.com
| Date of Received | 01 October 2019 |
| Date of Revision | 08 April 2020 |
| Date of Acceptance | 20 August 2021 |
| Indian J Pharm Sci 2021;83(4):838-847 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Chebulinic acid, a chemotaxonamic marker of Terminalia chebula has been described in Ayurveda, Unani and Homeopathy traditional medicinal system for the treatment of various fungal diseases. Almost 70-80 % of women are currently affected from vulvovaginal candidiasis and the overgrowth of Candida albicans is the major reason behind it. The aim of the present study was to determine the efficacy of chebulinic acid in inhibiting adhesion as well as growth of Candida albicans inside the host in comparison with currently available azole derivatives in the market. The inhibitory effect of chebulinic acid was determined by spectrophotometric growth curve analysis reflecting 10 % lower minimum inhibitory concentration value of fluconazole, ketaconazole, itraconazole and chebulinic acid. Virtual docking study was carried out at catalytic domain of agglutinin-like sequence protein 3 chain A and B respectively, to determine the binding affinity and its active binding sites. Chebulinic acid exhibited high sensitivity against Candida albicans with minimum inhibitory concentration of (0.025 μg/ml) followed by ketaconazole (0.05), itraconazole (0.048) and fluconazole (1.5). An extended lag phase and a decline in overall growth were observed in Candida albicans when compared to control when grown in presence of chebulinic acid. At ASN22, TYR23, TRP224, ASP169, TYR166, ASP162, ASN225 GLY297, ARG294 and ASP162, ASN160, ASP169, THR168, TYR166, TYR226, chebulinic acid binds with agglutinin-like sequence protein 3 chain A and B respectively with binding affinity of -10.4 and -9.8 kcal/mol. Our present study strengthens the evidences which stand in support of the fact that chebulinic acid can be used as potent inhibitor of Candida albicans than other commercially available synthetic anti-fungal drugs.
Keywords
Agglutinin-like sequence protein 3, azole derivatives, Terminalia chebula, vulvovaginal candidiasis
Vulvovaginal candidiasis (VVC) is caused by the overgrowth of Candida species. Curd like vaginal discharge, erythema and itching are the major characteristic features of this infection. Incidences of VVC have enormously increased in past 10 y. Candida albicans (C. albicans) alone itself contributes to 80- 90 % of vulvovaginal cases whereas other Candida species such as Non-albicans Candida, Candida krusei and Candida glabrata are rarely found to be involved in complicating the disease. Nearly about 5-10 million females seek advice from gynecologists to encounter this particular disease every year globally. Several azole derivatives are commonly available in market to tackle these types of issues but the population of females struggling with VVC is increasing rapidly[1-4]. Candida species have also started acquiring resistance against the available drugs. Azole derivatives have substantially affected patient’s quality of life as well. Systemic resistance and several side-effects associated with usage of azole derivatives have become a major issue of concern[5]. A search for a new substitute from medicinal plants has become the need of hour to overcome the issue. Several studies have documented the significant role of herbal remedy in treating the disease[6,7].
Terminalia chebula (T. chebula) is widely known to exhibit medicinal properties and it is used to treat several ailments[8,9]. T. chebula have been extensively used in Unani, Ayurveda and homeopathic medicinal system to treat geriatric diseases and acts as memory booster and improves the functioning of brain[10-14]. Efficacy of T. chebula to target C. albicans has not been explored yet. The present study aims to determine the potency of chemotaxonomic markers of T. chebula to inhibit C. albicans and to draw a comparative analysis of natural moieties and chemical analogues to target C. albicans.
Materials and Methods
Bacterial strains under study:
C. albicans was procured from Institute of Microbial Technology (IMTECH) Chandigarh with strain number microbial type culture collection (MTCC) 183 and was regularly grown in Sabouraud dextrose broth (SDB) at 25° and pH 7.
Chemicals and reagents:
SDB and agar was purchased from HiMedia and the medium was prepared according to manufacturer’s recommendations. Fluconazole, ketoconazole, itraconazole and chebulinic acid was availed from Sigma-Aldrich India wide Chemical Abstracts Service (CAS) number 86386-73-4 (fluconazole), 65277-42-1 (ketoconazole), 84625-61-6 (itraconazole) and 18942-26-2 (chebulinic acid).
Determination of antibiotic susceptibility and minimum inhibitory concentration (MIC):
MIC values were obtained for fluconazole, ketoconazole, itraconazole and chebulinic acid against C. albicans by using 96 well microdilution method as suggested by Jorgensen and Ferraro[15].
Determination of effect of flucanazole, ketoconazole, itraconazole and chebulinic acid over the growth of C. albicans:
To determine the effect of above mentioned drugs on the growth of C. albicans, culture was routinely grown in the presence of drugs at 10 % lower value of MIC in SDB at 25° and pH 7. Cell growth was monitored spectro-photometrically at 620 nm at regular intervals (20 h). The effect of drugs over the growth of C. albicans was compared with the control.
Molecular docking:
Preparation of the ligand library and protein: Agglutinin-like sequence protein 3 (ALS3) crystal structures were obtained from Research collaboratory for structural bioinformatics (RCSB) protein database bearing protein data bank (PDB) Id 4LE8. With the removal of non-relevant water molecules and the addition of hydrogen atoms to the receptor molecule, the protein structure was prepared by using Biovia discovery studio[16]. The protein structures were then saved as PDB file for further analysis. The ligand structures of fluconazole, ketoconazole, itraconazole and chebulinic acid were prepared using RPBS frog server with canonical smiles obtained from Pubchem (Table 1). The molecule structure format (mol2) ligand files were downloaded from the RPBS frog server and then converted into PDB file format using OpenBabel 2.4.1[17]. The energy minimization process (500 steps of steepest descent followed by 5000 steps of conjugant gradient for the receptor; 50 steps of steepest descent followed by 500 steps of conjugant gradient for the ligands) was carried out for removal of steric clashes on the receptor molecule and the ligands using UCSF Chimera suite[18] after adding H-atoms.
| Ligands | Pubchem compound identification number (CID) | Canonical smiles |
|---|---|---|
| Fluconazole | 3365 | C1=CC(=C(C=C1F)F)C(CN2C=NC=N2 (CN3C=NC=N3)O |
| Ketaconazole | 3823 | CC(=O)N1CCN(CC1)C2=CC=C(C=C2)OCC3COC(O3)(CN4C=CN=C4)C5=C(C=C(C=C5)Cl)Cl |
| Itraconazole | 55283 | CCC(C)N1C(=O)N(C=N1)C2=CC=C(C=C2)N3CCN(CC3)C4=CC=C(C=C4)OCC5COC(O5)(CN6C=NC=N6)C7=C(C=C(C=C7)Cl)Cl |
| Chebulinic acid | 72284 | C1=C(C=C(C(=C1O)O)O)C(=O)OCC2C3C(C(C(O2)OC(=O)C4=CC(=C(C(=C4)O)O)O)OC(=O)C5=CC(=C(C6=C5C(C(C(=O)O3)CC(=O)O)C(C(=O)O6)O)O)O)OC(=O)C7=CC(=C(C(=C7)O)O)O |
Table 1: Pubchem Id and Canonical Smiles of Fluconazole, Ketoconazole, Itraconazole and Chebulinic Acid
Evaluation of protein model: The protein model was uploaded to PDBsum and the Ramachandran plot was analyzed on a program to check the stereochemical quality of protein structures (PROCHECK)[19] for analysis of stereo-chemical properties of protein which was followed by validation using Verify three dimensional (3D) and protein structure analysis (ProSA) [20] plot. The amino acid bond angles (Ø & ?) in the protein conformation determined A and B protein helices as well as polypeptides backbone. The angles denoted in the Ramachandran plot were used for modelling and structural properties of the protein. Allowed and disallowed regions of the protein conformation were analyzed with high resolution. Statistical parameters displayed the distribution of amino acids in the secondary structures[21]. The hydropathy analysis of target protein exhibits the transmembrane or surface region in a hydropathy plot. This analysis assesses the hydrophilic or hydrophobic nature of the target protein.
Physico-chemical property analysis of the selected proteins: The selected proteins were analyzed for their physicochemical properties sequence wise using the ProtPARAM[22] server for their grand average of hydropathicity, aliphatic index, estimated half-life, atomic composition, extinction coefficient, amino acid structure, molecular weight and their atomic composition.
Cavity prediction and binding site analysis: After preparation of the protein, the active binding sites in the target protein were predicted using a web based server computed atlas of surface topography of proteins (CASTp) 3.0[23].
Screening and visualization: Prepared ligands were virtually screened and performed at the active sites of the ALS3 domain by using AutoDockVina software in PyRx platform[24]. Lamarckian genetic algorithm was selected for docking with standard protocol. Throughout the docking study, the ligand molecules were flexible, whereas macromolecule was kept rigid. Docking was performed to obtain a population of possible orientations and conformations for the ligands at the binding site and the lowest binding energy entities were chosen after the docking search was completed.
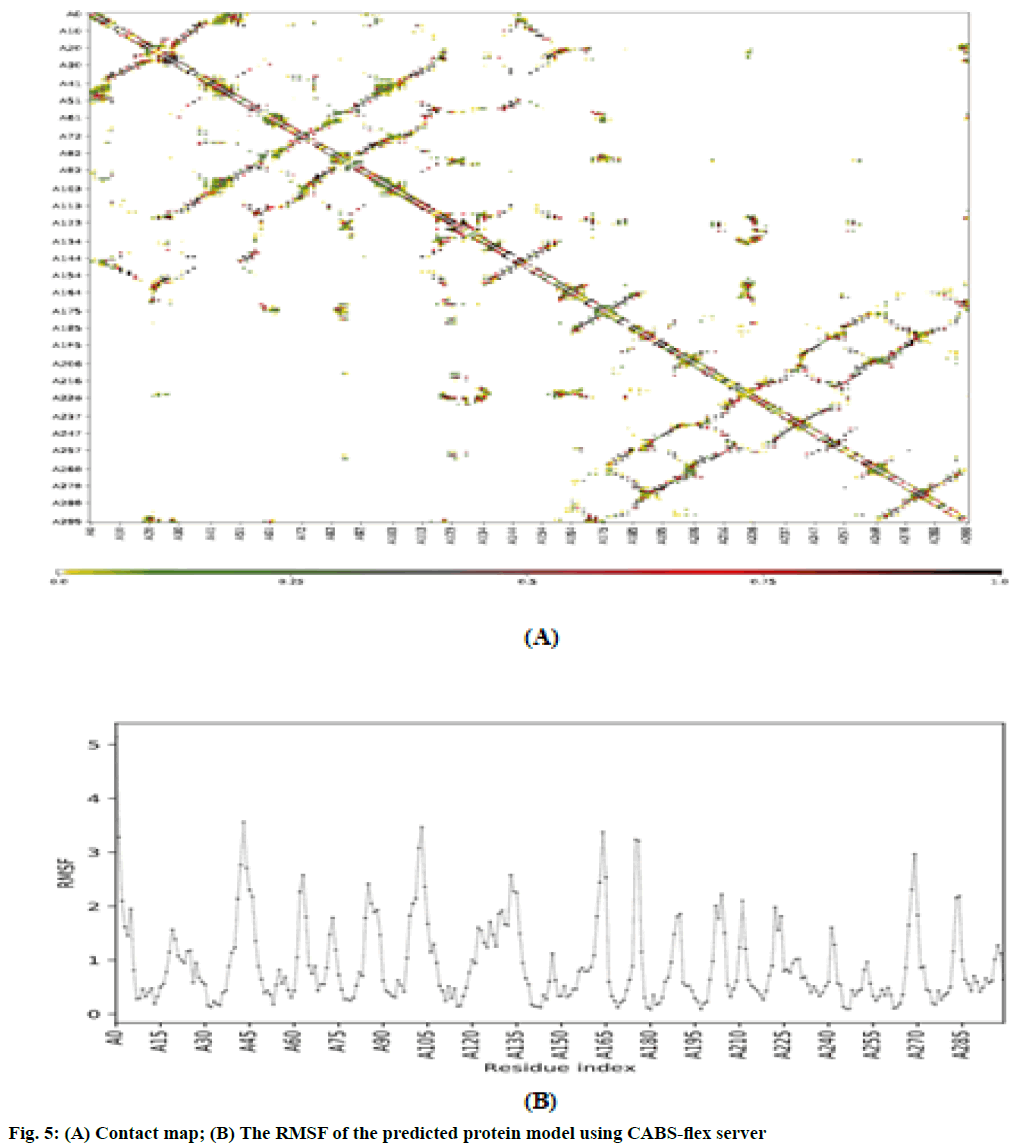
Molecular dynamic (MD) simulation, route means square fluctuation (RMSF) and radius of gyration (Rg) evaluation of the predicted protein: MD simulation was performed on CABS-flex 2.0 server[25] by 10 nanoseconds (ns) simulation and generated data are fairly similar to nuclear magnetic resonance (NMR) data[26]. RMSF was calculated by the following formula:
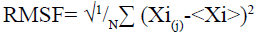
Here (Xi) is the average of entire trajectory and “i” is the particle with position X in “j” frame[26]. Rg was evaluated using supercomputing facility for bioinformatics & computational biology (SCFBio)[27] server. Following formula was used to calculate the Rg.
Rg= 0.395×N3/5+7.257
*Here N is the number of amino acids.
Results and Discussion
In first phase of the study, MIC was computed using microdilution method. Stock solution at concentration of 1 mg/ml of fluconazole, ketoconazole, itraconazole and chebulinic acid was prepared in dimethylsulfoxide (DMSO) and was used to determine the MIC. As expected, C. albicans was found to be more sensitive for ketoconazole and itraconazole in comparison to fluconazole. Ketoconazole and itraconazole exhibited almost equal potency in targeting C. albicans as determined from their MIC values. Contrastingly, chebulinic acid showed highest cell growth inhibition. C. albicans was found to be most sensitive for chebulinic acid followed by ketoconazole and itraconazole whereas fluconazole showed failure in inhibiting growth of C. albicans in comparison to other three. MIC values of fluconazole, ketoconazole, itraconazole and chebulinic acid against C. albicans are mentioned below in Table 2.
| Chebulinic acid (μg/ml) | Fluconazole (μg/ml) | Ketaconazole (μg/ml) | Itraconazole (μg/ml) | |
|---|---|---|---|---|
| C. albicans | 0.025 | 1.5 | 0.05 | 0.048 |
Table 2: MIC Determination of Fluconazole, Ketoconazole, Itraconazole and Chebulinic Acid against C. Albicans
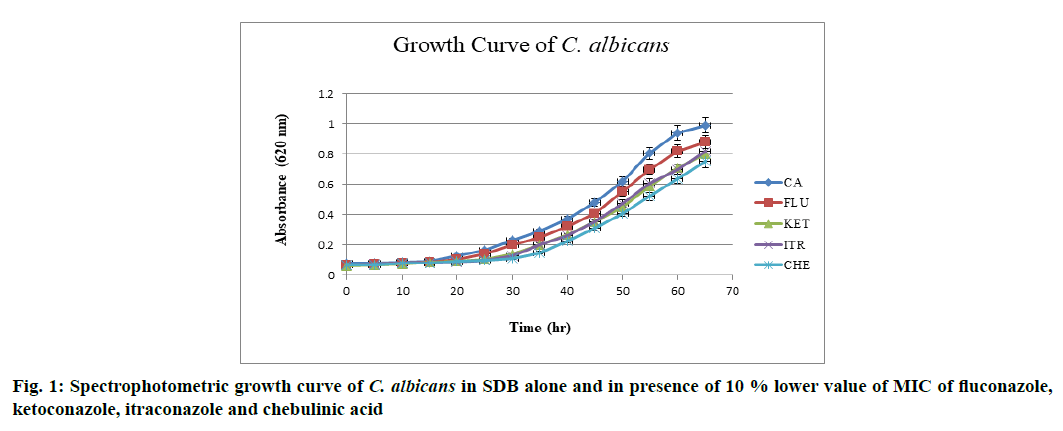
The effect of drugs over growth phase of C. albicans was determined. C. albicans when grown in SDB at 37° and pH 7 attained stationary phase by the end of 60th h after acquiring complete growth (absorbance (Abs)-0.990 nm). When grown in the presence of above mentioned drugs at 10 % lower value of MIC, a decline in overall growth was observed and lag phase was found to be prolonged, when in the presence of chebulinic acid. Ketoconazole and itraconazole showed almost equal potency in inhibiting the growth of C. albicans whereas fluconazole did not showed any significant effect in growth inhibition. Chebulinic acid proved to be a molecule of wonder since it not only declined the overall growth but also extended lag and log phase. Growth curve pattern of C. albicans in presence and absence of drugs are mentioned in fig. 1, which clearly indicated the potency of chebulinic acid to target C. albicans.
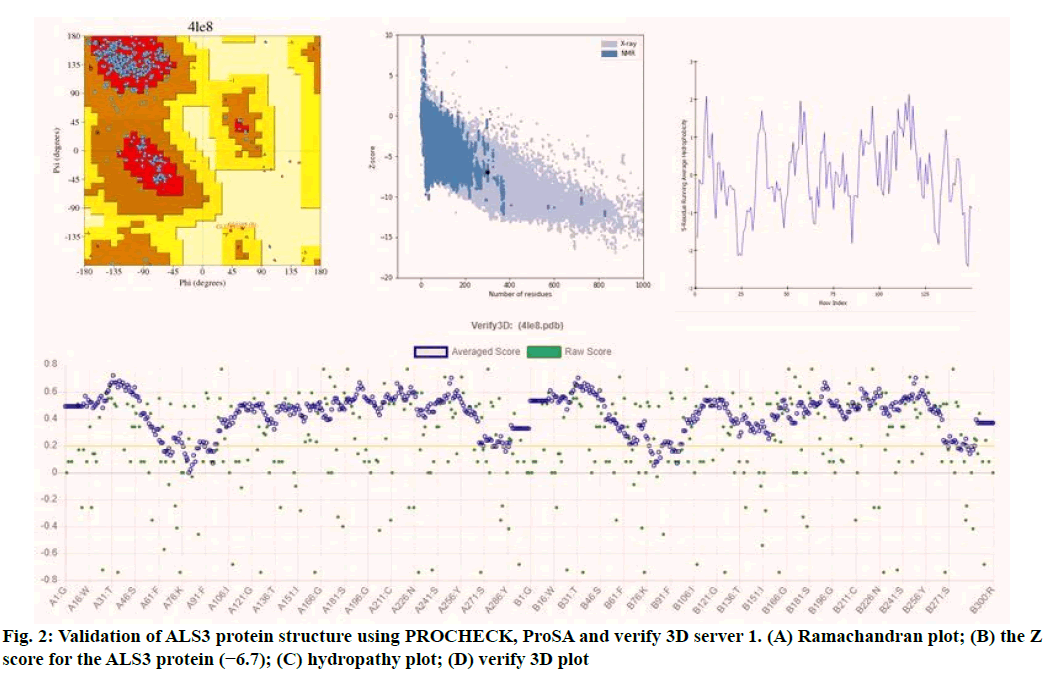
Validation of the model is very important in protein structural prediction since ultimately the modeled protein structure is used to understand the protein's biological function in order to design further experiments. The Ramachandran plot of ALS3 protein revealed amino acid residues in the favored region of about 97.8 % and the number of expected residues in the allowed region is 2.2 %. The plot represented the Ø & ? values of proteins (fig. 2). ProSA web z-scores of all protein chains in PDB were determined by X-ray crystallography (light blue) and NMR spectroscopy (dark blue) with respect to their length. The z-score of ALS3 was highlighted as large black dots in the light blue region representing the structures (X-ray crystallography structures) with a Z score value of -6.7. Z score plots elucidated that the structure of ALS3 possess model quality. Verify 3D plot of the modelled protein (fig. 2) showed pass and the 3D environment profile resulted in 91.83 % of the residues, having average 3D-1D score ≥0.2, suggesting that the modelled protein is structurally valid. The hydropathy analysis showed that there is no major peak in the hydropathy plot which is shown in fig. 2.
Different physiochemical parameters were computed by using ExPasy ProtParam tool and were shown in Table 3. The theoretical isoelectric point (PI) was found to be 6.14 which can be inferred that it is an acidic protein. It may be due to a large percentage of acidic amino acids in this protein. The instability index of this protein was found to be 20.72 which is less than 40, indicating that the protein is very stable in nature and it may be due to occurrence of less number of dipeptides in the composition. The aliphatic index that defines the thermal stability of the protein was found to be 63.03, suggesting that this protein is stable at a wide range of temperature. It might be due to the fact that a relative volume of this protein has occupied large numbers of aliphatic side chain amino acids such as alanine, valine and leucine. However the grand average of hydropathy (GRAVY) value was found to be -0.168 which is less than zero, that implies that this protein is more hydrophilic in nature which suggests a large number of polar amino acids on the surface of this protein. The CASTp online tool searches for probable active sites on the protein, predicted 91 pockets. The pocket which was selected for virtual screening have following amino acid residues ALA18, ALA19, THR20, TYR21, ASN22, TYR23, THR28, PRO29, THR30, LYS59, PHE60, THR61, THR62, SER63, GLN64, THR65, PHE81, GLN82, ALA 83, GLY84, GLU85, PHE87, SER91, THR92, SER159, ASN160, VAL161, LEU167, THR168, ASP169, SER170, VAL172, PRO174. SER175, ASN177, LEU182, TRP224, ASN225, TYR226, ALA265, THR266, ASP267, VAL268, ASN269, SER270, TYR271, THR292, LEU293, ARG294, TRP295, THR296, GLY297, TYR298, ARH299 in chain A and THR51, ASN53, PRO55, CYS56, PHE90, THR92, LYS132, CYS133, PHE134, THR135, THR138, ASN139, THR140, VAL141, THR142, PHE143, ASN144, GLU146, GLY147, LYS149, SER151 in chain B. The active site area covered by ALS3 molecules was 1239.096 and volume 1336.588.
| Serial No. | Physiochemical properties | |
|---|---|---|
| 1. | Number of amino acids | 300 |
| 2. | Molecular weight | 32375.02 |
| 3. | Theoretical pI | 6.14 |
| 4. | Total number of negatively charged residues (Asp+Glu) |
20 |
| 5. | Total number of positively charged residues (Arg+Lys) |
19 |
| 6. | Formula | C1446H2173N369O456S11 |
| 7. | Total number of atoms | 4455 |
| 8. | Extinction coefficients (M−1cm−1, at 280 nm measured in water) |
57340 (Abs 0.1 % (=1 g/l) 1.771, assuming all pairs of Cys residues form cystines) 56840 (Abs 0.1 % (=1 g/l) 1.756, assuming all Cys residues are reduced) |
| 9. | Estimated half-life (N-terminal of the sequence considered is G (Gly)) |
>20 h (yeast, in vivo). >10 h (Escherichia coli, in vivo). |
| 10. | Instability index | 20.72 |
| 11. | Aliphatic index | 63.03 |
| 12. | GRAVY | -0.168 |
Table 3: Physicochemical Properties of the ALS3 Protein
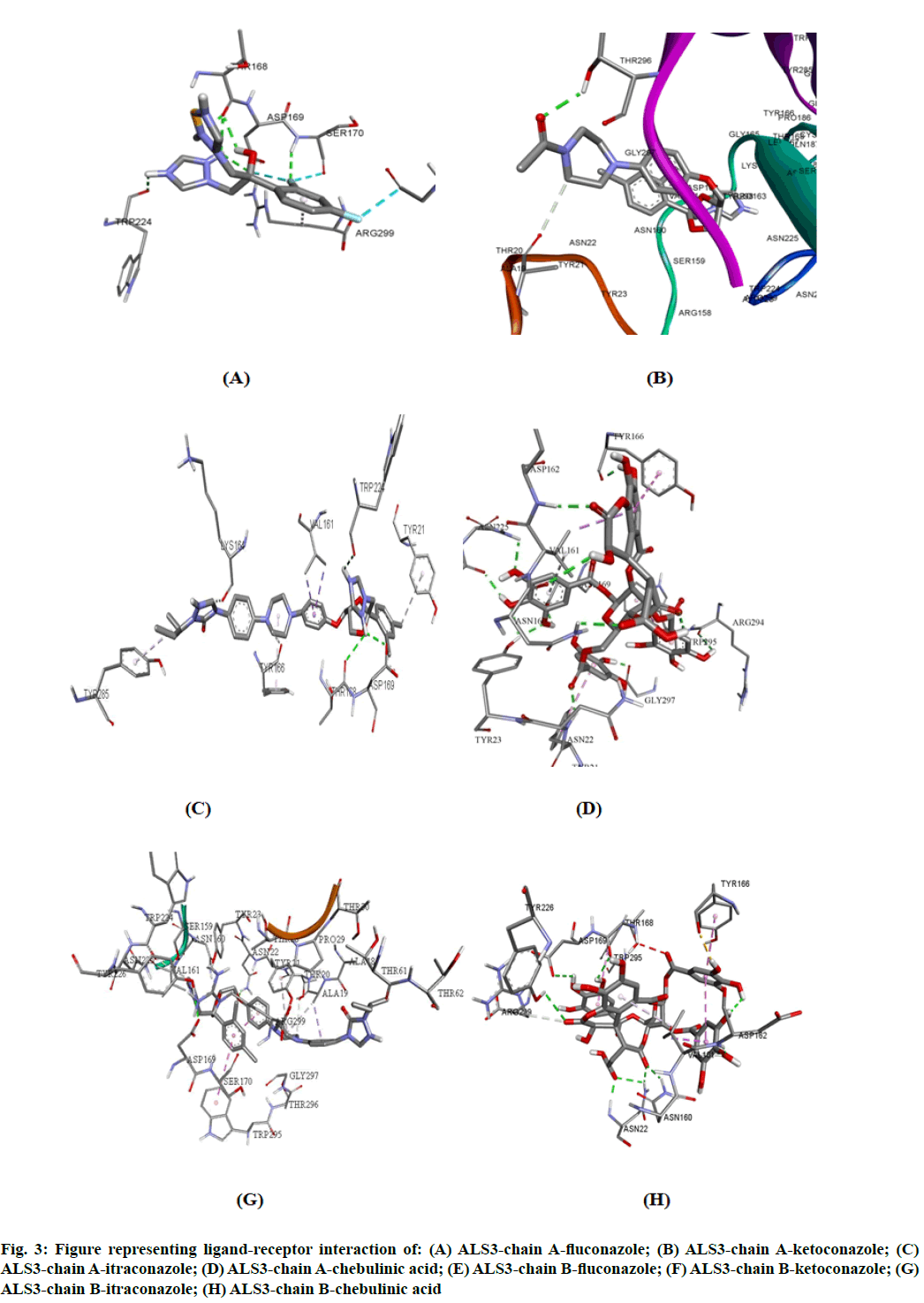
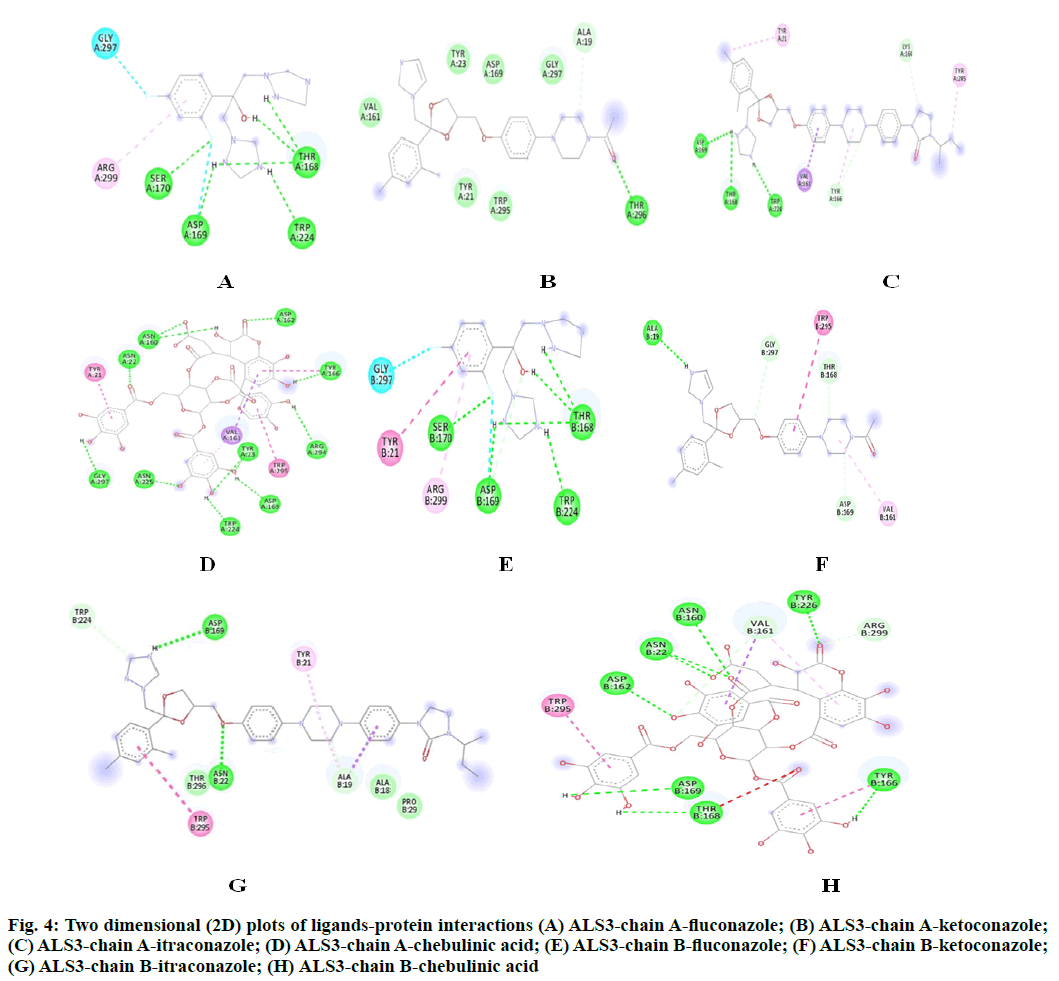
The potency of fluconazole, ketoconazole, itraconazole and chebulinic acid to target ALS3 and the particular site at which ALS3 is getting targeted was determined. To accomplish this, all the four mentioned drugs were docked with ALS3 using Autodock Vina in Pyrx platform. Chain A and B of ALS3 were docked separately with all the four ligands and their binding affinity and targeted site was determined. It was observed that fluconazole binds with ALS3 chain A at THR168, ASP169, SER170, TRP224 with -7.4 kcal/mol, ketoconazole binds at THR296 with -8.1 kcal/mol, itraconazole binds at ASP169, THR168, TRP224 with -6.9 kcal/mol whereas chebulinic acid binds at ASN22, TYR23, TRP224, ASP169, TYR166, ASP162, ASN225 GLY297, ARG294 with -10.4 kcal/ mol (fig. 2). When chain B was docked separately it was observed that fluconazole binds with chain B at SER170, ASP169 THR168, TRP224 with -7.3 kcal/ mol, ketoconazole binds at ALA19 with -7.8 kcal/mol, itraconazole binds ASN22, ASP169 at with -8.7 kcal/ mol whereas chebulinic acid binds at ASN22, ASP162, ASN160, ASP169, THR168, TYR166, TYR226 with -9.8 kcal/mol (fig. 3). From the results mentioned in Table 4, it can be further predicted that chebulinic acid showed highest potency to bind to chain A and B of ALS3 and inhibited it even more efficiently than other three chemical analogues which are currently available in market as a therapeutic measure against VVC (fig. 4).
| Receptor | Interaction | Binding energy (kcal/mol) | Binding site |
|---|---|---|---|
| ALS3-chain A | ALS3-chain A-fluconazole | -7.4 | THR168, ASP169, SER170, TRP224 |
| ALS3-chain A-ketaconazole | -8.1 | THR296 | |
| ALS3-chain A-itraconazole | -9.7 | ASP169, THR168, TRP224 | |
| ALS3-chain A-chebulinic acid | -10.4 | ASN22, TYR23, TRP224, ASP169, TYR166, ASP162, ASN225 GLY297, ARG294 | |
| ALS3-chain B | ALS3-chain B-fluconazole | -7.3 | SER170, ASP169 THR168, TRP224 |
| ALS3-chain B-ketaconazole | -7.8 | ALA19 | |
| ALS3-chain B-itraconazole | -8.7 | ASN22, ASP169 | |
| ALS3-chain B-chebulinic acid | -9.8 | ASN22, ASP162, ASN160, ASP169, THR168, TYR166, TYR226 |
Table 4: Representation of Binding Affinity of ALS3 Chain A and B Along with Fluconazole, Ketoconazole, Itraconazole and Chebulinic Acid Respectively and their Binding Sites
Fig. 3: Figure representing ligand-receptor interaction of: (A) ALS3-chain A-fluconazole; (B) ALS3-chain A-ketoconazole; (C) ALS3-chain A-itraconazole; (D) ALS3-chain A-chebulinic acid; (E) ALS3-chain B-fluconazole; (F) ALS3-chain B-ketoconazole; (G) ALS3-chain B-itraconazole; (H) ALS3-chain B-chebulinic acid
Fig. 4: Two dimensional (2D) plots of ligands-protein interactions (A) ALS3-chain A-fluconazole; (B) ALS3-chain A-ketoconazole; (C) ALS3-chain A-itraconazole; (D) ALS3-chain A-chebulinic acid; (E) ALS3-chain B-fluconazole; (F) ALS3-chain B-ketoconazole; (G) ALS3-chain B-itraconazole; (H) ALS3-chain B-chebulinic acid
The RMSF is used for analyzing local changes along with the protein chain residues and for analyzing changes in the ligand atom positions at specific temperature and pressure. The higher value of RMSF indicated that the structure has more flexible regions like turns and loop while the lower value of RMSF represented that the structure has a secondary structure like helix and sheets considered good as compared to the structure having high RMSF value. The contact map and fluctuation plot of the predicted protein model are shown in fig. 5. RMSF value ranges from 0.057 to 3.559. RMSF data revealed that the flexible regions are present within the random coils of the protein but the residues having high fluctuation did not form significant secondary structures. The Rg indicated the stability of native protein and the bound systems (protein-ligand complex) along the MD trajectories. The Rg value of our predicted model was 22.3544. All these data indicate good stability of our predicted protein. In conclusion, VVC is emerging out to be the major infectious diseases in women worldwide. Azole derivatives designed to target the disease have lost its sensitivity against its major causative agent i.e. C. albicans. It has started acquiring resistance against fluconazole, ketoconazole and itraconazole. Hence there is an urgent need to look for new therapeutic measure to combat the disease. Chebulinic acid has proved its efficacy in in vitro conditions to inhibit the proliferation and to hinder the binding of C. albicans to host organisms. But much more extensive research still needs to be carried out to bring this miraculous molecule in limelight to overcome VVC and to determine the possible mechanistic approach by which chebulinic acid may exert a potential inhibitory effect against VVC.
Acknowledgements:
Prashant Katiyar acknowledges Head, department of botany and microbiology, Gurukula Kangri (Deemed to be University) Haridwar and Indian Drugs and Pharmaceuticals Limited, Rishikesh for providing necessary facilities to perform the experiments conducted in this research. Kamal Kant Sharma acknowledges IIT, Roorkee for providing facilities to perform molecular docking studies.
Conflicts of interest:
The authors declared no conflict of interest.
References
- White DJ, Vanthuyne A. Vulvovaginal candidiasis. Sex Transm Infect 2006;82(4):28-30.
- Rathod SD, Klausner JD, Krupp K, Reingold AL, Madhivanan P. Epidemiologic features of Vulvovaginal Candidiasis among reproductive-age women in India. Infect Dis Obstet Gynecol 2012.]
- Denning DW, Kneale M, Sobel JD, Rautemaa-Richardson R. Global burden of recurrent vulvovaginal candidiasis: a systematic review. Lancet Infect Dis 2018;18(11):339-47.
- Arora B, Tayal P, Raghav H, Mittal R. A review on role of Candida albicans in causing complications of Vulvovaginal candidiasis. Bio Chem Acta 2016;1:77-86.
- Bag A, Bhattacharyya SK, Chattopadhyay RR. The development of Terminalia chebula Retz. (Combretaceae) in clinical research. Asian Pac J Trop Biomed 2013;3(3):244-52.
- Vasconcelos NG, Vaz MS, Radai JA, Kassuya CA, Formagio AS, Graciani FS, et al. Antimicrobial activity of plant extracts against carbapenem-producing Klebsiella pneumoniae and in vivo toxicological assessment. J Toxicol Environ Health A 2020;83(23-24):719-29.
- Kienzl M, Storr M, Schicho R. Cannabinoids and opioids in the treatment of inflammatory bowel diseases. Clin Transl Gastroenterol 2020;11(1):e00120.
- Afshari AR, Sadeghnia HR, Mollazadeh H. A review on potential mechanisms of Terminalia chebula in Alzheimer’s disease. Adv Pharmacol Sci 2016.
- Riyazzudin MD, Sharma M, Kaur G, Chaudhry N. A review on therapeutic potential of plant derived natural compounds. Bio Chem Acta 2016;1(1):30-4.
- Upadhyay A, Agrahari P, Singh DK. A review on the pharmacological aspects of Terminalia chebula. Int J Pharmacol 2014;10(6):289-98.
- Lu K, Chakroborty D, Sarkar C, Lu T, Xie Z, Liu Z, et al. Triphala and its active constituent chebulinic acid are natural inhibitors of vascular endothelial growth factor-a mediated angiogenesis. PLoS One 2012;7:e43934.
- Prabhat A, Navneet CA. Evaluation of antimicrobial activity of six medicinal plants against dental pathogens. Rep Opinion 2010;2(6):37-42.
- Liu Y, Filler SG. Candida albicans Als3, a multifunctional adhesin and invasin. Eukaryot Cell 2011;10(2):168-73.
- Phan QT, Myers CL, Fu Y, Sheppard DC, Yeaman MR, Welch WH, et al. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells PLoS Biol 2007;5(3):e64.
- Jorgensen JH, Ferraro MJ. Antimicrobial susceptibility testing: general principles and contemporary practices. Clin Infect Dis 1998:973-80.
- BIOVIA, D. S. Discovery Studio. San Diego: Dassault Systemes, California, USA; 2020.
- O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: An open chemical toolbox. J Cheminform 2011;3(1):1-4.
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, et al. UCSF Chimera-a visualization system for exploratory research and analysis. J Comput Chem 2004;25(13):1605-12.
- Laskowski RA, MacArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Crystallogr 1993;26(2):283-91.
- Wiederstein M, Sippl MJ. ProSA-web: interactive web service for the recognition of errors in three-dimensional structures of proteins. Nucleic Acids Res 2007;35(2):407-10.
- Lovell SC, Davis IW, Arendall III WB, De Bakker PI, Word JM, Prisant MG, et al. Structure validation by Cα geometry: ?, ψ and Cβ deviation. Protein Struct Funct Genet 2003;50(3):437-50.
- ProtParam E. ExPASy-ProtParam tool. ExPASy; 2017.
- Tian W, Chen C, Lei X, Zhao J, Liang J. CASTp 3.0: Computed atlas of surface topography of proteins. Nucleic Acids Res 2018;46(1):363-7.
- Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem 2010;31(2):455-61.
- Kuriata A, Gierut AM, Oleniecki T, Ciemny MP, Kolinski A, Kurcinski M, et al. CABS-flex 2.0: a web server for fast simulations of flexibility of protein structures. Nucleic Acids Res 2018;46(1):338-43.
- Bhattacharya M, Sharma AR, Patra P, Ghosh P, Sharma G, Patra BC, et al. Development of epitope?based peptide vaccine against novel coronavirus 2019 (SARS?COV?2): Immunoinformatics approach. J Med Virol 2020;92(6):618-31.
- Filters for globular protein evaluation: Radius of gyration. Supercomputing facility for bioinformatics and computational biology, IIT Delhi; 2016.