- *Corresponding Author:
- Y. J. Qi
Department of Chemical Engineering, Lanzhou, PR China
E-mail: qiajiao@163.com
| Date of Submission | 07 June 2016 |
| Date of Revision | 22 August 2016 |
| Date of Acceptance | 07 June 2016 |
| Indian J Pharm Sci 2016;78(4):512-524 |
This is an open access article distributed under terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows other the remix, tweak, and build up to the non-commercially, as long as the author is credited and the new creations are licensed under the identical terms.
Abstract
β-glucosidase is one of the critical enzymes to the type 2 diabetes, which belongs to a large family of glycoside hydrolases. In this article, we attempted to explore the binding modes between two drugs and β-glucosidases by comparing their bonding modes with β-glucosidases from different species. Results showed that the binding orientations and geometrical conformation of synthetic drug (miglitol) and natural product (quercetin) were all different in all active sites, which may be related to the flexibility of the molecules. Compared with the conformations obtained by density functional theory calculations at the B3LYP/6-31G* level, the docking conformations indicated that the -CH2CH2OH group of miglitol and the B ring of quercetin were the most critical groups to the stabilities in the active sites. Finally, protein sequence alignment was performed under default parameters. Although the sequence similarity is not very high between these β-glucosidases, the residues related to the active sites were conservative; especially the one involved in H-bond interactions between three species, namely soil metagenome, Micrococcus antarcticus and termite Neotermes koshunensis.
Keywords
β-glucosidase, binding mode, docking conformation, sequence alignment D-optimal design
Diabetes is rapidly becoming an enormous health burden by decreasing quality of life all over the world. Type 2 diabetes mellitus is a metabolic disease associated with micro- and macrovascular complications such as diabetic retinopathy, diabetic neuropathy and cardiovascular diseases. One of therapeutic approaches for managing increased blood glucose is to weaken the catalytic activity of key enzymes involved in hydrolytic cleavage of dietary oligosaccharides[1], and thus the blood glucose in the diabetics and a series of possible side effects will decrease. β-glucosidase is one of the key enzymes, which belongs to a large family of glycoside hydrolases. Many of β-glucosidases can also catalyze the synthesis of poly- or oligosaccharides, which have dual activities of hydrolysis and transglycosylation [2,3].
By delaying the absorption of digested carbohydrates from the small intestine, β-glucosidase inhibitors can also attenuate postprandial blood glucose fluctuations [4-6]. There are great number of studies about the inhibiting capacity of the glucosidase[7-10]. For example, inhibitor miglitol acted as therapeutic agent with high clinical efficacy for diabetes in combination with other drugs and/or dietary changes [11,12]. Although this synthetic drug has significant role in diabetes, it subsequently leads to various gastrointestinal side effects, such as flatulence, diarrhea and abdominal discomfort [13].
There are many doubts about whether nontoxic natural flavones have the similar inhibitory features as miglitol, which is very important to the treatment of diabetes. Recent research indicated that some natural products are associated with decreasing the blood glucose of the diabetics, such as multihydroxyl flavonoid compound, quercetin [14]. Studies demonstrated that quercetin has good anti-type 2 diabetes ability based on rat experiment and yeast α-glucosidase in vitro [15-19]. The detailed interaction between quercetin and α-glucosidase has also been reported [20]. However, the detailed inhibitory mechanisms between different quercetin and β-glucosidases are rare so far [21]. Molecular docking technique is a key tool in structural molecular biology and computer-assisted drug design [22-24]. In the present work, a comparative study of the interaction between β-glucosidase and synthetic drug (miglitol) or natural product (quercetin) was undertaken by molecular docking simulation methods. For the purpose of comparison, four enzymes from different species were also chosen, which hold the focus of the present study and belong to the β-glycosidase family. The results obtained from this analysis not only revealed the interaction mechanism between β-glycosidase and inhibitors, but also provided some valuable information for drug design, pharmaceutical research and sequence analysis based on comparative studies.
Materials and Methods
Ligand preparation
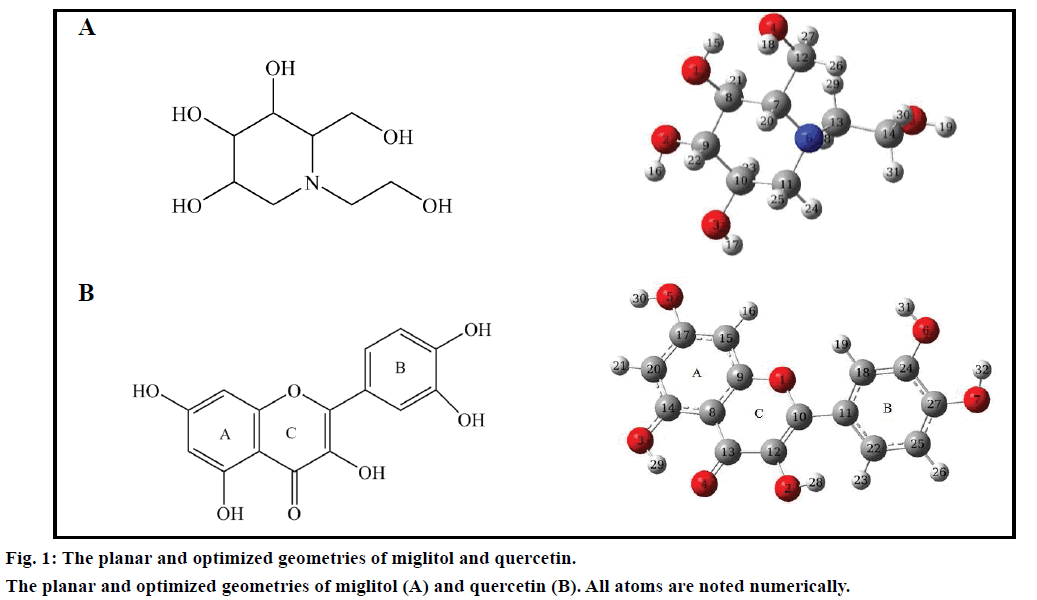
Three dimensional coordinates of all small molecules were obtained from the PubChem database (CIDs for miglitol and quercetin are 441314 and 5280343, respectively) and their geometries were further optimized to obtain the most stable conformation by using density functional theory (DFT) calculations at the B3LYP/6-31G* level [25,26], which can be executed via Gaussian 09 program package [27]. The stable conformation was used for further docking research.
Protein preparation
Crystal structure files of cocrystallized β-glycosidases used in this study were derived from the Protein Data Bank (PDB). The β-glycosidase from soil metagenome was noted as Sm-β-glu, β-glucosidase from Micrococcus antarcticus was noted as Ma-β-glu, β-glucosidase from termite Neotermes koshunensis was noted as Tnk-β-glu, and β-glucosidase from Homo sapiens was noted as Hu-β-glu. All crystallographic water molecules were removed, polar hydrogens were added, Gasteiger charges were assigned to the proteins by AutoDockTools (ADT) 1.5.6 [28] and were then converted into AutoDock readable file format.
Molecular docking
Docking and the corresponding results were carried out by using ADT 1.5.6 package and Pymol Molecular Visualization package [29,30]. The ligands were automatically prepared by detecting the flexible torsions and assigning gasteiger charges under default settings. Lamarckian genetic algorithm (LGA) was used for docking by using the following settings: maximum number of 25 000 000 energy evaluation, number of generation=27 000, mutation rate=0.02, crossover rate=0.80, elitism value of 1 and maximum number of iterations=300. The grid size was 60×60×60 Å with a spacing of 0.375 Å and was centered on the binding site showed in crystal structure of β-glucosidase cocrystallized structure.
The binding sites in these proteins were found to be the putative binding sites for the ligands based on the reference in the complex substance from the PDB database. Exact dimensions of the grid centre of Sm-β-glu, Ma-β-glu, Tnk-β-glu and Hu-β-glu were (x=34.253; y=1.006; z=-3.937), (x=17.849; y=20.669; z=24.342), (x=-26.809; y=82.413; z=13.836) and (x=-29.711; y=10.91; z=1.156), respectively. After 50 independent docking runs, the docking results were clustered with root mean square deviation tolerance of 2.0 Å. The top output poses were ranked by their calculated binding affinities.
Protein sequence analysis
The sequences of proteins were downloaded from the National Center for Biotechnology Information (NCBI) website. Multiple sequence alignment is performed by Clustal software. The composition analysis was carried out by Lasergene software, and the sequence similarity analysis was performed by MegAlign software with Lipman-Pearson method.
Results
Molecular structure and protein information
By inhibiting the ability of the patient to break down complex carbohydrates into glucose, miglitol, is an oral antidiabetic drug [31-33]. The molecular structures of small molecules used in this study were given in fig. 1. The most stable conformation was used for further docking research after DFT calculations. β-glucosidase plays an important role in degradation of cello-oligosaccharides, which influence the liberation of glucose from cellulose [34]. Basic information of four β-glucosidases was listed in Table 1. The X-ray crystallographic structures of the β-glucosidases from soil metagenome (PDB ID: 3CMJ) at a resolution of 1.60 Å, Micrococcus antarcticus (PDB ID: 3W53) at a resolution of 2.20 Å, termite Neotermes koshunensis (PDB ID: 3VIL) at a resolution of 1.15 Å and Homo sapiens (PDB ID: 3VKK) at a resolution of 2.00 Å were obtained from the RCSB Protein Data Bank. All of the protein lengths are about 500 aa. The composition ratio of Sm-β-glu, Ma-β-glu, Tnk-β-glu and Hu-β-glu are relatively similar to each other. It accounts for a large proportion for hydrophobic amino acid and polar one, sum of which own 58.06, 57.11, 58.52 and 62.26% for total number of Sm-β-glu, Ma-β-glu, Tnk-β-glu and Hu-β-glu residues, respectively. Sequence similarity analysis indicated that Hu-β-glu has similarities of 36.10, 30.70 and 45.70% with Sm-β-glu, Ma-β-glu and Tnk-β-glu, respectively.
| Name | PDB ID | Length (aa) |
Amino acids composition | Organism | |||
|---|---|---|---|---|---|---|---|
| S-Basic | S-Acidic | Hydrophobic | Polar | ||||
| Sm-β-glu | 3CMJ | 465 | 40 | 54 | 158 | 112 | Soil metagenome |
| Ma-β-glu | 3W53 | 506 | 41 | 67 | 176 | 113 | Micrococcus antarcticus |
| Tnk-β-glu | 3VIL | 487 | 41 | 72 | 164 | 121 | Neotermeskoshunensis |
| Hu-β-glu | 3VKK | 469 | 46 | 57 | 167 | 125 | Homo sapiens |
Table 1: Structural Information Of β-Glucosidase For Sm-β-Glu, Ma-β- Glu, Tnk-β-Glu And Hu-β-Glu
Analysis of the binding sites of miglitol and β-glucosidases
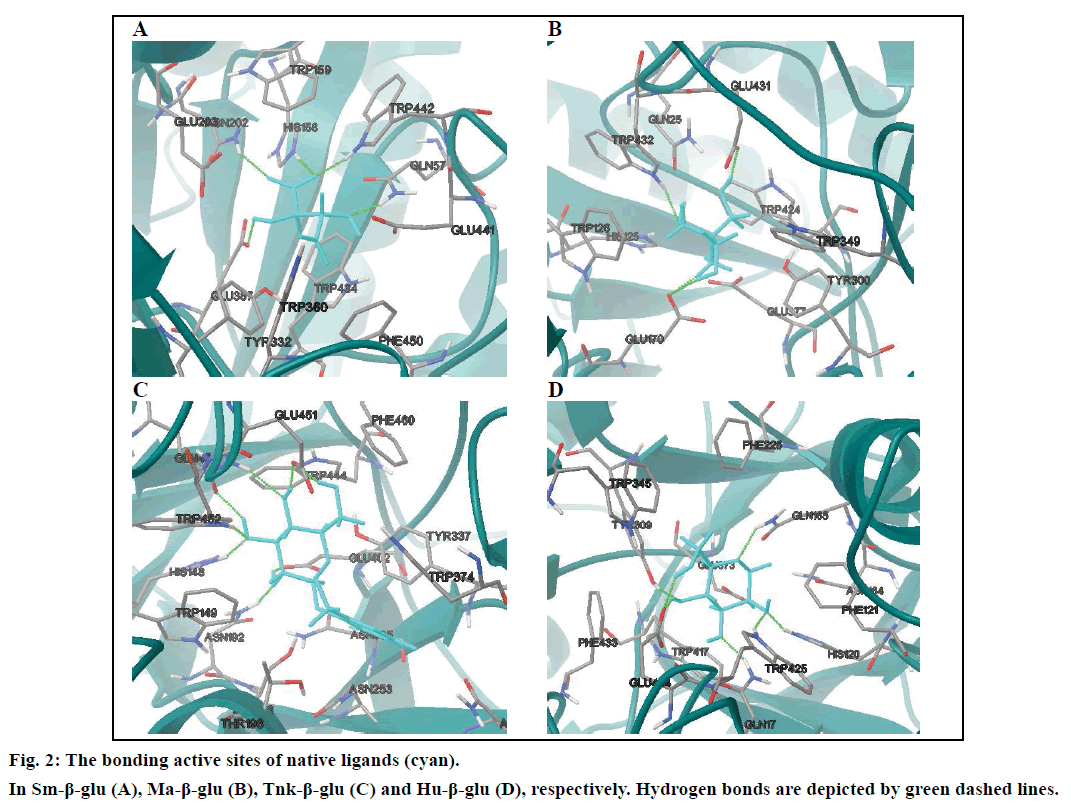
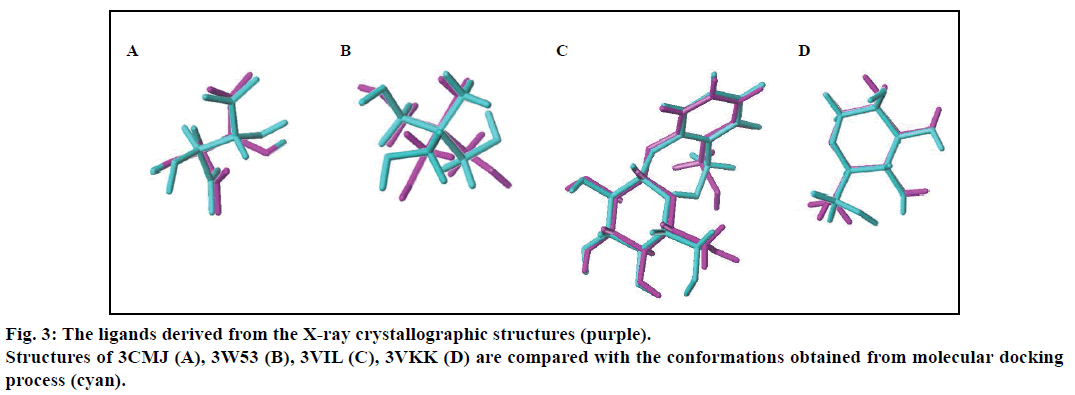
In order to prove the reliability of the docking process, the native ligands derived from the X-ray crystallographic structures were redocked into the active sites of the four β-glucosodases using the same parameters described in this text (fig. 2). Results indicated that the active site of the Sm-β-glu (PDB ID: 3CMJ) was composed of amino acids Gln57, His158, Trp159, Asn202, Glu203, Tyr332, Glu337, Trp360, Glu441, Trp434 and Phe450, which were consistent with the published data [35]. Similar to the information reported in PDB, amino acids surrounded the active site of the Ma-β-glu (PDB ID: 3W53) were Gln25, His125, Trp128, Glu170, Cys173, Tyr300, Trp349, Glu377, Glu431, Trp432 and Phe440. From the fig. 2, it can also be seen that amino acids located in the active site of the Tnk-β-glu (PDB ID: 3VIL) were Gln45, His148, Trp149, Thr196, Asn253, Asn335, Tyr337, Trp374, Glu402, Trp444, Glu451, Trp452, Phe460 and Phe480. These were extremely identical with the reported data [36]. Similarly, amino acids Gln17, His120, Phe121, Asn164, Gln165, Phe225, Tyr309, Trp345, Glu373, Glu424, Trp425 and Phe433 were identical with data on the Protein Data Bank about the Hu-β-glu (PDB ID: 3VKK). The values of the binding free energies of the native ligands were –6.63,–8.18,–8.91 and –7.13 kcal/mol for Sm-β-glu, Ma-β-glu, Tnk-β-glu and Hu-β-glu, respectively. It was noticed that the docked structures derived from the molecular docking process in the text were very consistent with the original ligands located in the crystallographic complexes structures (fig. 3). Their similarities concluded by spin alignment method were 0.889, 0.976, 0.916 and 0.898 for Sm- β-glu, Ma-β-glu, Tnk-β-glu and Hu-β-glu, respectively (This value is between 0~1, the larger the value, the higher the similarity of the molecules). All of the root- mean-square deviation (RMSD) values were less than 2.0 Å, which suggested good models [37].
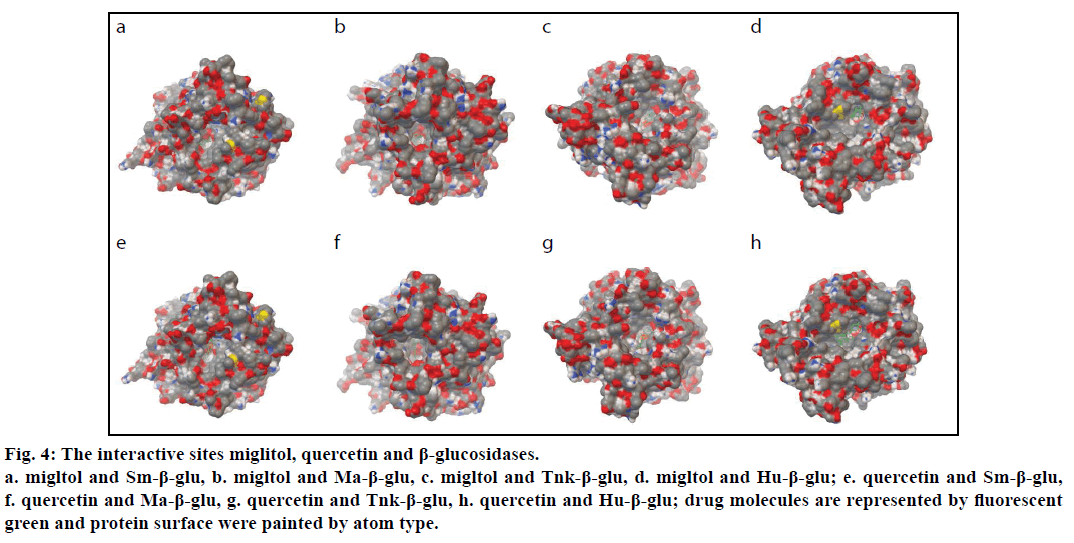
To explore the geometrical shape complementation, the poses of the miglitol in the binding pockets of the four β-glucosidases were shown (fig. 4). The values of the binding free energies of the miglitol were –7.53,–6.44,–9.09 and –4.43 kcal/mol for Sm-β-glu, Ma-β-glu, Tnk-β-glu and Hu-β- glu, respectively. In the three complexes, all small molecule poses were stuck onto the walls of the pockets and embedded into the active sites, which comprised by random coil structure, showing very good geometric complementation. In order to further reveal the key residues involved in the binding sites, the detailed interactions between the molecules and β-glucosidases were analyzed.
Figure 4:The interactive sites miglitol, quercetin and β-glucosidases.
a. migltol and Sm-β-glu, b. migltol and Ma-β-glu, c. migltol and Tnk-β-glu, d. migltol and Hu-β-glu; e. quercetin and Sm-β-glu,
f. quercetin and Ma-β-glu, g. quercetin and Tnk-β-glu, h. quercetin and Hu-β-glu; drug molecules are represented by fluorescent
green and protein surface were painted by atom type.
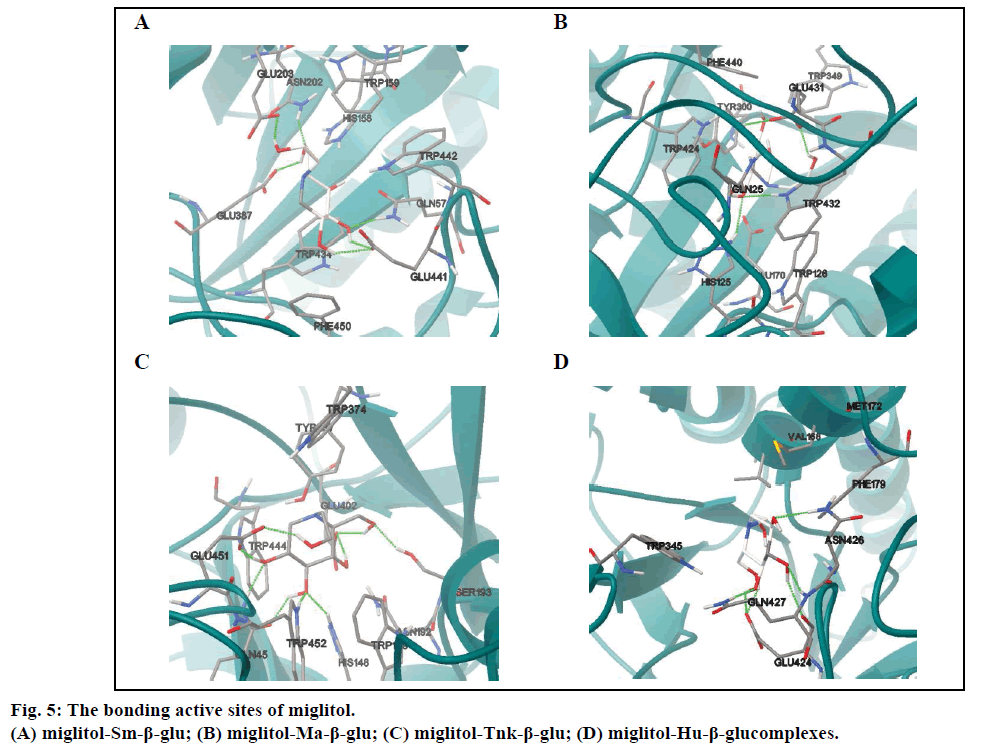
Molecular docking results indicated that miglitol was located in a cavity formed by random coil structure of Sm-β-glu. The active site was surrounded by Gln57, His158, Trp159, Asn202, Glu203, Glu387, Glu441, Trp442 and Phe450 (fig. 5A). It was shown that there existed non-bonded interactions between the small molecule and the protein, including H-bonds bonding, van der Waals and electrostatic forces. There are eleven residues around the active site of Ma-β-glu, namely Gln25, Trp126, Glu170, His125, Cys173, Tyr300, Trp349, Trp432, Glu431, Phe440 and Trp424. The polar hydrogen atoms H19, H18, H16 and H17 could interact with nearby OE1 of acidic amino acids Glu by H-bonds except H15 (Table 2). The O4 and O1 also form two H-bonds with Asn202 (1.621 Å) and Gln57 (1.836 Å), respectively. The docking result (fig. 5B) indicated that exocyclic N-C bond rotates clockwise about 100° from the optimized stable conformation, which could lead to variation of the spatial positions for other polar hydrogens. Compared with fig. 5A,obviously, it was shown that H15 can interact with acid Glu431 (2.214 Å) through H-bond (Table 3).
| Ligand name | No. of H-bonds | Ligand atom | Residue name | Residue atom | Length/Å |
|---|---|---|---|---|---|
| Miglitol | 1 | H19 | Glu203 | OE1 | 1.777 |
| 2 | O4 | Asn202 | HD21 | 1.621 | |
| 3 | H18 | Glu387 | OE1 | 2.061 | |
| 4 | O1 | Gln57 | HE22 | 1.836 | |
| 5 | H16 | Glu441 | OE1 | 1.71 | |
| 6 | H17 | Glu441 | OE1 | 2.183 | |
| Quercetin | 1 | O6 | Trp442 | HE1 | 1.991 |
| 2 | O7 | Gln57 | HE22 | 1.848 | |
| 3 | H32 | Glu441 | OE1 | 2.081 | |
| 4 | O3 | Asn262 | HD21 | 2.095 | |
| 5 | O5 | Phe261 | HN | 2.124 | |
| 6 | H30 | Phe261 | O | 1.781 |
Table 2: H-Bonds Between Two Drugs And β-Glucosidase From Soil Metagenome
| Ligand name | No. of H-bonds | Ligand atom | Residue name | Residue atom | Length/Å |
|---|---|---|---|---|---|
| Miglitol | 1 | H19 | Glu431 | OE2 | 2.035 |
| 2 | O4 | Trp432 | HE1 | 2.005 | |
| 3 | O4 | His125 | HE2 | 2.245 | |
| 4 | H18 | Gln25 | OE1 | 2.024 | |
| 5 | H15 | Glu431 | OE1 | 2.214 | |
| Quercetin | 1 | O6 | Tyr300 | HN | 2.201 |
| 2 | H31 | His301 | O | 1.999 | |
| 3 | H32 | His301 | O | 1.961 | |
| 4 | H30 | Glu377 | OE1 | 2.011 |
Table 3: H-Bonds Between Two Drugs And β-Glucosidase From Micrococcus Antarcticus
Results showed that the binding pocket of Tnk-β-glu was mainly formed by Gln45, His148, Trp149, Asn192, Ser193, Tyr337, Trp374, Glu402, Trp432, Trp444 and Glu451 (fig. 5C). Miglitol was stabilized by the Tnk- β-glu residues via hydrogen bonds, van der Waals and electrostatic forces. And the most number of H-bonds were observed in the four β-glu-miglitol complexes, involving residues Glu451 Ser193 Glu402 Gln45 Tyr452 and His148 (Table 4). It can be seen from fig. 5C, that the location of -CH2CHs2OH group was greatly different from the one in fig. 5B, and other polar hydrogen atoms all changed their position accordingly. Polar hydrogen atoms were prone to interact with acid Glu by hydrogen bonds except the H16 atom.
| Ligand name | No. of H-bonds | Ligand atom | Residue name | Residue atom | Length/Å | |
|---|---|---|---|---|---|---|
| Miglitol | 1 | H19 | Glu451 | OE2 | 1.922 | |
| 2 | O4 | Ser193 | HG | 1.904 | ||
| 3 | H18 | Glu402 | OE1 | 2.105 | ||
| 4 | H15 | Glu402 | OE2 | 1.871 | ||
| 5 | H16 | Gln45 | OE1 | 1.893 | ||
| 6 | O2 | Tyr452 | HE1 | 1.735 | ||
| 7 | O2 | His148 | HE2 | 2.1 | ||
| 8 | H17 | Glu451 | OE1 | 1.733 | ||
| 9 | O3 | Gln45 | HE21 | 1.918 | ||
| Quercetin | 1 | H31 | Asn255 | OD1 | 2.151 | |
| 2 | O6 | Asn255 | HD22 | 1.995 | ||
| 3 | H32 | Asn255 | OD1 | 1.924 | ||
| 4 | H28 | Thr196 | OG1 | 2.208 | ||
| 5 | O2 | Ser193 | HG | 2.186 | ||
| 6 | H29 | Glu402 | OE1 | 2.239 | ||
| 7 | O3 | His148 | HE2 | 1.996 | ||
| 8 | H30 | Glu451 | OE1 | 1.798 | ||
Table 4: H-Bonds Between Two Drugs And The Tnk-β-Glu
The number of hydrogen bonds formed between the Hu-β-glu and miglitol molecule were the least (Table 5). Among the residues involved in the active sites, there were residues Val168 and Met172 in a small α helix, which played no significant role in the stability for both antidiabetic drugs (figs. 5D and 6D). Surrounding the binding pocket of Hu-β-glu, five residues, namely Trp345, Phe179, Glu424, Asn426 and Gln427 (fig. 5D). Miglitol interacted with adjacent residues through six H-bonds, namely Glu424, Asn426 and Gln427 (Table 5).
| Ligand name | No. of H-bonds | Ligand atom | Residue name | Residue atom | Length/Å |
|---|---|---|---|---|---|
| Migltol | 1 | H18 | Glu424 | OE2 | 1.712 |
| 2 | H15 | Glu424 | OE2 | 1.74 | |
| 3 | O1 | Gln427 | HE21 | 1.911 | |
| 4 | H16 | Glu424 | O | 2.225 | |
| 5 | O2 | Gln427 | HN | 1.984 | |
| 6 | O3 | Asn426 | HD21 | 2.11 | |
| Quercetin | 1 | H31 | Glu424 | O | 2.179 |
| 2 | O6 | Gln427 | HN | 2.057 | |
| 3 | H32 | Glu424 | OE2 | 1.77 | |
| 4 | O7 | Gln427 | HE21 | 1.689 |
Table 5: H-Bonds Between Two Drugs And The Hu-β-Glu
Analysis of quercetin and β-glucosidase binding sites
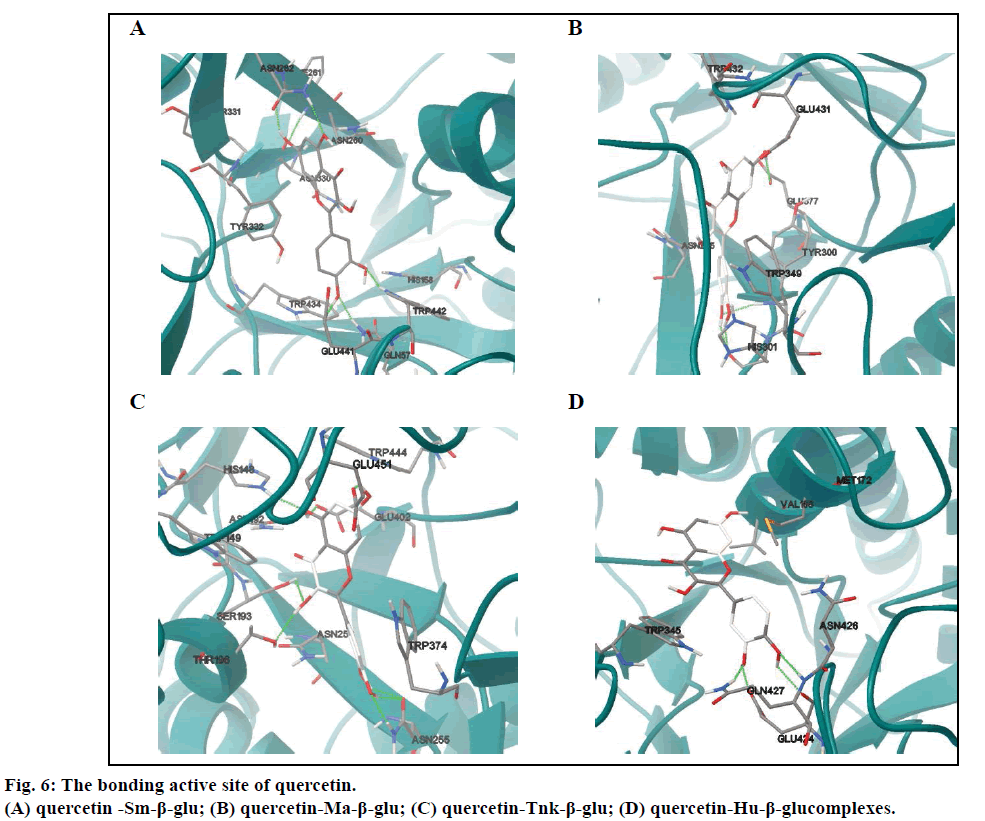
In the four complexes (figs. 4E-H), all small molecules were also embedded into the active sites, displaying a very good geometric complementation. Compared with miglitol, the quercetin showed lower binding free energies for Sm-β-glu, Ma-β-glu, Tnk-β-glu and Hu- β-glu, which were –9.0, –8.62, –9.38 and –6.55 kcal/ mol, respectively, which suggested stronger inhibitory ability. This was consistent with the reported results [38]. As for Sm-β-glu, the quercetin was fixed in the binding pocket composed of residues Gln57, His158, Asn260, Phe261, Asn262, Asn330, Tyr331, Tyr332, Trp434, Glu441 and Trp442. There were mainly hydrogen bonds and hydrophobic interactions (fig. 4A) between the complexes. The residues involved in H-bonds were mainly polar amino acids, such as Trp442, Gln57 and Asn262 (Table 2).
It was shown that residues in the binding site of Ma-β-glu-quercetin complexes was different from the one between Ma-β-glu and miglitol, whereas the ligands were all located in a cavity surrounded by random coil structure composed of Asn225, Tyr300, His301, Trp349, Glu377, Glu431 and Trp432. It could be easily observed that the most obvious variation of quercetin was the variation of the B ring compared with fig. 4A. This resulted in its H31 and H32 forming moderate H-bonds with the O atom of basic His301, which had similar length of 1.999Å and 1.961Å, respectively (Table 3). The docking results of Tnk-β-glu complexes showed that quercetin and miglitol had similar binding pockets, which composed of residues Asn192, Ser193, His148, Trp149, Thr196, Asn253, Asn255, Trp374, Glu402, Trp444 and Glu451. Most of the residues involved in H-bonds interactions were identical, such as Glu451, Glu402, Ser193 and His148. Residue Asn255 played a considerably important role in the stability of Tnk-β-glu-quercetin complex, of which the OD1 and HD22 can form three H-bonds with H31, H32 and O7 of quercetin (Table 4). Quercetin remained the greatest number of H-bonds interactions with the Tnk-β-glu among all of the β-glucosidases, which might lead to the lowest binding affinity correspondingly.
However, there was the least number of H-bonds interactions when quercetin docked into the binding pocket of the Hu-β-glu. Residues composed by the H-bonds interactions were identical to miglitol, namely Glu424 and Gln427 (Table 5). Besides, there were Asn426, Val168, Met172 and Trp345 also involved in the binding site. In summary, there were two antidiabetic drugs miglitol and quercetin can form H-bonds with these β-glucosidases via some common residues, such as Glu441 and Gln57 for Sm-β-glu, Glu402, Ser193, His148 and Glu451 for Tnk-β-glu, Glu424 and Gln427 for Hu-β-glu. It can be speculated that these amino acids are extraordinarily critical to the binding of the two ligands with glucosidase enzyme, suggesting that quercetin is a good hypoglycemic agents with low IC50 value [39-42].
Geometrical conformation analysis of docking molecules
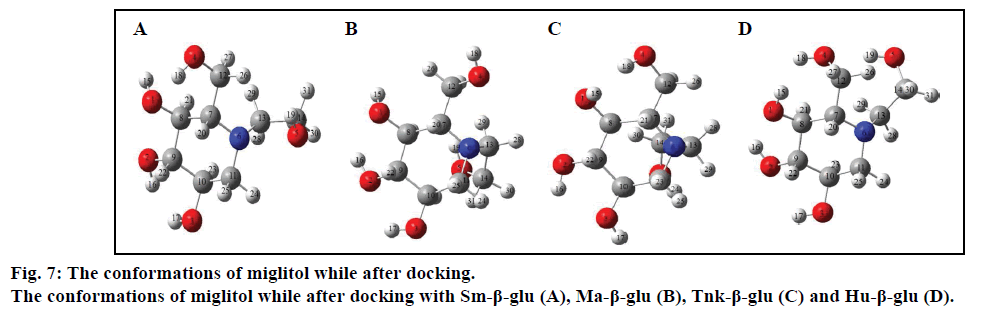
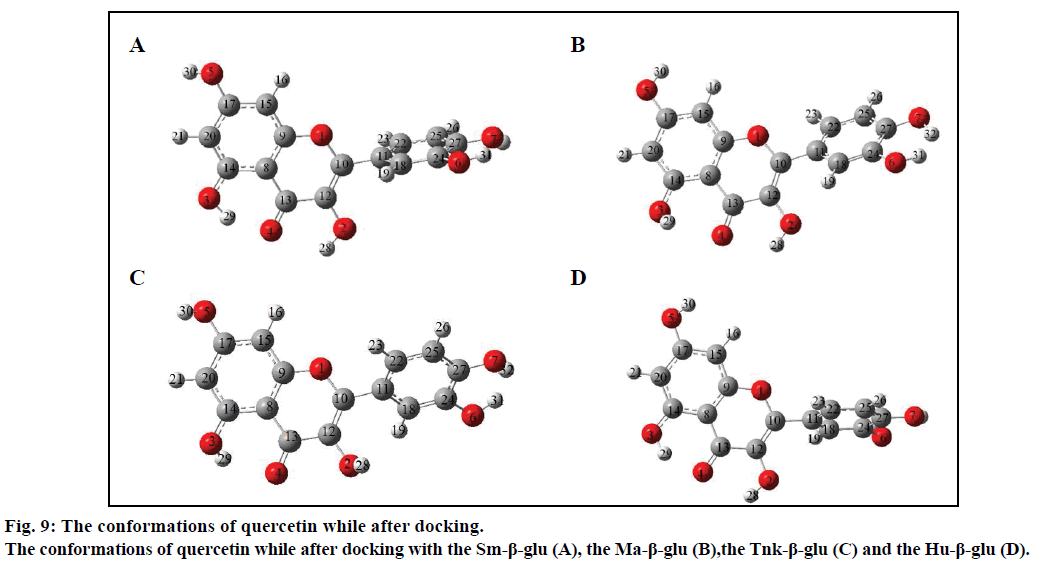
In order to further analyze the inhibitory characteristics of miglitol, the conformations were listed before or after interacting with the β-glucosidases from different sources (fig. 7). Compared with the confirmation before docking, it could be seen from the figure that the most variation occurred was the -CH2CH2OH group, which connected with N-atom via covalent interaction. Especially the one after docking with Tnk- β-glu, -CH2CH2OH turned backward to the hexatomic ring, which made H1 interacting with residue Glu451 by H-bond. Actually, experimental results showed that their inhibition would be greatly increased when hydroxyl groups connected behind the miglitol ring [43]. Moreover, multiple hydroxyl groups that connected with the N atom could not be ignored [44,45].
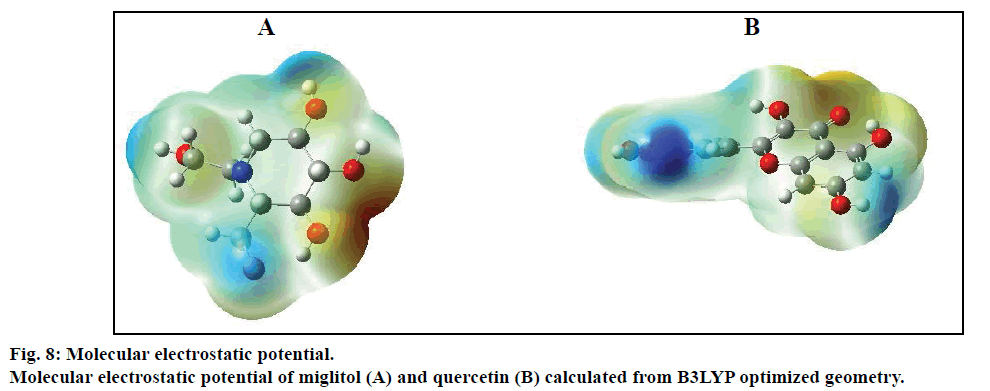
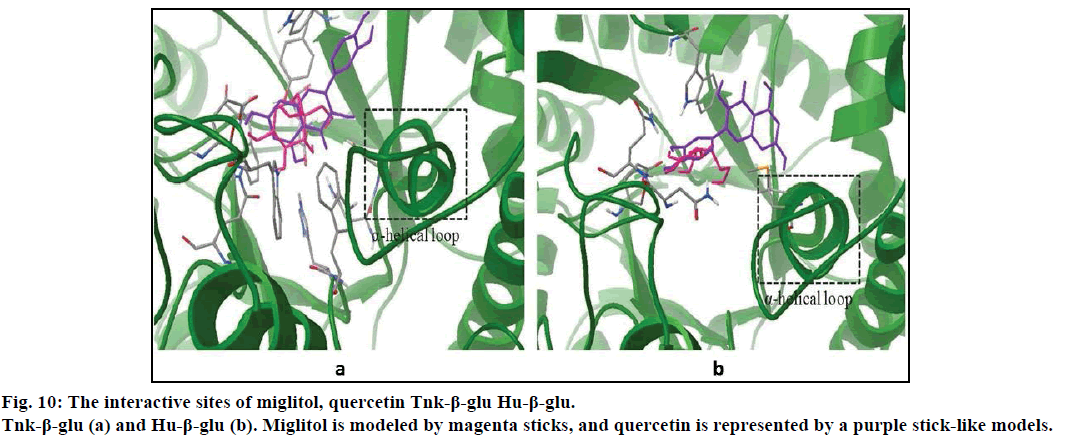
Compared with previous docking conformation, it was shown that the –O4H18 hydroxyl group changed its geometrical conformation evidently, which might be mainly influenced by the -CH2CH2OH group. Compared to previous docking 61.1°, the dihedral angles of C7— C12—O4—H18 were –81.7°, –1.5°, 10.2° and 43.2° while after docking with the Sm-β-glu, the Ma-β-glu, the Tnk-β-glu and the Hu-β-glu, respectively. This may lead H18 to interact with acid residue Glu by H-bonds easily in all the four β-glucosidases-miglitol complexes. From the molecular electrostatic potential, which was calculated from B3LYP optimized geometry, it could be seen that the negative (red and yellow) regions were related to electrophilic reactivity and the positive (blue) regions to nucleophilic reactivity (fig. 8A). The negative charge covers the H18 atom, which is the most reactive part in the miglitol molecule. In addition, there was a pair of H-bond between O4 and HG of Ser193 while –O4H18 group turned anticlockwise about 6.8~198.4° compared with previous docking conformations (fig. 9). Residue Ser193 (Thr196) help the miglitol stabilizing its conformation by H-bonds interactions, which was in the front (inner) of an alpha helix formed by additional nine residues from Pro194 to Ala202 (fig. 10A).
Other hydroxyls also changed their initial conformations into their better orientations, and thus were convenient for forming H-bonds with adjacent residues, such as H4 and H5. For example, H4 changed its position nearly 90°, which lead to three H-bonds interactions between H4, O4 and Gln45, Tyr452 and His148 (Table 4). Actually, multiple hydroxyl groups played a critical role in the stabilization of the miglitol at the catalytic sites of β-glucosidases [46-49].
The geometrical conformations of quercetin were also listed after interacting with the β-glucosidases from different sources (fig. 9). It can be seen that the position of B ring changed greatly, which might have some influence on variation of other hydroxyl groups. Compared with the conformation before docking, the B ring of quercetin shifted clockwise by (a)→(b)→(c)→(d)→(e) order sequentially (fig. 9). However, this did not affect the H-bond numbers between surrounding amino acids and H1, O1, H2, O2 atoms, which were just located in the positive (blue) regions and acted as acceptors while forming H-bonds with other residues (fig. 8B). This was in accordance with the experiments, from which it can be speculated that most critical and significant role in the inhibition is contributed by the B ring of quercetin derivatives [50].
It can be observed, from figs. 6B-D that O4 can hardly form H-bonds with other residues except the Asn262 of the Sm-β-glu. Compared with fig. 9A, -OH3 group could remain the best geometrical conformation when rotating clockwise of about 30~45° centered by the C atom that connected with -OH3. For example, H3 and O3 interacted with Thr196 and Ser193 by forming H-bonds via OG1 and HG, respectively. When -OH3 group was toward ahead the B ring, the steric hindrance caused by B ring would greatly decrease. This effect was more significant while H3 was substituted by larger groups [51,52]. It was noteworthy that the B ring plays a very important role in the stability of β-glucosidases-quercetin complex. This was consistent with the α-glucosidases that the inhibition power greatly apparently increased with the replacement of hydroxyl groups of the flavones B-ring [53]. Hydrophobic interactions existed between A ring and residues Val168, which was located in a short α helix loop comprised by eleven amino acids residues from Gln165 to Asp175 (fig. 10).
Sequence alignment analysis
Finally, β-glucosidase protein sequence alignment was employed under the default settings. The results (fig. 11) revealed that although the similarities between these sequences are not very high, most of the amino acids residues related to the non-bonded interactions are conserved in the four β-glucosidases. The other three species are more conservative except the Hu-β- glu, especially the amino acids involved in forming H-bonds (indicated by blue marked letters). From the residues of the active sites, it was shown that 64.3% of the amino acids between Thk-β-glu and Sm-β-glu (Ma- β-glu) are identical, although the sequence similarities between them were only 34.9% and 33.3%, respectively. Although the sequence similarity between Thk-β-glu and Hu-β-glu was the highest (45.7%), residues that related the active sites had just only 14.3% identity.
Discussion
It is critical to study the glucosidase inhibitors for further recognizing the type II diabetes. However, there have been a lot of reports about the inhibitors of α-glucosidase [54,55], the interactions between β-glucosidase and inhibitors were reported scarcely until now. In this article, molecular docking was employed to evaluate the binding modes between two drugs (synthetic and extraction from natural product) and four β-glucosidases from different species. Results showed that the binding orientations and geometrical conformation of miglitol (quercetin) were all different in all complexes, which may be related to the differences in binding affinities.
Compared with the conformations obtained at the B3LYP/6-31G* level, two molecules show different interactions in the active sites of four β-glucosidases. Compared with the optimized structures by DFT calculations, all of the conformations more or less altered after interacting with these β-glucosidases. From the two molecular electrostatic potentials, it can be seen that the negative (red and yellow) regions are related to electrophilic reactivity and the positive (blue) regions to nucleophilic reactivity. This is consistent with the molecular docking results. From the molecular structural point of view, actually, multiple hydroxyl groups are very important to the stabilization of the miglitol at the catalytic sites of β-glucosidases. In addition, hydroxyl groups of the quercetin B-ring are vital for its inhibition power. Finally, protein sequence alignment was performed under default parameters. Although the sequence similarity is not very high between these β-glucosidases, most of the amino acid residues related to the active sites were conservative, especially the one involved in H-bond interactions.
Based on bioinformatics and molecular docking technology, the interactions between β-glucosidases and antidiabetic drugs are not only significant to further understand of the β-glucosidases, but also directive to design new inhibitors.
Financial support and sponsorship
This work was supported by the State Ethnic Affairs Commission Research Projects (14XBZ021), the Fundamental Research Funds for the Central Universities (zyz2011064) and scientific research project of Gansu Province (2014B-006).
Conflicts of interest
There are no conflicts of interest.
References
- Ceriello A. Postprandial hyperglycemia and diabetes complications: is it time to treat? Diabetes 2005;54:1-7.
- Bohlin C, Praestgaard E, Baumann MJ, Borch K, Praestgaard J, Monrad RN, et al. A comparative study of hydrolysis and transglycosylation activities of fungal β-glucosidases. ApplMicrobiolBiotechnol 2013;97:159-69.
- Ketudat Cairns JR, Esen A. β-Glucosidases. Cell Mol Life Sci 2010;67:3389-405.
- Scheen A. Is there a role for α-glucosidase inhibitors in the prevention of type 2 diabetes mellitus? Drugs 2003;63:933-51.
- Bischoff H. Pharmacology of alpha-glucosidase inhibition. Eur J Clin Invest 1994;24:3-10.
- Scott LJ, Spencer CM. Miglitol: a review of its therapeutic potential in type 2 diabetes mellitus. Drugs 2000;59:521-49.
- Wubshet SG, Moresco HH, Tahtah Y, Brighente IM, Staerk D. High-resolution bioactivity profiling combined with HPLC-HRMS-SPE-NMR: α-Glucosidase inhibitors and acetylated ellagic acid rhamnosides from Myrciapalustris DC. (Myrtaceae). Phytochemistry 2015;116:246-52.
- Trapero A, Egido-Gabás M, Bujons J, Llebaria A. Synthesis and evaluation of hydroxyl methylaminocyclitols as glycosidase inhibitors. J Org Chem 2015;80:3512-29.
- Li KY, Jiang J, Witte MD, Kallemeijn WW, Donker-Koopman WE, Boot RG, et al. Exploring functional cyclophellitol analogues as human retaining beta-glucosidase inhibitors. Org BiomolChem 2014;12:7786-91.
- Panwar H, Calderwood D, Grant IR, Grover S, Green BD. Lactobacillusstrains isolated from infant faecespossess potent inhibitory activity against intestinal alpha- and beta-glucosidases suggesting antidiabetic potential. Eur J Nutr 2014;53:1465-74.
- Mooradian AD, Thurman JE. Drug therapy of postprandial hyperglycaemia. Drugs 1999;57:19-29.
- Keating GM. Alogliptin: a review of its use in patients with type 2 diabetes mellitus. Drugs 2015;75:777-96.
- Lebovitz HE. Alpha-glucosidase inhibitors. EndocrinolMetabClin North Am 1997;26:539-51.
- Chen L, Wang JJ, Zhang GG, Song HT, Qin LP. A new cerebroside from Gynuradivaricata. Nat Prod Res 2009;23:1330-6.
- Mahmoud MF, Hassan NA, El Bassossy HM, Fahmy A. Quercetin protects against diabetes-induced exaggerated vasoconstriction in rats: effect on low grade inflammation. PLoS One 2013;8:e63784.
- Yin Z, Zhang W, Feng F, Zhang Y, Kang W. α-Glucosidase inhibitors isolated from medicinal plants. Food Sci Hum Wellness 2014;3:136-74.
- Figueiredo-González M, Grosso C, Valentão P, Andrade PB. α-Glucosidase and α-amylase inhibitors from Myrciaspp.: a stronger alternative to acarbose? J Pharm Biomed Anal 2016;118:322-7.
- Flores-Bocanegra L, Pérez-Vásquez A, Torres-Piedra M, Bye R, Linares E, Mata R. α-Glucosidaseinhibitors from Vauqueliniacorymbosa. Molecules 2015;20:15330-42.
- Nurul Islam M, Jung HA, Sohn HS, Kim HM, Choi JS. Potent α-glucosidase and protein tyrosine phosphatase 1B inhibitors from Artemisia capillaris. Arch Pharm Res 2013;36:542-52.
- Jhong CH, Riyaphan J, Lin SH, Chia YC, Weng CF. Screening alpha-glucosidase and alpha-amylase inhibitors from natural compounds by molecular docking in silico. Biofactors 2015;41:242-51.
- Sesink AL, Arts IC, Faassen-Peters M, Hollman PC. Intestinal uptake of quercetin-3-glucoside in rats involves hydrolysis by lactase phlorizin hydrolase. J Nutr 2003;133:773-6.
- deRuyck J, Brysbaert G, Blossey R, Lensink MF. Molecular docking as a popular tool in drug design, an in silicotravel. AdvApplBioinformChem 2016;9:1-11.
- Ferreira LG, Dos Santos RN, Oliva G, Andricopulo AD. Molecular docking and structure-based drug design strategies. Molecules 2015;20:13384-421.
- Grinter SZ, Zou X. Challenges, applications, and recent advances of protein-ligand docking in structure-based drug design. Molecules 2014;19:10150-76.
- Pathak AK, Mukherjee T, Maity DK. Microhydration shell structure in Cl2*-.nH2O clusters: A theoretical study. J ChemPhys 2006;125:074309.
- Lee C, Yang W, Parr RG. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys Rev B Condens Matter 1988;37:785-9.
- Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, et al. Gaussian 09. Gaussian, Inc., Wallingford CT; 2009.
- Morris GM, Huey R, Olson AJ. Using AutoDock for ligand-receptor docking. CurrProtoc Bioinformatics 2008;8:8-14.
- Sanner MF. Python: A programming language for software integration and development. J Mol Graph Model 1999;17:57-61.
- Seeliger D, de Groot BL. Ligand docking and binding site analysis with PyMOL and Autodock/Vina. J Comput Aided Mol Des 2010;24:417-22.
- Sels JP, Huijberts MS, Wolffenbuttel BH. Miglitol, a new alpha-glucosidase inhibitor. Expert OpinPharmacother 1999;1:149-56.
- Venditti A, Maggi F, Vittori S, Papa F, Serrilli AM, Di CeccoM, et al. Antioxidant and α-glucosidase inhibitory activities of Achilleatenorii. Pharm Biol 2015;8:1-6.
- Imai C, Saito M, Mochizuki K, Fuchigami M, Goda T, Osonoi T. Cotreatment with the α-glucosidase inhibitor miglitol and DPP-4 inhibitor sitagliptin improves glycemic control and reduces the expressions of CVD risk factors in type 2 diabetic Japanese patients. Metabolism 2014;63:746-53.
- Liu L, Zeng Z, Zeng G, Chen M, Zhang Y, Zhang J, et al. Study on binding modes between cellobiose and β-glucosidases from glycoside hydrolase family 1. Bioorg Med ChemLett 2012;22:837-43.
- Nam KH, Kim SJ, Kim MY, Kim JH, Yeo YS, Lee CM, et al.Crystal structure of engineered beta-glucosidase from a soil metagenome. Proteins 2008;73:788-93.
- Jeng WY, Wang NC, Lin CT, Chang WJ, Liu CI, Wang AHJ. High-resolution structures of Neotermeskoshunensisbeta-glucosidase mutants provide insights into the catalytic mechanism and the synthesis of glucoconjugates. ActaCrystallogr Sect D 2012;68:829-38.
- Dalton JA, Jackson RM. Homology-modelling protein-ligand interactions: allowing for ligand-induced conformational change. J MolBiol 2010;399:645-61.
- Pereira DF, Cazarolli LH, Lavado C, Mengatto V, Figueiredo MS, Guedes A, et al.Effects of flavonoids on α-glucosidase activity: potential targets for glucose homeostasis. Nutrition 2011;27:1161-7.
- Pham AT, Malterud KE, Paulsen BS, Diallo D, Wangensteen H. α-Glucosidase inhibition, 15 –lipoxygenaseinhibition, and brine shrimp toxicity of extracts and isolated compounds from Terminaliamacropteraleaves. Pharm Biol 2014;52:1166-9.
- Li YQ, Zhou FC, Gao F, Bian JS, Shan F. Comparative evaluation of quercetin, isoquercetin and rutin as inhibitors of alpha-glucosidase. J Agric Food Chem 2009;57:11463-8.
- Sun H, Xue Y, Lin Y. Enhanced catalytic efficiency in quercetin-4'-glucoside hydrolysis of Thermotogamaritimaβ-glucosidase A by site-directed mutagenesis. J Agric Food Chem 2014;62:6763-70.
- Lee YS, Huh JY, Nam SH, Kim D, Lee SB. Synthesis of quercetin-3-O-glucoside from rutin by Penicilliumdecumbensnaringinase. J Food Sci 2013;78:C411-5.
- Kato A, Zhang ZL, Wang HY, Jia YM, Yu CY, Kinami K, et al. Design and synthesis of labystegines, hybrid iminosugars from LAB and calystegine, as inhibitors of intestinal α-Glucosidases: binding conformation and interaction for ntSI. J Org Chem 2015;80:4501-15.
- Mohan S, Sim L, Rose DR, Pinto BM. Probing the active-site requirements of human intestinal N-terminal maltase-glucoamylase: Synthesis and enzyme inhibitory activities of a six-membered ring nitrogen analogue of kotalanol and its de-O-sulfonated derivative. Bioorg Med Chem 2010;18:7794-8.
- Natori Y, Imahori T, Murakami K, Yoshimura Y, Nakagawa S, Adachi I, et al.The synthesis and biological evaluation of 1-C-alkyl-L-arabinoiminofuranoses, a novel class of α-glucosidase inhibitors. Bioorg Med ChemLett 2011;21:738-41.
- Aoki K, Kamiyama H, Masuda K, Kamiko K, Noguchi Y, Tajima K, et al. Effects of miglitol, vildagliptin, or their combination on serum insulin and peptide YY levels and plasma glucose, cholecystokinin, ghrelin, and obestatin levels. Endocr J 2014;61:249-56.
- Kishimoto M, Noda M. Additive effects of miglitol and anagliptin on insulin-treated type 2 diabetes mellitus: a case study. Clin Drug Investig 2015;35:141-7.
- Aoki C, Suzuki K, Yanagi K, Satoh H, Niitani M, Aso Y. Miglitol, an antidiabetic drug, inhibits oxidative stress-induced apoptosis and mitochondrial ROS over-production in endothelial cells by enhancement of AMP-activated protein kinase. J PharmacolSci 2012;120:121-8.
- Roy D, Kumar V, Acharya KK, Thirumurugan K. Probing the binding of Syzygium-derived α-glucosidase inhibitors with N- and C-terminal human maltase glucoamylase by docking and molecular dynamics simulation. ApplBiochemBiotechnol 2014;172:102-14.
- Yoshida K, Hishida A, Iida O, Hosokawa K, Kawabata J. Flavonolcaffeoylglycosides as alpha-glucosidase inhibitors from Spiraeacantoniensisflower. J Agric Food Chem 2008;56:4367-71.
- Kim JS, Kwon YS, Sa YJ, Kim MJ. Isolation and identification of sea buckthorn (Hippophaerhamnoides) phenolics with antioxidant activity and α-glucosidase inhibitory effect. J Agric Food Chem 2011;59:138-44.
- Datta BK, Datta SK, Sarker SD. Quercetin 3-O-(6"-caffeoyl)-beta-D-galactopyranoside from Polygonumviscosum.Fitoterapia 2000;71:459-460.
- Matsui T, Kobayashi M, Hayashida S, Matsumoto K. Luteolin, a flavone, does not suppress postprandial glucose absorption through an inhibition of alpha-glucosidase action. BiosciBiotechnolBiochem 2002;66:689-92.
- Niaz H, Kashtoh H, Khan JA, Khan A, Wahab AT, Alam MT. Synthesis of diethyl 4-substituted-2,6-dimethyl-1,4-dihydropyridine-3,5-dicarboxylates as a new series of inhibitors against yeast α-glucosidase. Eur J Med Chem 2015;95:199-209.
- Trapero A, Llebaria A. A prospect for pyrrolidineiminosugars as antidiabetic α-glucosidase inhibitors. J Med Chem 2012;55:10345-6.