- *Corresponding Author:
- K. R. Iyer
Department of Pharmaceutical Chemistry, Bombay College of Pharmacy, Kalina, Santacruz (E), Mumbai 400098, India
E-mail: krishna.iyer@bcp.edu.in
| Date of Received | 14 October 2021 |
| Date of Revision | 14 May 2023 |
| Date of Acceptance | 11 October 2023 |
| Indian J Pharm Sci 2023;85(5):1524-1529 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Rat, mouse, guinea pig and rabbit liver microsomes were prepared by both conventional differential centrifugation (ultracentrifugation) method and calcium aggregation method. The isolated microsomes were compared in terms of their spectral cytochrome P450 content, protein content, specific spectral cytochrome P450 content and cytochrome P450 family 2 subfamily E member 1 catalytic activity. The specific spectral cytochrome P450 content (nmol/mg protein) was generally higher in microsomes isolated by the calcium aggregation method. Likewise, the catalytic activity expressed as nmol of p-nitrocatechol formed/min/ mg protein was also higher in microsomes isolated by calcium aggregation. However, the catalytic activity expressed as nmol of p-nitrocatechol formed/min/nmol cytochrome P450 was statistically different (less) only is mouse microsomes isolated by calcium aggregation. Overall, the two microsome isolation methods were comparable with ultracentrifugation appearing to yield more catalytically incompetent protein than calcium aggregation.
Keywords
Hepatic microsomes, calcium aggregation, ultracentrifugation
Liver microsomes are one of the most popular in vitro models for studies of drug metabolism[1,2]. This subcellular component is composed of endoplasmic reticulum and contains most of the important drug metabolizing enzymes, such as Cytochrome P450 (CYP450), flavin monooxygenases, glucuronosyltransferases, epoxide hydrolases, and esterases[1,2]. The isolation of these hepatic microsomes can be carried out using two different methods[1]. The conventional method for isolation of liver microsomes as a source of drug metabolizing enzymes involves differential centrifugation technique (ultracentrifugation method)[3-5]. In this technique, the liver homogenate is first centrifuged at low speeds (13 000 ×g for 30 min) to separate the cell debris, nuclei and mitochondria. Further, in the second step, ultracentrifugation of the supernatant at 100 000 ×g for 1 h, yields microsomes. In an alternative method (calcium aggregation method) isolation of microsomes is achieved without the use of an ultracentrifuge. This method involves the use of calcium for aggregation of microsomes, yielding a complex with higher density, and consequently better sedimentation properties[6-16]. We have used calcium aggregation method previously for the isolation of microsomes from rat, mice, guinea pig and rabbit but had not undertaken a direct comparison between calcium aggregation and ultracentrifugation method[16]. This study extends the previous studies and compares the activity of microsomes isolated by conventional ultracentrifugation and calcium aggregation method. Carbon monoxide gas (reagent purity) was obtained from Alchemi Gases and Chemicals Pvt. Ltd., Mumbai. Trizma-base was purchased from Sigma Chemical Co., USA. Commassie Brilliant Blue dye (gift from Dr. Bhuvaneshwari Krishna, C. H. M. College, Ulhasnagar) was used for protein estimation. p-Nitrocatechol (PNC) (Cat. No. 1208) was purchased from Lancaster Chemicals, Mumbai. Salicylamide (Cat. No. 1947160) and NADPH (Cat. No. 144935) were obtained from SRL Chemicals Ltd., Mumbai. Acetonitrile and methanol (High-Performance Liquid Chromatography (HPLC) grade) was obtained from Qualigens Ltd., Mumbai. Trifluoroacetic Acid (TFA) AR, p-nitrophenol (Cat. No. 30008G25), diethyl ether LR, ammonium sulfate AR, sucrose AR, ethylene diamine tetraacetic acid AR (Cat. No. 88021), Triton X-100, sodium dithionite purified, dextrose AR, disodium hydrogen orthophosphate dihydrate AR (Na2HPO4.2H2O), sodium dihydrogen orthophosphate AR (NaH2PO4.2H2O) were purchased from SD fine-Chem. Ltd., Mumbai. All other chemicals were of analytical grade. Rat, mice, guinea pig and rabbit livers were obtained from Department of Pharmacology, Bombay College of Pharmacy, Kalina. The animals used in this study were those that were sacrificed as part of other experiments approved by the Institutional Animal Ethics Committee. It should be noted that, the potential of these experiments to alter liver function (and consequently cytochrome P450 content), although a possibility, was not taken into consideration, since the intention of this study was a relative comparison of the two isolation methods. The livers obtained were stored at -70° until further use. Since the study involved a comparison of two microsomal isolation methods[16], the two procedures were carried out simultaneously for a particular animal species to avoid any undue variability. The entire microsomal isolation was carried out at 0-4° in an ice bath. The excised liver (pooled liver samples, 10 g per batch and 3 such batches) was thawed, finely chopped with a pair of scissors and homogenized with four times the weight in volume (40 ml) of 10 mM Tris-HCl buffer containing 0.25 M sucrose, pH 7.4, in a Potter Elvejhem glass homogenizer equipped with Teflon pestle. The homogenate was centrifuged at 13 000 ×g for 30 min at 4° in a refrigerated centrifuge (Biofuge Stratos) and the precipitate was discarded. Supernatant (S9 fraction) from each batch was split into two parts; each half was individually subjected to either calcium precipitation or ultracentrifugation for microsomal isolation[16]. In calcium precipitation method[16], calcium chloride was added to the supernatant to yield a final concentration of 10 mM. The solution was stirred for 15-20 min with a glass rod and then centrifuged at 25 000 ×g for 10 min at 4° (Biofuge Stratos). In case of ultracentrifugation method, the S9 fraction was directly centrifuged at 100 000 ×g for 1 h at 4° in a Beckmann ultracentrifuge[3-5]. The firmly packed pellets of microsomes obtained from both the methods were resuspended (20 ml per 10 g liver) by homogenization in 100 mM Tris-HCl buffer containing 20 % w/v of glycerol and 10 mM EDTA, pH 7.4. The microsomes were stored at -70° until use. The isolation experiment was performed in triplicate for each animal liver. Thus 6 microsomal samples (3 by calcium precipitation and 3 by ultracentrifugation) were obtained for each animal liver. Thus, a total of 24 microsomal samples from 4 species of animals were prepared.
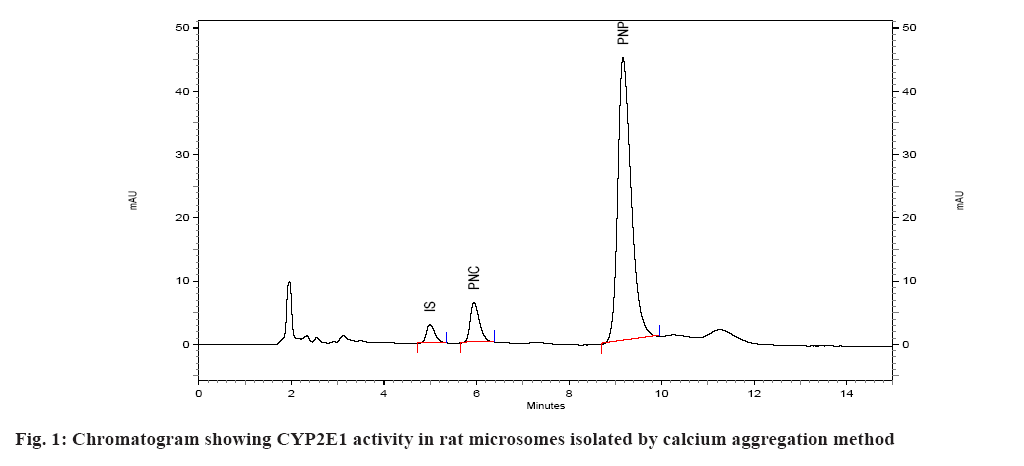
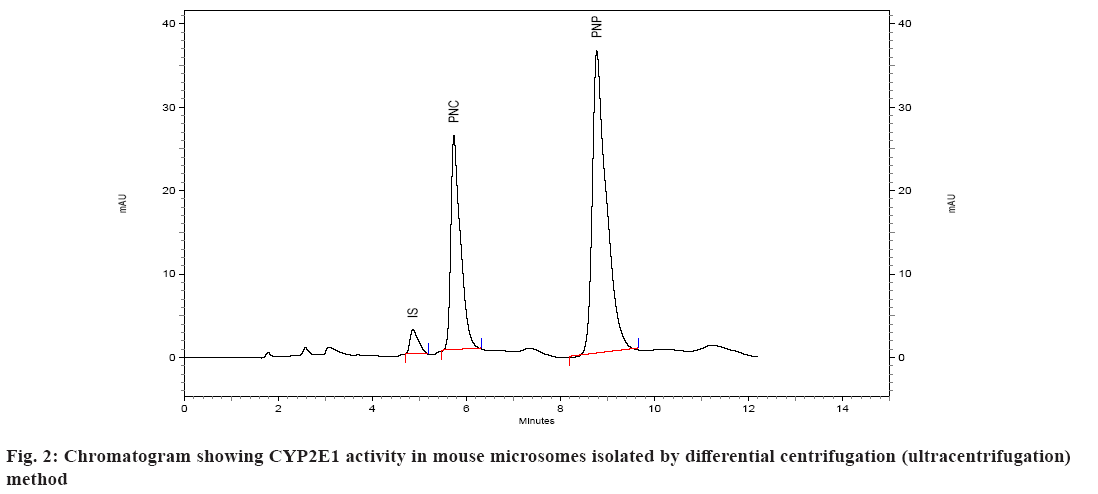
Spectral CYP450 content of microsomes was determined by the method of Omura et al.[17]. The microsomes were diluted in the ratio 1:4 with 0.1 M potassium phosphate buffer, pH 7.4, containing 0.5 % Triton X 100 and 1 mM EDTA. The solution was stirred thoroughly and divided into two 3 ml quartz cuvettes. The sample and reference cuvettes containing the microsomal preparations were saturated with 30 to 40 bubbles of carbon monoxide at the rate of 1 bubble/s. After the samples were equilibrated with carbon monoxide, the baseline was recorded. Sodium dithionite was added only to the sample cuvette and the sample was scanned from 400 to 500 nm to obtain a reduced carbon monoxide vs. oxidized carbon monoxide difference spectrum. An extinction coefficient of 106 mM-1cm-1 was used for the determination of CYP450 content. CYP450 content of all the samples was determined using this method and expressed in nmoles/ml (µM)[18]. Protein estimation of the microsomal preparation was done using Commassie Brilliant Blue dye method. Calibration curve/standard protein plot was prepared using Bovine Serum Albumin (BSA) as protein standard. The optical density was read on the spectrophotometer at 595 nm. All of the standards as well as samples were prepared in duplicate for the estimation[19]. The catalytic competency of the isolated liver microsomes was evaluated by quantifying the Cytochrome P450 Family 2 Subfamily E Member 1 (CYP2E1) activity of isolated microsomes[16]. This was done by monitoring the ability of microsomes to convert p-nitrophenol to PNC. Incubations were performed as described previously[16,20,21]. Briefly, incubations were performed in a total volume of 0.5 ml of 0.1 M sodium phosphate buffer containing 0.001 M EDTA, pH 7.4 and containing 100 µl microsomes, p-nitrophenol at a final concentration of 40 µM, and 0.6 mM Nicotinamide Adenine Dinucleotide Phosphate Hydrogen (NADPH). Incubations were initiated with NADPH and were continued at 37° for 30 min. After incubation, the reactions were stopped with 250 µl of 0.6 M perchloric acid. After stoppage of the incubation, 50 µl of 350 µg/ml solution of salicylamide (IS) was added as internal standard, followed by addition of 500 mg of ammonium sulfate. The sample was then extracted with (4 ml) diethyl ether, the ether layer separated and evaporated under nitrogen at 40°. The residue was reconstituted in mobile phase and subjected to HPLC. A standard curve of PNC was prepared in the range of 0.2 to 10 µM in 0.1 M sodium phosphate buffer, pH 7.4. The standard (calibration) curve was plotted using area ratio of PNC to internal standard on ordinate, and concentration on the abscissa. Linear regression (Microsoft Excel XP) was performed, to obtain the regression equation, and the unknown concentrations of PNC in the incubations were obtained by interpolation. The results of the two methods were evaluated statistically. The existence of significant difference between the two methods was determined using single factor Analysis of Variance (ANOVA) (Microsoft Excel). A p value>0.05 represents no significant difference between the two methods of isolation of microsomes. The microsomes isolated by the two methods were evaluated on the basis of several parameters. In the first case, spectral CYP450 content was determined (which reflects the mixture of various isoforms of P450). In the second case, the microsomal protein content was determined and compared. In the third case, specific spectral P450 content (nmoles of CYP450 per mg of protein) of microsomes obtained by dividing spectral P450 content with protein content was calculated and compared. The results of this analysis are given in Table 1. Isolation of the microsomal fraction from liver tissue (or any other tissue) generally is conventionally accomplished by differential ultracentrifugation method[3-5]. The major drawback of this procedure is the requirement of an ultracentrifuge. Ultracentrifuges are fairly expensive and may not be within the means of many research laboratories. An alternative is to use the calcium-aggregation method, which uses a cooled high-speed centrifuge rather than an ultracentrifuge[6-16]. A previous study on isolation of microsomes by calcium aggregation has been carried out in our laboratory[16]. In this study, we have tried to extend this work by performing a head-on comparison of the activity of microsomes isolated by both the methods i.e. calcium aggregation and ultracentrifugation. In our study, the spectral CYP450 content per ml was found to be higher in microsomes isolated by calcium aggregation in case of rat, mice and guinea pig than in case of rabbit where a higher spectral CYP450 content was observed in ultracentrifugation method. In case of rat, mice and guinea pig the observed differences i.e. less than 0.3 nmol/ml between two methods, were not statistically significant. Statistically, a significant difference was observed between the two methods only in the case of rabbit liver microsomes. Protein content of rat, mice and guinea pig microsomes isolated by ultracentrifugation method was found to be higher than calcium precipitation method. The mean specific activity was found to be higher in microsomes isolated by calcium precipitation method in case of rat, mice and guinea pig than in case of rabbit, where a higher specific activity was observed in ultracentrifugation method. In case of mice and guinea pig microsomes, significant difference in specific activity was observed between the two methods. This is a direct reflection of the high protein content (but not CYP content) observed in case of microsomes isolated by ultracentrifugation method (Mice: protein content of 4.80 mg/ml by calcium precipitation and 9.69 mg/ml by ultracentrifugation method, Guinea pig: protein content of 3.17 mg/ml by calcium precipitation and 9.08 mg/ml by ultracentrifugation method). The mean rate of formation of metabolite PNC per nmol of CYP450 by ultracentrifugation method appeared to be more than calcium precipitation method across all the four species (fig. 1 and fig. 2). However, statistical significance was observed between the two methods only in case of mice liver microsomes (Table 2). The mean rate of formation of metabolite PNC per mg of protein in rat, mice, guinea pig and rabbit microsomes was found to be 0.0607, 0.3664, 0.4565 and 0.2008 nmol/min/mg of protein, respectively, by calcium precipitation method and 0.0480, 0.1834, 0.1782 and 0.4802 nmol/min/mg of protein respectively, by ultracentrifugation method. Thus, overall rate of formation PNC per mg of protein in case of rat, mice and guinea pig microsomes was found to be more by calcium precipitation method. Statistically, data from all the four animals shows that the two methods are significantly different when reported in terms of protein content. This is because of the fact that the protein content of microsomes isolated by ultracentrifugation is higher than calcium precipitation method. In studies carried out for the liver esterase enzyme content estimation using calcium aggregation method as well as ultracentrifugation method, the activity of the esterases in the microsomes prepared by ultracentrifugation and by calcium aggregation was not significantly different although the ultracentrifugation method resulted in a higher microsomal protein (30.9 mg/g liver in case of ultracentrifugation and 18.4 mg/g of calcium aggregation method)[12]. The UDPglucuronosyl activity was also comparable in microsomes obtained by calcium aggregation or ultracentrifugation. However, the specific activity of the hepatic benzopyrene hydroxylase of the benzopyrene treated animals in the calcium harvested microsomes was 55 per cent of that in the ultracentrifuged microsomes[15]. Brain microsomes prepared by the usual calcium aggregation method resulted in the loss of cytochrome P450. A modification of the calcium aggregation method for the rapid preparation of rat and mouse brain microsomes has been reported. This involves the incorporation of glycerol, dithiothreitol, and EDTA in the preparation of microsomes[22]. In conclusion, the data obtained in this study suggests that evaluation of the two methods for isolation of hepatic microsomes is not straightforward. In some cases calcium precipitation is seen to be better than ultracentrifugation but at the same time the coefficient of variation (i.e. the variation observed between different batches A, B, C in the same animal species) is also seen to be higher for calcium precipitation. The protein content of the microsomes isolated by ultracentrifugation method was found to be higher than calcium precipitation method. From our perspective, reporting the activity of microsomes in terms of CYP450 content (nmol/min/nmoles of P450) seems more reasonable than reporting in terms of protein content (nmol/min/mg of protein) since the normalization factor is the enzyme rather than total protein present in the microsomes. Overall, it appears that the two methods of microsome isolation yield similar catalytic capabilities in the microsomal fraction. However, it appears that more catalytically incompetent protein is present in microsomes isolated by ultracentrifugation. One can thus suggest that the present study indicates that both methods are comparable in terms of catalytic activity of the isolated microsomes but that the calcium aggregation method yields a cleaner microsomal preparation in terms of the catalytic active CYP450 protein.
| Animal | Isolation Method | Spectral P450 (µM) | Protein Content (mg/ml) | Specific CYP450 Content (nmol/mg protein) |
|---|---|---|---|---|
| Rat | Calcium | 2.5050±0.5232 | 5.7899±1.2993 | 0.4393±0.0726 |
| Ultra | 2.2240±0.7070 | 6.6822±0.4564 | 0.3295±0.1036 | |
| Mice | Calcium | 2.7320±0.5321 | 4.7960*±1.9284 | 0.6224*±0.1777 |
| Ultra | 2.4427±0.1576 | 9.6931±0.8152 | 0.2536±0.0297 | |
| Guinea Pig | Calcium | 2.7777±0.5855 | 3.1737*±1.1501 | 0.9642*±0.3735 |
| Ultra | 2.5400±0.3240 | 9.0816±3.5263 | 0.3115±0.1209 | |
| Rabbit | Calcium | 1.5969*±0.0980 | 2.7831±0.8632 | 0.5960*±0.1176 |
| Ultra | 2.1487±0.2695 | 2.1609±0.6491 | 1.2898±0.7520 |
Note: *Statistical difference at p<0.05
Table 1: Spectral P450 Content, Protein Content and Specific activity of Microsomes Isolated by Calcium Precipitation and Ultracentrifugation Method
| Animal | Isolation Method | CYP2E1 Activity (nmol/min/nmol P450) | CYP2E1 Activity (nmol/min/mg protein) |
|---|---|---|---|
| Rat | Calcium | 0.1405±0.320 | 0.0607*±0.107 |
| Ultra | 0.1532±0.349 | 0.0480 ± 0.0058 | |
| Mice | Calcium | 0.5775*±0.854 | 0.3664*±0.1320 |
| Ultra | 0.7210±0.486 | 0.1834±0.0277 | |
| Guinea Pig | Calcium | 0.5030±0.1088 | 0.4565*±0.0989 |
| Ultra | 0.5793±0.0901 | 0.1782±0.0550 | |
| Rabbit | Calcium | 0.3382±0.0168 | 0.2008*±0.0325 |
| Ultra | 0.3828±0.0491 | 0.4802±0.2213 |
Note: *Statistical difference at p<0.05
Table 2: CYP2E1 activity of the Isolated Microsomes in Terms Of Spectral P450 Content and Protein Content
Conflict of interests:
The authors declared no conflict of interests.
References
- Munir P, Kevin BP. Metabolic models of cytotoxicity. In: Woolf TF, editor. Handbook of drug metabolism. New York: Marcel Dekker Inc; 1999. p. 443-68.
- Paul R, Ortiz de M. The cytochrome P450 oxidative system. In: Woolf TF, editor. Handbook of drug metabolism. New York: Marcel Dekker Inc; 1999. p. 109-15.
- Ozols J. Preparation of membrane fractions. Methods Enzymol 1990;182:225-35.
[Crossref] [Google Scholar] [PubMed]
- Moldéus P, Högberg J, Orrenius S. Isolation and use of liver cells. Methods Enzymol 1978;52:60-71.
[Crossref] [Google Scholar] [PubMed]
- Judy L, Jerome M. Isolation of P450 enzymes from human liver. Methods Enzymol 1958;52:577-8.
- Burchell A. A simple method for purification of rat hepatic microsomal cytochrome b5. Biochem J 1985;226(1):339-41.
[Crossref] [Google Scholar] [PubMed]
- Schenkman JB, Cinti DL. Preparation of microsomes with calcium. Methods Enzymol 1978;52:83-9).
[Crossref] [Google Scholar] [PubMed]
- Arion WJ, Canfield WK, Callaway ES, Burger HJ, Hemmerle H, Schubert G, et al. Direct evidence for the involvement of two glucose 6-phosphate-binding sites in the glucose-6-phosphatase activity of intact liver microsomes: Characterization of T1, the microsomal glucose 6-phosphate transport protein by a direct binding assay. J Biol Chem 1998;273(11):6223-7.
[Crossref] [Google Scholar] [PubMed]
- Bhagwat SV, Boyd MR, Ravindranath V. Rat brain cytochrome P450. Reassessment of monooxygenase activities and cytochrome P450 levels. Drug Metab Dispos 1995;23(6):651-4.
[Google Scholar] [PubMed]
- Balanehru S, Nagarajan B. Intervention of adriamycin induced free radical damage. Biochem Int 1992;28(4):735-44.
- Bodd EG, Gadeholt G, Christensson PI, Mørland J. Mechanisms behind the inhibitory effect of ethanol on the conjugation of morphine in rat hepatocytes. J Pharmacol Exp Thera 1986;239(3):887-90.
[Google Scholar] [PubMed]
- Yeh SY. Localization and characterization of meperidine esterase of rats. Drug Metab Dispos 1982;10(4):319-25.
[Google Scholar] [PubMed]
- Roders MK, Glende Jr EA, Recknagel RO. NADPH dependent lipid peroxidation of calcium bound microsomes. Res Commun Chem Pathol Pharmacol 1976;15(2):393-6.
- Ecobichon DJ. Preparation of guinea pig hepatic microsomes: A comparison of three techniques. Res Commun Chem Pathol Pharmacol 1976;14(3):515-25.
[Google Scholar] [PubMed]
- Aitio A, Vainio H. UDPglucuronosyltransferase and mixed function oxidase activity in microsomes prepared by differential centrifugation and calcium aggregation. Acta Pharmacol Toxicol 1976;39(5):555-61.
[Crossref] [Google Scholar] [PubMed]
- Walawalkar P, Serai P, Iyer K. Isolation and catalytic competence of different animal liver microsomal fractions prepared by calcium-aggregation method. Indian J Pharm Sci 2006;68(2):262-5.
- Omura T, Sato R. The carbon monoxide-binding pigment of liver microsomes. I. Evidence for its hemoprotein nature. J Biol Chem 1964;239(7):2370-8.
[Google Scholar] [PubMed]
- Guengerich FP. Analysis and characterization of enzymes. In: Hayes AW, editor. Principles and methods of toxicology, 3rd ed. New York: Raven Press; 1994. p. 1259-1313.
- Macart M, Gerbaut L. An improvement of the Coomassie Blue dye binding method allowing an equal sensitivity to various proteins: Application to cerebrospinal fluid. Clin Chim Acta 1982;122(1):93-101.
[Crossref] [Google Scholar] [PubMed]
- Lavhekar S, Lohade A, Coutinho E, Iyer K. Estimation of microsomal CYP1A2 activity by high performance liquid chromatography. Indian J Pharm Sci 2006;68(2):258-61.
- Tassaneeyakul W, Birkett DJ, Veronese ME, McManus ME, Tukey RH, Quattrochi LC, et al. Specificity of substrate and inhibitor probes for human cytochromes P450 1A1 and 1A2. J Pharmacol Exp Ther 1993;265(1):401-7.
[Google Scholar] [PubMed]
- Ravindranath V, Anandatheerthavarada HK. Preparation of brain microsomes with cytochrome P450 activity using calcium aggregation method. Anal Biochem 1990;187(2):310-3.
[Crossref] [Google Scholar] [PubMed]