- *Corresponding Author:
- Y. He
Department of CardiovescularDisease, Renmin Hospital of Wuhan University, No. 238 Jiefang Road, Wuhan, Hubei 430060, China
E-mail: Lkwang999@163.com
| This article was originally published in a special issue,“Biomedical Research in Clinical and Preclinical Pharmaceutics” |
| Indian J Pharm Sci 2020:82(5)Splissue7;139-144 |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
Clustered regularly interspaced short palindromic repeats/Cas9 gene editing technique was used to construct the human liver Lo2 stable cell line with low density lipoprotein receptor gene deletion. According to the design principle of clustered regularly interspaced short palindromic repeats/Cas9 target, the single guide RNA specifically target low density lipoprotein receptor gene was designed and the recombinant plasmid lenticlustered regularly interspaced short palindromic repeats v2 was used to construct the recombinant plasmid which can express this single guide RNA and Cas9 protein. After sequencing identification, the recombinant plasmid was transferred into Human embryonic kidney 293T cells together with retroviral packaging plasmids VSVG and PAX2. 48 h after transfection, the virus supernatant was collected and directly infected with human liver Lo2 cell. Puromycin was used to screen low density lipoprotein receptor-deficient Lo2 cells, which was verified by polymerase chain reaction, western blotting, and immunofluorescence staining. Then polymerase chain reaction and western blotting were used to verify the effects of low density lipoprotein receptor knockout on the function, lipid metabolism, glucose metabolism pathway, and insulin resistance of the Lo2 cell line. In this paper, the clustered regularly interspaced short palindromic repeats/Cas9 plasmid targeting low density lipoprotein receptor was successfully constructed. The results of polymerase chain reaction and western blotting showed that the human liver Lo2 cell with a stable knockout of low density lipoprotein receptor was obtained. The results of immunofluorescence staining showed that compared with the control group, the content of prealbumin in low density lipoprotein receptor knockout cells decreased significantly, while the content of diacylglycerol increased. At the same time, low density lipoprotein receptor gene knockout can increase the expression level of fatty acid synthase, peroxisome proliferator-activated receptor-γ, sterol regulatory element binding protein 1c, and inhibit the expression of carnitine palmitoyltransferase 1a. In addition, compared with the control group, there was no significant difference in mRNA expression and protein level of mammalian target of rapamycin and phosphatidylinositol-3-kinase, and there was no significant change in epsilon isoform of protein kinase C and insulin receptor substrates 1 expression level. The Lo2 cell line with low density lipoprotein receptor gene knockout was constructed by clustered regularly interspaced short palindromic repeats/Cas9 technique, and the role of low density lipoprotein receptor in lipid metabolism and glucose metabolism was proved by Polymerase chain reaction and Western blotting. The low density lipoprotein receptor knockout polyclonal and monoclonal cells constructed in this study can provide a useful tool for future research.
Keywords
Cas9/single guide RNA technology, low density lipoprotein receptor, Liver cell, Lipid metabolism, Glucose metabolism
Low density lipoprotein receptor (LDLR) was a kind of transmembrane protein, which was distributed in the liver, arterial wall smooth muscle cells, and vascular endothelial cells. It was mainly responsible for the removal of LDL particles in blood [1]. The LDLR gene was located on the human 19th chromosome and consists of 18 exons and 17 introns. The total length of the gene was about 45.5 kb, encoding 839 amino acid residues. At present, there were more than 1700 kinds of LDLR gene mutations, including insertion, deletion, nonsense, and missense mutations, with the most reported mutations in 4th exon [2]. LDLR gene mutations can lead to dyslipidemia, such as hypercholesterolemia and familial hypercholesterolemia [3], and can also induce hepatocyte degeneration and promote the development of non-alcoholic fatty liver disease (NAFLD) [3].
Some studies have shown that the abnormal expression of LDLR may be related to insulin resistance. Free cholesterol accumulation can activate the expression of proteins related to insulin resistance in the liver [4,5]. LDLR has been shown to play a key role in mediating cholesterol and LDL-induced β-cell dysfunction and impaired insulin secretion [6]. NAFLD was an independent risk factor for type 2 diabetes and cardiovascular disease [7]. In the general population, the prevalence of NAFLD was about 30 %, while in patients with type 2 diabetes, the prevalence rate has tripled. Studies have shown that the abnormal expression of LDLR can associate NAFLD with liver insulin resistance. Therefore, LDLR may be a key signal molecule in the insulin resistance pathway and participate in the pathogenesis of NAFLD.
In this study, Clustered regularly interspaced short palindromic repeats (CRISPR)/Cas9 genome editing technique was used to establish LDLR gene knockout liver cell line. It was found that the intracellular lipid content of the cell line increased significantly. The gene expression levels of the key proteins [fatty acid synthase (FAS), peroxisome proliferator-activated receptor-γ (PPARγ), sterol regulatory element binding protein 1c (SREBP-1c), carnitine palmitoyltransferase 1a (CPT1a), long chain 3-hydroxyacyl-CoA dehydrogenase deficiency α (HADHα)] involved in lipid metabolism pathway were analyzed, and the relationship between LDLR and the key proteins in insulin resistance pathway [epsilon isoform of protein kinase C (PKCε), insulin receptor substrates 1 (IRS-1)] and gluconeogenesis pathway [rapamycin (mTOR), phosphatidylinositol-3-kinase (PI3K/Akt)] was evaluated.
Materials and Methods
Cell culture:
The human liver Lo2 cell line was purchased from the China Center for Type Culture Collection (Wuhan, China) and cultured in Dulbecco’s Modified Eagle’s medium (DMEM) (37°, 5 % CO2, 10 % fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin). About 1×105 Lo2 cells (37°, 24 h) were treated with 10 μM LDLR inhibitor.
Design the single guide RNA (sgRNA) sequence to identify the target site:
The LDLR gene sequence was obtained by National Center for Biotechnology Information (NCBI). According to the CRISPR-Cas9 target design principle, the online Guide RNA (gRNA) design tool (http://chopchop.cbu.uib.no/) was used to design the sgRNA sequence that meets the requirements of the experiment. The Off-Targeting effect of sgRNA sequence was evaluated. After excluding the other homologous sequences, sgRNA interference sequence was obtained. The BsmB I restriction site (CACCG/AAAC) was added to the 5’ end of the sense and antisense chains of the sequence. The forward sequence of sgRNA: 5’-AGGAGACGTGCTTGTCTGTC-3 and the reverse sequence: 5’-CTGAGCCGTTGTCGCAGT-3’. The designed single-stranded sgRNA was synthesized by the company and annealed by gradient Polymerase chain reaction (PCR) to form a double-stranded sgRNA, named LDLR-sg. At the same time, a general negative control was designed, named LDLR-sgcon.
Construction and identification of the recombinant plasmid:
The synthesized LDLR-sg and LDLR-sgcon were done denaturation at 95° for 10 min, and cooled to room temperature for annealed into double strands. The 2 μg empty plasmid lentiCRISPR v2 was digested with BsmB I, then agarose gel electrophoresis was performed and the gel was recovered. The double-stranded sgRNA product was ligated with the gel recovery product, and the ligating system was as follows: 1 μL doublestranded sgRNA, 100 ng digested plasmid, 1 μL T4 ligase and 2 μL 10×T4 ligase buffer, adding water to 20 μL and ligating overnight at 16°. The ligated products were transformed into DH5α competent cells, and the positive clones were screened by Luria broth (LB) plate (ampicillin resistance) and cultured overnight at 37°. The monoclonal bacterial colony was selected and the plasmids were extracted after enlarged culture, and then sequenced to identify the positive clones with lentiCRISPRv2-LDLR-sg and lentiCRISPRv2-LDLRsgcon.
All the positive clone vectors of sgRNA were transfected into Human embryonic kidney (HEK) 293AT cells by three-plasmid packaging system. When the fusion degree of HEK293T cells was 70 %-80 %, 10 μg recombinant vector GlentiCRISPRv2-LDLR-sg and 10 μg packaging plasmids psPAX and 5 μg pMD2.G were added to every 10 cm petri dish. The complete culture medium was changed 6 h after transfection, and the recombinant lentivirus LVLDLR-sgRNA was collected after 48 h and 72 h, respectively. After mixing the virus collected twice, the virus was filtered with a 0.45μm filter and stored at -80°. The titer of the virus was detected, and the recombinant lentiviruses of LVLDLR- sg and LV-LDLR-sgcon were obtained.
Construction and detection of Lo2 cell line with LDLR gene knockout:
Human liver Lo2 cells in logarithmic growth phase were inoculated in 6-well plate (37°, 5 % CO2, 10 % fetal bovine serum, 100 U/ml penicillin and 100 μg/ml streptomycin) at a density of 2×105 cells per well. It was digested with 0.25 % trypsin solution every 2-3 d for routine passage. When the cells grow to a fusion degree of 30 %-40 %, 5 μg/mL polybrene was added, and the cells were infected with Cas9 overexpression lentivirus (LVCas9-Puro), and the multiplicity of infection (MOI) was 10. After 24 h of infection, the DMEM medium was changed and cultured for 2 d, and the cells were screened with 2 μg/mL puromycin. Finally, the Lo2 cells which could stably express the Cas9 protein were obtained named Cas9/Lo2 cells.
Cas9/Lo2 cells were infected with the packaged LVLDLR- sgRNA lentivirus (LV-LDLR-sg) and control lentivirus (LV-LDLR-sgcon). Cas9/Lo2 cells in the logarithmic growth phase were inoculated on a 6-well plate at a density of 2×105 cells per well, and were infected with LV-LDLR-sgRNA lentivirus when the cells grew to a fusion degree of 30 %-40 %, as described before. After 24 h of infection, changing the culture medium and continuing to culture, LDLR gene knockout Lo2 polyclonal cells (Lo2 LDLR-sg) and negative control Lo2 polyclonal cells (Lo2 LDLR-sgcon) which could express the Cas9 protein and LDLR-sgRNA bacterial colony were obtained. Because the recombinant lentiCRISPRv2-LDLR-sgNA vector contains enhanced green fluorescent protein (EGFP) gene sequence, Lo2 cells infected with LV-LDLR-sgRNA lentivirus will emit green fluorescence. Therefore, the virus infection was observed under the fluorescence microscope and the photos were collected, and then the photos were collected with the phase contrast microscope. Finally, the cells in each group were digested and the percentage of green fluorescent cells was detected by Fluorescein isothiocyanate (FITC) channel of flow cytometry (FCM), which was called infection efficiency.
Oil Red O staining and determination of optical density (OD) value:
WT Lo2 cells and LDLR knockout Lo2 cells grown in 24-well plates were harvested. As mentioned earlier [8], the cells were stained with oil red O and the steatosis based on oil red O was quantified. The cell nuclear were stained with hematoxylin for 15 sec and washed with saturated Lithium carbonate (Li2CO3) solution. The image was captured using a Leica DFC 420C microscope. The experiment was repeated three times.
Reverse transcriptase-Quantitative polymerase chain reaction (RT-qPCR) analysis:
The total RNA (1 μg) extracted by RNeasy Mini kit was used to synthesize complementary DNA (cDNA), and then the target gene was amplified at 25 μl reaction volume. The reaction system consists of 150 ng cDNA, 0.2 μM primers, 12.5 μl 2X SYBR buffer (10 mM dNTP and 1 U Taq polymerase). All tests were repeated three times with Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as internal reference and quantified using 2-ΔΔCq method [9].
Western blotting analysis:
As mentioned before, the proteins were extracted from cells using Radio-Immunoprecipitation Assay (RIPA) buffer [8] and quantified using Pierce bicinchoninic acid kits, which (80 μg) were then used for 10 % sodium dodecyl sulphate–polyacrylamide gel electrophoresis (SDS-PAGE) separation and transferred to polyvinylidene fluoride (PVDF) membranes. At room temperature, the PVDF membrane was blocked with Phosphate-buffered saline (PBS) buffer containing 3 % bovine serum albumin and 0.5 % v/v Tween-20 for 1 h. The membrane was then incubated with the primary antibodies targeting FAS (1:500), CPT1a (1:300), PPARγ (1:500), mTOR (1:500), IRS1 (1:500;), p-IRS1 (1:300), LDLR (1:500), SREBP-1c (1:500), AKT (1:500), and p-AKT (1:300) at 37° for 1 h. Then, the goat anti-rabbit immunoglobulin G (IgG) polyclonal antibody labeled with horseradish peroxidase (HRP) was incubated together. After incubation, the PVDF membrane were placed at 37° for 1 h and observed on Tanon 5500 chemiluminescence imaging system. According to the manufacturer’s procedures, the protein level was visualized using the Supersignal West Pico chemiluminescence detection system. The protein level was measured by ImageCal software.
Detection of Phosphatidic acid (PA) and diacylglycerol (DAG):
Lo2 cells infected with LV-LDLR-sgRNA lentivirus and negative control cells were cultured for 48 h, and the cells were lysed with RIPA buffer. RIPA buffer contains 50 mM Tris (pH 7.5), 150 mM Sodium chloride (NaCl), 1 % nonidet P-40, and 0.1 % sodium dodecyl sulfate (SDS), and 1 mM phenylmethanesulfonyl fluoride. The total lipids in the lysate were collected after centrifugation at 4°, 10 000 rpm/min for 10 min. According to the manufacturer’s instructions, total PA kit was used to quantify PA. The human DAG ELISA kit was used to determine the amount of DAG in each sample.
Statistical analysis:
The quantitative analysis of gene expression was repeated 3 times in each group. All the data were expressed by x±s and processed by SPSS 10.0. The difference between the two samples was analyzed by t-test. p<0.05 indicates that the difference is significant.
Results and Discussion
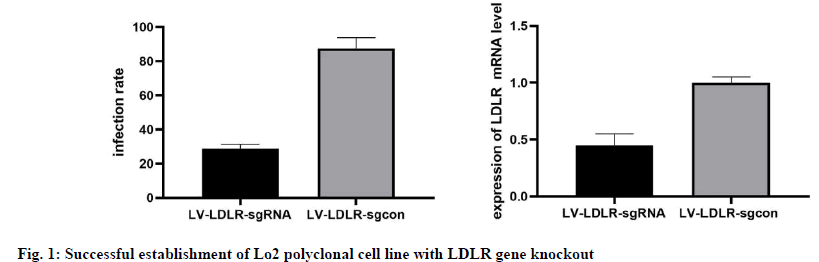
The results of phase contrast microscope and fluorescence microscope observation and FCM analysis showed that both the LV-LDLR-sgRNA lentivirus targeting LDLR gene and the negative control LVLDLR- sgcon lentivirus successfully infected Cas9/ Lo2 cells with an infection efficiency of more than 80 % (fig. 1A). The results of quantitative analysis also showed that the expression level of LDLR protein in LDLR knockout group was significantly decreased (p≤0.01, fig. 1B). The above results showed that all the sgRNA designed in this study were effective, and the Lo2 polyclonal cell line with LDLR gene knockout was successfully constructed.
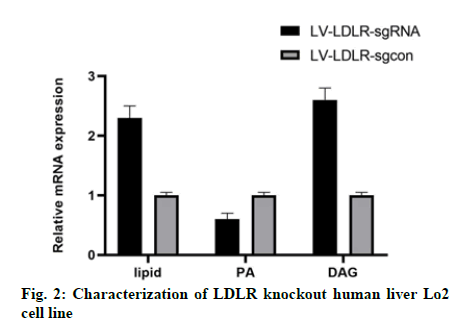
In this study, we analyzed the effect of LDLR gene knockout on the function of liver cells. Oil red O staining showed that the intracellular lipid content of Lo2 cells with LDLR gene knockout was 40 % higher than that of the control group (fig. 2). As expected, the content of PA in LDLR knockout cells was significantly lower than that in the control group cells (fig. 2), while the content of DAG was increased (fig. 2).
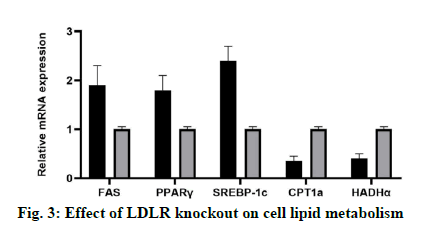
In this study, the expression levels of genes related to lipid synthesis (FAS, PPAR γ and SREBP-1c) and genes related to lipolysis (CPT1a and Hydroxyacyl- Coenzyme A dehydrogenase alpha (HADHα)) in LDLR knockout Lo2 cells were detected at mRNA and protein levels. The results showed that compared with the control group, LDLR gene knockout could increase the expression of FAS, PPARγ and SREBP-1c, and inhibit the expression of CPT1a (fig. 3).
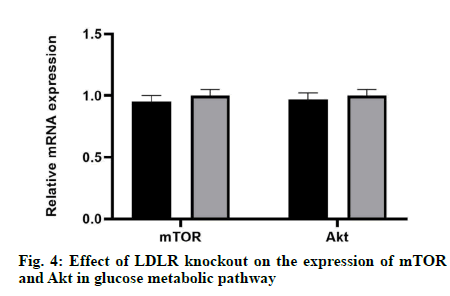
The results showed that there was no significant difference in mRNA expression and protein level between mTOR and Akt compared with the control group, which indicated that LDLR gene knockout did not affect neither the mRNA expression nor the protein level of mTOR and Akt (fig. 4).
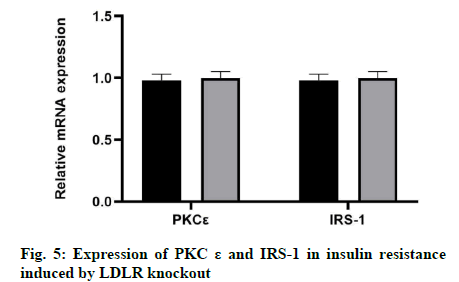
In this study, we examined whether the knockout of LDLR would affect the expression of PKCε and IRS-1 in insulin resistance. The results showed that there was no significant change in the expression levels of PKCε and IRS-1 by LDLR knockout at the mRNA or protein level (fig. 5).
In this study, LDLR gene knockout liver Lo2 cell line was successfully constructed by CRISPR/Cas9 technique, and the intracellular lipid accumulation was significantly increased. By evaluating the expression of genes related to lipid and glucose metabolism in this cell line, it was confirmed that the established cell line could be used to study the NAFLD and related liver insulin resistance.
After continuous improvement and development since its appearance, CRISPR/Cas9 gene knockout technology has successfully realized the single-gene, double-gene, and multi-gene knockout in animals, plants, and cells [10-14]. The confirmation of the infection efficiency was a key step in the construction of stable cell lines by CRISPR/Cas9 technology. In this study, the infection efficiency was determined by fluorescence observation and FCM analysis. If the infection efficiency was low, the cells would be infected with lentivirus again. According to past experience, continuous infection of lentivirus for 2 times can improve the infection efficiency effectively. Another key step in the construction of stable cell lines by CRISPR/Cas9 was the identification of positive cell lines. In this study, U87 cell lines with LDLR gene knockout were identified by restriction endonuclease digestion of PCR products. Because the BsmB I endonuclease site was located on the target of sgRNA, this enzyme can be used to detect the mutation efficiency of this enzyme site, to preliminarily determine whether the monoclonal DNA strands have mutations in the double strands or not.
In this study, LDLR gene knockout Lo2 polyclonal cells were cloned by the limited dilution method, and a negative control monoclonal cell line and an LDLR gene knockout monoclonal cell line were obtained, which laid a good foundation for later study of the function and mechanism of the target genes. In this study, WB and DNA sequencing were used to determine the knockout efficiency of LDLR gene at the protein and gene level, which ensured the reliability of the results.
The results showed that the expression of all three lipid synthesis related genes (FAS, PPAR γ and SREBP-1c) in LDLR knockout liver cells increased, while the expression of lipolysis related gene CPT1a decreased, which may be the reason for the increase of intracellular lipid content. Notably, a study reported that the use of sgRNA to knock down the LDLR gene led to a decrease in SREBP-1c expression. However, this was carried out in the human adrenocortical cells [15], and its lipid metabolism pathway may be different from that of human liver cells. Studies have shown that the overexpression of FAS not only promotes the adipogenesis, but also promotes the growth of breast cancer cells [16]. This may explain why LDLR knockout liver cells show an increased growth rate. The increase of FAS and the decrease of CPT1a in LDLR knockout liver cells may be related to the increased activity of SREBP-1c. It was reported that SREBP-1 can activate FAS transcription [17] and down-regulate the expression of the lipase gene [18].
In previous studies, LDLR can regulate the cellular DAG and PA, PA further regulate the expression of DAG-sensitive proteins associated with liver insulin resistance (PKCε) [19], and the expression of PA-sensitive proteins (mTOR and Akt) [20-23]. The results of this study showed that the level of p-PKC ε expressed by LDLR knockout cells increased, which may be related to the increase of intracellular DAG and the decrease of p-IRS-1 level. In this study, LDLR knockout cells expressed low levels of p-mTOR and p-AKT, which may be caused by the decrease of PA.
In this study, LDLR gene knockout Lo2 cell line was constructed by CRISPR/Cas9 system, and the role of LDLR in lipid metabolism and glucose metabolism was proved by PCR and WB. The LDLR gene knockout polyclones and monoclonal cells constructed in this study can provide a useful tool for future research.
Acknowledgment
This work was supported by the Natural Science Foundation of Shandong Province (No. ZR2014HM081); the Shandong Province medical and health science and technology development plan (No. 2015WS0378); the National Natural Science Foundation of China (No. 81800444), the Natural Science Foundation of Hubei Province (No. 2018CFB415); Yi Yu He and Li Kun Wang are considered co-corresponding authors in this work.
Conflict of Interests
The authors declared no conflict of interest.
References
- Esmaillzadeh A, Azadbakht L. Food intake patterns may explain the high prevalence of cardiovascular risk factors among Iranian women. J Nutr 2008;138:1469-75.
- Im KH, Choi J, Baek SA, Lee TS. Hyperlipidemic inhibitory effects of Phellinus pini in rats fed with a high fat and cholesterol diet. Mycobiology 2018;46:159-67.
- Li L, Zhang GF, Lee K, Lopez R, Previs SF, Willard B, et al. A Western diet induced NAFLD in LDLR−/− mice is associated with reduced hepatic glutathione synthesis. Free Radic Biol Med 2016;96:13-21.
- Garcia-Jaramillo M, Spooner MH, Löhr CV, Wong CP, Zhang W, Jump DB. Lipidomic and transcriptomic analysis of western diet-induced nonalcoholic steatohepatitis (NASH) in female Ldlr-/-mice. Plos one 2019;14:e0214387.
- Lytle K A, Wong C P, Jump D B. Docosahexaenoic acid blocks progression of western diet-induced nonalcoholic steatohepatitis in obese Ldlr-/- mice. PLoS one 2017;12:e0173376.
- Li L, Bebek G, Previs SF, Smith JD, Sadygov RG, McCullough AJ, et al. Proteome dynamics reveals pro-inflammatory remodeling of plasma proteome in a mouse model of NAFLD. J Proteome Res 2016;15:3388-404.
- Liu J, Ma KL, Zhang Y, Wu Y, Hu ZB, Lv LL, et al. Activation of mTORC1 disrupted LDL receptor pathway: A potential new mechanism for the progression of non-alcoholic fatty liver disease. Int J Biochem Cell Biol 2015;61:8-19.
- Xie L, Gu Y, Wen M, Zhao S, Wang W, Ma Y, et al. Hydrogen sulfide induces Keap1 S-sulfhydration and suppresses diabetes-accelerated atherosclerosis via Nrf2 activation. Diabetes 2016;65:3171-84.
- Bazinet M, Pantea V, Cebotarescu V, Cojuhari L, Jimbei P, Vaillant A. Significant reduction of HBsAg and HDV RNA by the nucleic acid polymer REP 2139 in Caucasian patients with chronic HBV/HDV co-infection. J Hepatol 2015;62:S257-8.
- Yuan T, Zhong Y, Wang Y, Zhang T, Lu R, Zhou M, et al. Generation of hyperlipidemic rabbit models using multiple sgRNAs targeted CRISPR/Cas9 gene editing system. Lipids Health Dis 2019;18:1-9.
- Jia H, Xu J, Orbović V, Zhang Y, Wang N. Editing citrus genome via SaCas9/sgRNA system. Front Plant Sci 2017;8:2135.
- Xie S, Shen B, Zhang C, Huang X, Zhang Y. sgRNAcas9: a software package for designing CRISPR sgRNA and evaluating potential off-target cleavage sites. PloS one 2014;9:e100448.
- KANG N, CHEN H, GAO J. Efficient generation of multiple-gene knockout pig parthenogenetic embryos by using CRISPR/Cas9 system and rapid estimation of knockout efficiency by single blastocyst genotyping. Sci Sin Vitae 2016;46:304-13.
- Van Rooyen DM, Larter CZ, Haigh WG, Yeh MM, Ioannou G, Kuver R, et al. Hepatic free cholesterol accumulates in obese, diabetic mice and causes nonalcoholic steatohepatitis. Gastroenterology 2011;141:1393-403.
- Min HK, Kapoor A, Fuchs M, Mirshahi F, Zhou H, Maher J, et al. Increased hepatic synthesis and dysregulation of cholesterol metabolism is associated with the severity of nonalcoholic fatty liver disease. Cell Metab 2012;15:665-74.
- Musso G, Gambino R, Cassader M. Cholesterol metabolism and the pathogenesis of non-alcoholic steatohepatitis. Prog lipid res 2013;52:175-91.
- Tomita K, Teratani T, Suzuki T, Shimizu M, Sato H, Narimatsu K, et al. Free cholesterol accumulation in hepatic stellate cells: mechanism of liver fibrosis aggravation in nonalcoholic steatohepatitis in mice. Hepatology 2014;59:154-69.
- Liu B, Xu H, Miao J, Zhang A, Kou X, Li W, et al. CRISPR/Cas: a faster and more efficient gene editing system. J Nanosci Nanotechnol 2015;15:1946-59.
- Mehta KD, Radominska-Pandya A, Kapoor GS, Dave B, Atkins BA. Critical role of diacylglycerol-and phospholipid-regulated protein kinase Cε in induction of low-density lipoprotein receptor transcription in response to depletion of cholesterol. Mol Cell Biol 2002;22:3783-93.
- Saparbay J, Tanaka Y, Yano T, Ohdan H. mTOR Inhibitors Enhance Antitumor Activity of Liver Resident NK Cells. Transplantation 2018;102:S282.
- Naito T, Kuma A, Mizushima N. Differential Contribution of Insulin and Amino Acids to the mTORC1-Autophagy Pathway in the Liver and Muscle. J Biol Chem 2013;288:21074-81.
- Wang Q, Holst J. L-type amino acid transport and cancer: targeting the mTORC1 pathway to inhibit neoplasia. Am J Cancer Res 2014;5:1281.
- Rizell M, Lindner P. Inhibition of mTOR suppresses experimental liver tumours. Anticancer Res 2005;25:789-93.