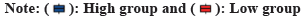- *Corresponding Author:
- Q. Guo
Department of Stomatology, Deyang Stomatological Hospital, Deyang, Sichuan Province 618000, China
E-mail: qingquan-1975@163.com
| This article was originally published in a special issue, “Drug Development in Biomedical and Pharmaceutical Sciences” |
| Indian J Pharm Sci 2023:85(5) Spl Issue “166-172” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
The pivotal role of cuproptosis in the beginning and progression of various cancers is evident. Yet, the influence of cuproptosis-related genes on oral squamous cell carcinoma has not been thoroughly explored. In the present study, we determine distinct oral squamous cell carcinoma classifications and formulate a new prognostic indicator for oral squamous cell carcinoma. Transcriptomic data and patient outcomes were gathered from the cancer genome atlas repository and then 12 cuproptosis-related genes were identified and analyzed. Thereafter, a least absolute shrinkage and selection operator-based risk score signature involving 7 cuproptosis-related genes was constructed. All patients were classified into high and low-risk groups, followed by analyses of the immune landscape and sensitivity to different therapies in different groups. In addition, column line graphs were plotted to predict outcomes based on different clinic pathological features. Based on the identified 12 cuproptosis-related genes, 2 subtypes of cuproptosis were identified. Patients in the high-risk group had advanced clinical stages and worse overall survival. Furthermore, the immune response and function were significantly suppressed in patients of the high-risk group, which may be an important contributor to their poor prognosis. Oral squamous cell carcinoma patients were arranged into high and low-risk subgroups based on risk scores. The results exhibited that the survival probability of patients was markedly higher in the low-risk group than in the high-risk group (p<0.001). Following that, a precise bar line chart was devised to augment the clinical usefulness of the risk score, exhibiting strong predictive capabilities and calibration. Patients with both low and high risk displayed notable immune cell penetration and modifications in immune checkpoints. Further analysis of risk scores revealed that low-risk patients were sensitive to immunotherapy and multiple chemotherapeutic agents. In this study, we identify 2 cuproptosis subgroups and construct a new prognostic model, thus providing new insight into the prognostic assessment of oral squamous cell carcinoma subtypes and guidance for the development of more effective treatment options.
Keywords
Cuproptosis, oral squamous cell carcinoma, immune infiltration, prognosis
Oral cancer is one of the most frequent head and neck cancers, with more than 370 000 new cases and approximately 170 000 deaths worldwide every year[1,2]. In addition, the mortality rate of oral cancer has increased at an annual rate of 0.5 % over the past 10 y despite substantial advances in surgical techniques, radiotherapy and chemotherapy[3]. Oral Squamous Cell Carcinoma (OSCC) is the most common type of oral cancer, which accounts for about 90 % of oral cancer[4]. Moreover, the Overall Survival (OS) rate of OSCC has remained stable at around 50 % in the past three decades with insignificant improvement[5,6]. Accordingly, the construction of an appropriate and effective prognostic model is a necessity for the evaluation, diagnosis and treatment of OSCC patients, which can improve the early diagnosis and survival of OSCC.
Cuproptosis is one of the hot topics in cancer research. A new type of cell death, copper induced cell death (also termed cuproptosis), was discovered by a team of researchers from the Massachusetts Institute of Technology and the Broad Institute of Harvard University, which differs markedly from the identified pyroptosis, apoptosis, ferroptosis and necroptosis. Cuproptosis occurs in human cells and has a strong connection with hindered mitochondrial respiration. This process involves an excessive amount of intracellular copper being transported to mitochondria through ion carriers. Once inside, the copper binds to fatty acrylate components within the mitochondrial Tricarboxylic Acid (TCA) cycle, it disrupts iron-sulfur clusters. These events contribute to the aggregation of fatty acylated proteins, loss of iron-sulfur clusters and protein toxicity stress. Ultimately, this leads to cell death[7]. Previous studies have shown that the content of copper ions is higher in the serum and tumor tissues of patients with various human malignancies, including breast cancer, than in normal serum and tissues[8]. Moreover, copper ion accumulation has also been elucidated to be associated with proliferation, growth, angiogenesis and metastasis in tumors[9,10]. However, little is known about the mechanism of the high expression of Cuproptosis-Related Genes (CRGs) in oral cancer tissues, the carcinogenic role of these genes through the activation of cuproptosis-mediated carcinogenic pathways, and their role in tumor immunotherapy. Therefore, the exploration and development of a prognostic model involving CRGs are helpful in accurately understanding the prognosis of OSCC patients.
In the present research, OSCC sample transcriptomic information was acquired from the Xena repository, while CRGs were gathered from separate investigations. Based on this foundation, a predictive risk model was developed to assess the level of prognostic risk for individuals with OSCC. The results revealed that this model not only accurately predicted the survival of patients, but also effectively evaluated the immune status of OSCC patients and the sensitivity of patients with different risk values to immunologic drugs and chemicals. In summary, the prognostic risk model provides a new perspective on the management of OSCC patients.
Materials and Methods
Datasets and preprocessing of OSCC:
The Ribonucleic Acid sequencing (RNA-seq) data (HTSeq counts) of OSCC were downloaded from the Xena website (https://xena.ucsc.edu/) of the University of California, Santa Cruz. Clinical pathologic data were also collected, including age, gender, pathological grade, tumor stage, tumor-node-metastasis stage, vital status and survival time. CRGs were obtained from the study of Tsvetkov et al.[7]. For survival analysis, raw counts of RNA-seq data were normalized with the Fragments Per Kilobase Million (FPKM) method and Log2-based conversion (log2FPKM).
Construction of a prognostic signature of CRGs with Least Absolute Shrinkage and Selection Operator (LASSO) regression:
LASSO regression was applied to choose predictor variables and avoid overfitting. Following that, comprehensive Cyclooxygenase (COX), regression assessments were performed to determine the concluding elements incorporated within the risk profile. Prognostic predictors of OSCC patients were identified by developing a risk signature as per cuproptosis-related messenger RNA (mRNAs). The formula employed to determine the risk score is as follows:
Risk Score=∑I=1N(Expi×Coei)
Within this equation, N symbolizes the count of cuproptosis-associated mRNAs in the prognostic risk profile, Expi denotes the expression level of each mRNA, and Coei signifies the regression coefficient for each mRNA in the multivariable COX regression evaluation. Based on the median risk score, patients were categorized into high and low-risk cohorts. Subsequently, risk scores were calculated with regression coefficients for identifying the prognostic signature of OS. Patients were divided into high and low-risk classifications using the median risk score. The R package "ggsurvplot" was employed to generate a Kaplan-Meier survival chart for comparing OS between the high and low-risk groups. Building on this aspect, the Receiver Operating Characteristic (ROC) curve was created using the R package "survival ROC" to assess prediction accuracy.
Assessment of cellular infiltration within the Tumor Microenvironment (TME):
The single-sample Genomic Enrichment Analysis (ssGSEA) method was employed to gauge variations in the enrichment of 28 immune cells within an OSCC cluster. Subsequently, the R software package was used for conducting differential abundance evaluations to identify significant alterations across distinct cell infiltration clusters in the TME.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis:
The GO analysis was conducted for assessing the enrichment of GO terms associated with CRGs. All of these genes were mapped directly to the database for the KEGG analysis. The ggplot2 R software package was used to identify the major biological processes and pathways of these genes.
Construction of a line graph:
The R packages "survival" and "Regression Modeling Strategies (RMS)" were harnessed to build a diagram merging risk scores with clinic pathological attributes for estimating and scrutinizing the 1 y, 3 y and 5 y survival outcomes of OSCC patients. Calibration curves were applied to ascertain if the projected survival rates corresponded to the genuine survival rates.
Drug sensitivity analysis:
The R package "pRRophetic" was used to assess drug sensitivity in various risk groups, predicting the Half Maximal Inhibitory Concentration (IC50) for commonly employed chemotherapeutic drugs in OSCC therapy. The Wilcoxon signed rank test was applied to detect disparities between the groups.
Statistical analysis:
The R Statistics Program (version 4.1.0) was utilized for analysis and plotting in our research. As necessary, data analysis was performed using the student t-test or Mann-Whitney U test for comparing two groups, or the Kruskal-Wallis test for comparisons involving three or more groups. OS curves were assessed with the Kaplan-Meier technique, and differences between groups were analyzed using log-rank tests. Forest plots were generated using the "ggforest" function in the "survminer" and "survival" R packages. Somatic mutation information from The Cancer Genome Atlas (TCGA)-OSCC was examined and visualized using the maftools R package. Risk scores and outcomes were appraised through multivariate and univariate Cox proportional hazard regression analyses. Differences with p values below 0.05 were deemed statistically significant.
Results and Discussion
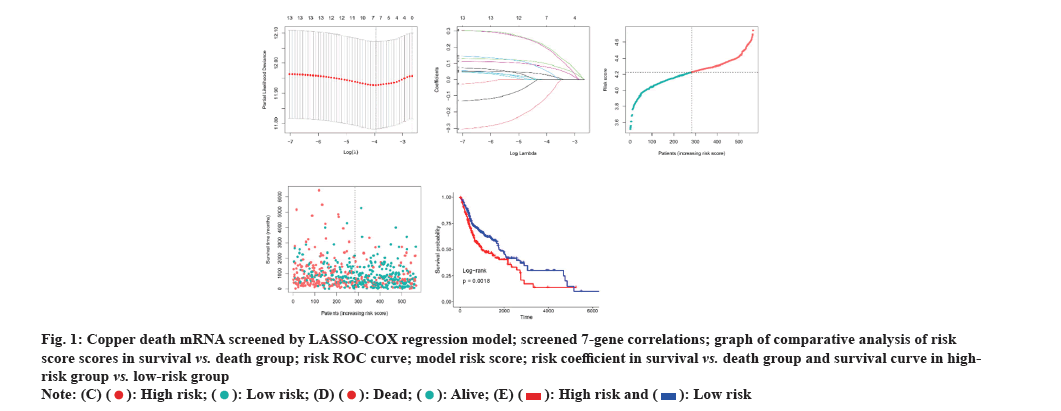
The LASSO-Cox regression approach was employed to establish the risk signature, eventually identifying seven genes based on the optimal Gamma (λ) value (fig. 1A). The risk score was determined by utilizing the coefficient of each gene. Risk score=-0.099×Pyruvate Dehydrogenase E1 Subunit Beta (PDHB) (exp)+0.095×ATP7B (exp)+0.189×Dihydrolipoamide S-Succinyltransferase (DLST) (exp)+0.027×SLC31A1 (exp)+0.198×FDX1 (exp)+0.049×Lipoyltransferase 1 (LIPT1) (exp)+0.077×Glycine Cleavage System Protein H (GCSH) (exp). Utilizing the median risk score, 283 patients were allocated to the low-risk group and 282 to the high-risk group. Additionally, this risk formula was also utilized to calculate the risk score for each patient, and the correlation among the obtained seven genes in each cohort was plotted (fig. 1B). Mortality in OSCC patients increased with increasing risk (fig. 1C). It was observed from the risk profile that survival rates were lower and survival times were shorter in high-risk patients than in low-risk patients (fig. 1D). Similarly, Kaplan-Meier curves revealed that high-risk patients exhibited substantially shorter OS times and lower survival probability than low-risk patients (fig. 1E) (Hazard Ratio (HR): 2.338, 95 % Confidence Interval (CI) 95 %: 1.75-3.11, p<0.001).
Fig. 1: Copper death mRNA screened by LASSO-COX regression model; screened 7-gene correlations; graph of comparative analysis of risk
score scores in survival vs. death group; risk ROC curve; model risk score; risk coefficient in survival vs. death group and survival curve in highrisk
group vs. low-risk group.
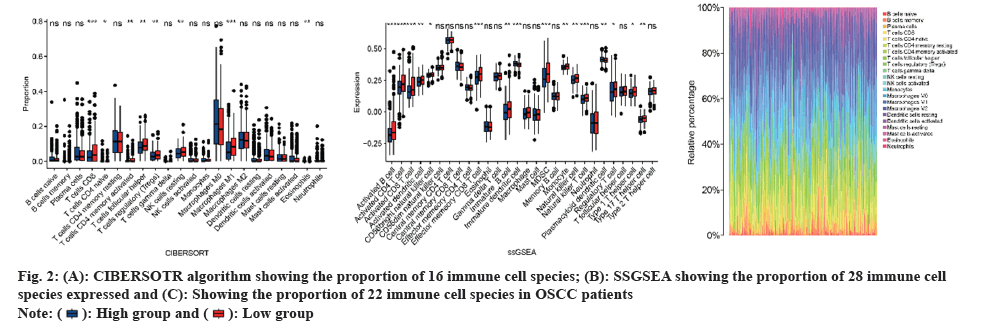
CIBERSORT and ssGSEA were performed to assess differences in immune function. Moreover, the CIBERSORT analysis revealed a greater proportion of Cluster of Differentiation (CD), T cells in the high-risk group compared to the low-risk group (fig. 2A). The ssGSEA findings indicated that the high-risk group had a higher representation of 16 immune cell subtypes, including activated B cells, activated CD4 T cells, activated CD8 T cells, activated dendritic cells, natural killer cells, and natural killer T cells (fig. 2B). Heatmaps were generated to display the overall status of 28 immune cell subtypes across both groups (fig. 2C). The observations showed that the high-risk group had more pronounced immune infiltration than the low-risk group, particularly in terms of CD8 T cell-related infiltration.
GO and KEGG enrichment analyses were carried out to identify the biochemical roles of the seven core genes. The most prominent GO term associated with these core genes was the negative regulation of the immune system. Furthermore, the KEGG enrichment analysis displayed that the core genes were mainly involved in the TCA cycle, metabolic pathways, carbon metabolism and platinum drug resistance. (fig. 3A). Afterward, protein-protein interactions were analyzed to identify the correlation among genes, which showed a strong correlation among seven genes (fig. 3B).
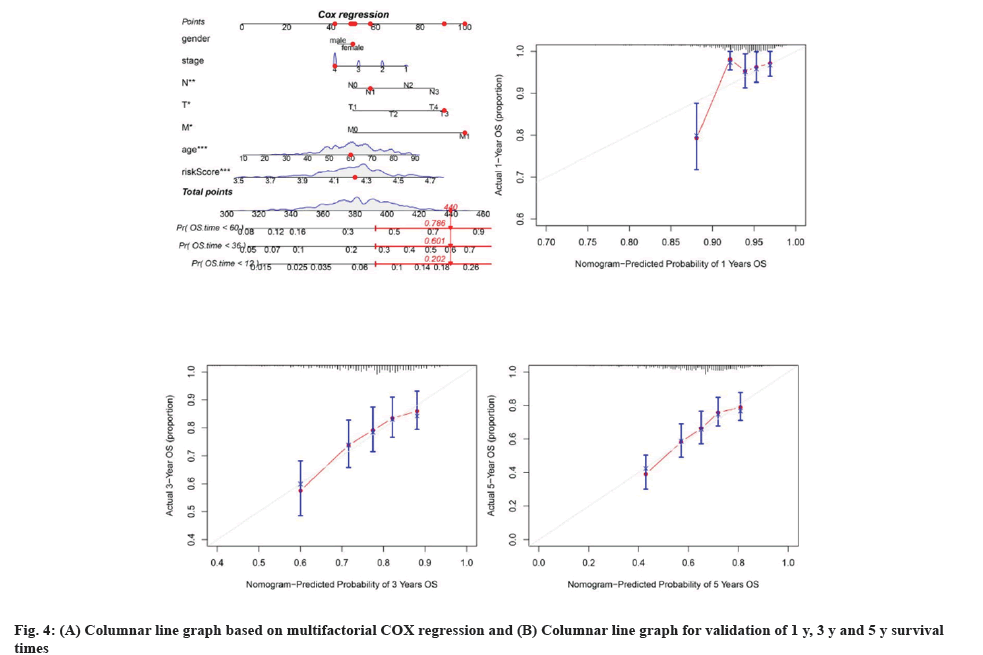
Clinical data and genetic features of patients in TCGA were combined to enhance the predictive model's clinical utility, leading to the creation of a multivariate COX regression model and a corresponding column chart. The column chart showed statistical significance for T stage, N stage, M stage, age, and risk scores (fig. 4A). The R package "regplot" was used to produce a graph depicting the risk score and additional clinical variables for the cuproptosis-related prognostic model in OSCC patients. Calibration curves for the cuproptosis risk score and other clinical factors in OSCC patients were generated using 1000 bootstrap methods to assess bias-corrected estimates between predicted and observed values and were analyzed with the R package "rms". The 1 y, 3 y, and 5 y calibration curves aligned with the estimated values, suggesting that the nomogram created with multivariate COX regression was reliable (fig. 4B-fig. 4D). Analysis of chemotherapy response with a risk model based on seven genetic signatures.
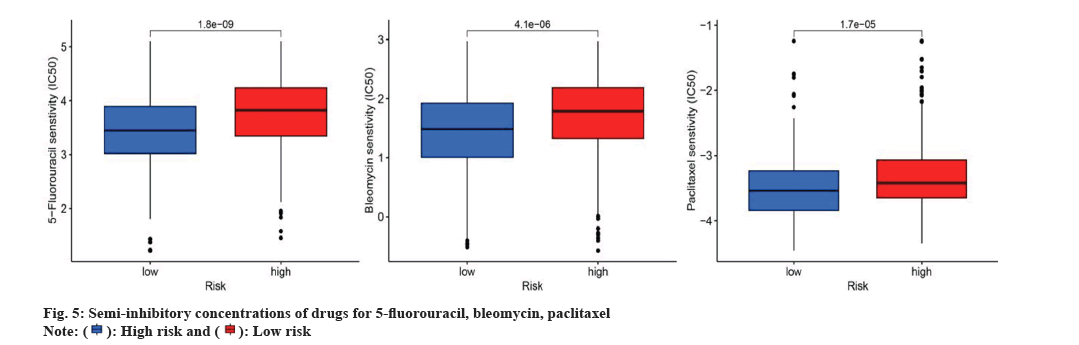
The response to chemotherapy drugs was assessed in both elevated and diminished-risk categories using the R package "pRRophetic". Fig. 5 demonstrates the results of three chemotherapeutic agents commonly utilized for OSCC management. In particular, the IC50 values for 5-Fluorouracil, Bleomycin and Paclitaxel were considerably lower for the reduced-risk category compared to the elevated-risk category, indicating that patients with low-risk OSCC were more susceptible to these treatments as shown in fig. 5A-fig. 5C.
This research examined variations in expression and mutation co-occurrence of CRGs between OSCC and healthy samples, as well as the association between these gene expressions and survival outcomes and prognosis. We noted that CRGs were, to a certain degree, associated with the initiation and progression of OSCC. Therefore, a 7-gene risk prognostic model was established for OSCC based on CRGs with LASSO regression.
Our findings highlighted significant disparities in immune infiltration between high and low-risk groups. The immune infiltration analysis revealed a higher occurrence of immune desert phenotypes in low-risk patients, while increased immune cell infiltration was observed in the high-risk group. Furthermore, the primary infiltrating cells in high-risk patients included activated B cells, activated CD4T, CD8T, natural killer T cells and Myeloid-Derived Suppressor Cells (MDSCs).
Activated B cells, a type of signal transduction mediator, convert Transforming growth factor beta1-Activated kinase 1-Binding protein 2 (TAB2) into multiple biological processes. Of note, a prior study unraveled the potential pathogenic role of TAB2 in OSCC through activated B cells[11]. Furthermore, there is a notable association between the quantity of CD4+ T helper cells (Th1) and the concentration of CD8+ T cells within the TME[12]. Activated T cells express Programmed Death-Ligand 1 (PD-L1), a typical target in immunotherapy, on their surfaces[13]. Different from physiologically differentiated myeloid cells, MDSCs are distinctly characterized by relatively weak phagocytic function, immature phenotype and morphology and anti-inflammatory and immunosuppressive functions. Mounting evidence has reported MDSCs as a basic characteristic of malignancies and a potential target for tumor therapy.
The scoring model generated in our research could quantify the degree of cuproptosis-induced risk in each OSCC patient. By closely studying immune infiltration in groups with varying levels of risk, personalized treatment and prognosis assessment can be provided for OSCC patients. Reportedly, predicting future tumor events is crucial for both patients and clinicians[14]. Programmed cell death is involved in many biological functions, such as the growth, movement and spread of cancer cells, and the TME is being explored as a new way to predict outcomes and possibly target therapies[15]. As a new model of copper-induced cell death, copper poisoning is a promising therapeutic prospect for tumor patients[7]. Wang et al.[16] established a prognostic model of OSCC after immunogenic cell death was classified, providing a new approach to immunotherapy.
In our study, the genes involved in the model shared a relatively strong correlation with infiltrating immune cells and immune checkpoint genes, thus providing a new perspective for clinical immunotherapy. Our study has identified seven hub genes, of which two, ATP7B and SLC31A1, have been found to be linked to Head and Neck Squamous Cell Carcinoma (HNSCC). ATP7B, which encodes the copper transporter, participates in the release of cisplatin by head and neck cancer cells into the extracellular space, thereby enabling cancer cells to escape cisplatin-induced cell death. In the research by Ryumon et al.[17], the application of cisplatin increased the level of ATP7B, a cisplatin effector transporter. Accordingly, targeting ATP7B could be a promising approach in the treatment of HNSCC. Cluster analyses by Warta et al.[18] unraveled that simultaneous upregulation of SLC31A1, ATP Binding Cassette Subfamily C Member 2 (ABCC2), and ATP Binding Cassette Subfamily G Member 2 (ABCG2) was associated with the lower survival rate of HNSCC patients, concordant with our results. Among the remaining 5 genes, LIPT1 has been confirmed as a novel therapeutic target for hepatocellular carcinoma[19]. LIPT1 can modulate lipoic acid metabolism by transferring the lipoyl group of lipoyl adenylate to the GCSH and the 2-oxyacid dehydrogenase E2 subunit[7]. Liu et al.[20] found from recent studies that CRGs are regulated by multiple microRNAs (miRs), among which the most important miRs are hsa-miR-185-5p and hsa-miR-98-5p. Furthermore, hsa-miR-98-5p mediates PDHB, and hsa-miR-576-5p regulates ATP7B and LIPT1 simultaneously. DLST is an E2 component of the Alpha-Ketoglutarate (αKG) dehydrogenase complex that regulates the entrance of glutamine into the TCA cycle to undergo oxidative decarboxylation. This conversion is irreversible and leads to the production of succinyl-CoA, which in turn generates Nicotinamide Adenine Dinucleotide (NAD) for oxidative phosphorylation. Anderson et al.[21] observed from studies on high-risk patients with neuroblastoma that DLST upregulation elevated tumor cell invasion, whereas DLST knockout hindered the occurrence and development of this disease.
Acknowledgments:
We acknowledge TCGA and GEO database for providing their platforms and contributors for uploading their meaningful datasets.
Data availability:
The RNA sequencing (RNA-seq) data (HTSeq counts) of OSCC were downloaded from the Xena website (https://xena.ucsc.edu/) of the University.
Funding:
This work was supported by Deyang city social development key research and development project (No. 2022SZ057).
Authors’ contributions:
Qingquan Guo and Xiaoying Dai designed experiments; Xiaoying Dai, Chunmei Liu and Yimeng Fan carried out experiments; Xiaoying Dai analyzed experimental results and analyzed sequencing data and developed analysis tools. Xiaoying Dai assisted with Illumina sequencing. Qingquan Guo and Xiaoying Dai wrote the manuscript.
Conflict of interests:
The authors declared no conflict of interests.
References
- Li Q, Ouyang X, Chen J, Zhang P, Feng Y. A review on salivary proteomics for oral cancer screening. Curr Issues Mol Biol 2020;37(1):47-56.
[Crossref] [Google Scholar] [PubMed]
- Pare A, Joly A. Oral cancer: Risk factors and management. Press Med 2017;46(3):320-30.
[Crossref] [Google Scholar] [PubMed]
- Panarese I, Aquino G, Ronchi A, Longo F, Montella M, Cozzolino I, et al. Oral and Oropharyngeal squamous cell carcinoma: Prognostic and predictive parameters in the etiopathogenetic route. Expert Rev Anticancer Ther 2019;19(2):105-19.
[Crossref] [Google Scholar] [PubMed]
- Chamoli A, Gosavi AS, Shirwadkar UP, Wangdale KV, Behera SK, Kurrey NK, et al. Overview of oral cavity squamous cell carcinoma: Risk factors, mechanisms, and diagnostics. Oral Oncol 2021;121:105451.
[Crossref] [Google Scholar] [PubMed]
- Janssen EM, Dy SM, Meara AS, Kneuertz PJ, Presley CJ, Bridges JF. Analysis of patient preferences in lung cancer-estimating acceptable tradeoffs between treatment benefit and side effects. Patient Prefer Adherence 2020;14:927-37.
[Crossref] [Google Scholar] [PubMed]
- Vos JL, Elbers JB, Krijgsman O, Traets JJ, Qiao X, van der Leun AM, et al. Neoadjuvant immunotherapy with nivolumab and ipilimumab induces major pathological responses in patients with head and neck squamous cell carcinoma. Nat Commun 2021;12(1):7348.
[Crossref] [Google Scholar] [PubMed]
- Tsvetkov P, Coy S, Petrova B, Dreishpoon M, Verma A, Abdusamad M, et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 2022;375(6586):1254-61.
[Crossref] [Google Scholar] [PubMed]
- Jiang Y, Huo Z, Qi X, Zuo T, Wu Z. Copper-induced tumor cell death mechanisms and antitumor theragnostic applications of copper complexes. Nanomedicine 2022;17(5):303-24.
[Crossref] [Google Scholar] [PubMed]
- Cui L, Gouw AM, LaGory EL, Guo S, Attarwala N, Tang Y, et al. Mitochondrial copper depletion suppresses triple-negative breast cancer in mice. Nat Biotechnol 2021;39(3):357-67.
- da Silva DA, de Luca A, Squitti R, Rongioletti M, Rossi L, Machado CM, et al. Copper in tumors and the use of copper-based compounds in cancer treatment. J Inorg Biochem 2022;226:111634.
[Crossref] [Google Scholar] [PubMed]
- Liu H, Zhang H, Fan H, Tang S, Weng J. TAB2 promotes the biological functions of head and neck squamous cell carcinoma cells via EMT and PI3K pathway. Dis Markers 2022;2022:1217918.
[Crossref] [Google Scholar] [PubMed]
- Fridman WH, Pagès F, Sautès-Fridman C, Galon J. The immune contexture in human tumours: Impact on clinical outcome. Nat Rev Cancer 2012;12(4):298-306.
[Crossref] [Google Scholar] [PubMed]
- Mei Z, Huang J, Qiao B, Lam AK. Immune checkpoint pathways in immunotherapy for head and neck squamous cell carcinoma. Int J Oral Sci 2020;12(1):16.
- Correa AF, Jegede O, Haas NB, Flaherty KT, Pins MR, Messing EM, et al. Predicting renal cancer recurrence: Defining limitations of existing prognostic models with prospective trial-based validation. J Clin Oncol 2019;37(23):2062-71.
[Crossref] [Google Scholar] [PubMed]
- Huang J, Xu Z, Yuan Z, Cheng L, Zhou C, Shen Y. Identification of cuproptosis-related subtypes and characterization of the tumor microenvironment landscape in head and neck squamous cell carcinoma. J Clin Lab Anal 2022;36(9):e24638.
[Crossref] [Google Scholar] [PubMed]
- Wang X, Wu S, Liu F, Ke D, Wang X, Pan D, et al. An immunogenic cell death-related classification predicts prognosis and response to immunotherapy in head and neck squamous cell carcinoma. Front Immunol 2021;12:781466.
[Crossref] [Google Scholar] [PubMed]
- Ryumon S, Okui T, Kunisada Y, Kishimoto K, Shimo T, Hasegawa K, et al. Ammonium tetrathiomolybdate enhances the antitumor effect of cisplatin via the suppression of ATPase copper transporting beta in head and neck squamous cell carcinoma. Oncol Rep 2019;42(6):2611-21.
[Crossref] [Google Scholar] [PubMed]
- Warta R, Theile D, Mogler C, Herpel E, Grabe N, Lahrmann B, et al. Association of drug transporter expression with mortality and progression-free survival in stage IV head and neck squamous cell carcinoma. PLoS One 2014;9(10):e108908.
[Crossref] [Google Scholar] [PubMed]
- Yan C, Niu Y, Ma L, Tian L, Ma J. System analysis based on the cuproptosis-related genes identifies LIPT1 as a novel therapy target for liver hepatocellular carcinoma. J Transl Med 2022;20(1):1-8.
[Crossref] [Google Scholar] [PubMed]
- Liu H. Pan-cancer profiles of the cuproptosis gene set. Am J Cancer Res 2022;12(8):4074-30.
[Google Scholar] [PubMed]
- Anderson NM, Qin X, Finan JM, Lam A, Athoe J, Missiaen R, et al. Metabolic enzyme DLST promotes tumor aggression and reveals a vulnerability to OXPHOS inhibition in high-risk neuroblastoma. Cancer Res 2021;81(17):4417-30.
[Crossref] [Google Scholar] [PubMed]