- *Corresponding Author:
- Lei ChengDepartment of Neurosurgery, Tianjin Huanhu Hospital, Jinnan, Tianjin 300350, China E-mail: petre1000@163.com
| This article was originally published in a special issue, “Current Trends in Pharmaceutical and Biomedical Sciences” |
| Indian J Pharm Sci 2022:84(5) Spl Issue “117-124” |
This is an open access article distributed under the terms of the Creative Commons Attribution-NonCommercial-ShareAlike 3.0 License, which allows others to remix, tweak, and build upon the work non-commercially, as long as the author is credited and the new creations are licensed under the identical terms
Abstract
To explore the function of C-X-C motif chemokine ligand as a prospective early biomarker for recurrent glioblastoma multiforme is the objective of the study. The differentially expressed genes for glioblastoma multiforme patients were systematically explored; the gene ontology and Kyoto encyclopedia of genes and genomes enrichment analysis for potential hub gene was established; the functional network was developed by protein-protein interaction method. The glioblastoma multiforme cell line U251 was established for the in vitro functional experiments; the wound healing assay as well as transwell assay were conducted to measure tumor properties in U251 cells. The expression of C-X-C motif chemokine ligand 6 in recurrent patients of glioblastoma multiforme was significantly higher than in non-recurrent patients (p<0.05), suggesting C-X-C motif chemokine ligand 6 was an independent prognostic indicator for glioblastoma multiforme recurrence. Based on the gene ontology and Kyoto encyclopedia of genes and genomes enrichment analysis as well as a protein-protein interaction network for the C-X-C motif chemokine ligand 6, programmed death ligand-1 and signal transducer and activator of transcription 3 signaling were primarily associated with C-X-C motif chemokine ligand 6. C-X-C motif chemokine ligand 6 over-expression treatments induced markedly enhanced expression levels for programmed death ligand-1 and phospho-signal transducer and activator of transcription 3. At the same time, the regulation of C-X-C motif chemokine ligand 6 for U251 cells tumor properties was programmed death ligand-1 dependent since silencing of programmed death ligand-1 obviously attenuated the functions of C-X-C motif chemokine ligand 6 over-expression. Moreover, it could be implied that the expression levels of programmed death ligand-1 and phospho-signal transducer and activator of transcription 3 were linked to the recurrent status of glioblastoma multiforme patients, as their expression was remarkably accelerated for recurrent glioblastoma multiforme patients. This integrated study provided a potential biomarker and offered beneficial references for future clinical administration as well as evaluation of recurrent glioblastoma multiforme.
Keywords
Glioblastoma multiforme, C-X-C motif chemokine ligand 6, programmed death ligand-1, phospho-signal transducer and activator of transcription 3
Gliomas is the most common form of brain tumor in which Glioblastoma Multiforme (GBM) is the major malignant form, comprising for more than 3 % of cancer-related deaths[1]. The GBM has been defined as a grade IV cancer which is characterized as malignant, mitotically active and predisposed to necrosis by World Health Organization[2]. The worldwide incidence of GBM is heterogeneous because of the variable prevalence of the risk factors[3]. Even with years of study, the knowledge of the genetic basis in GBM is still far from satisfactions. Currently, several reports support that epigenetic change of Epidermal Growth Factor Receptor (EGFR) especially the overexpression of EGFR contribute to a considerable part of GBM cases[4]. At the same time, the well-known tumor suppressor proteins such as p53 as well as Phosphatase and Tensin Homolog deleted on Chromosome 10 (PTEN) are also closely associated with the formation and development of GBM[5,6]. The standard procedure for GBM treatment in clinic is pursued by surgical resection of the tumor with the subsequent combination of radiotherapy and chemotherapy[7]. One striking aspect of GBM is the significant poor prognosis, with approximately a 5 y survival rate of 4 %-5 % and a median survival time of 12.6 mo[8]. One potential reason is the fact that nearly 50 % of all GBM patients are of ages 65 y and above[9]. Moreover, the recurrent nature of GBM has greatly hampered its resilience. Despite decades of research to develop an effective biomarker for detection of recurrent GBM, only few have yielded significant commercial results[10].
Chemokines refers to a superfamily of inducible, secreted, heparin-binding proteins, which play a fundamental role in inflammation as well as immune response[11]. The receptor for chemokines is characterized as a superfamily of seven transmembrane spanning proteins coupled to G-Protein-Coupled-Receptors (GPCRs). The family of receptor is subdivided into four groups according to the pattern of cysteine residues, which are CXC, CC, C, as well as CX3C[12]. Generally speaking, the C stands for cysteine and X for non-cysteine amino acids[13], whereas, the CXCL refers to the C-X-C Motif Chemokine Ligand (CXCL) family. Previously, several studies have suggested the tight connection between CXCL and tumor cell proliferation as well as metastasis. For example, the chemokine axis such as CXCL12/C-X-C Motif Chemokine Receptor 4 (CXCR4) has been shown to be involved in the invasiveness and metastasis of lung cancer[14]. More importantly, the targeted drug for this direction is under development[15]. Meanwhile, serum CXCL8 has been approved as a prospective biomarker for colorectal cancer progression, which is comparable with classical tumor marker Carcinoembryonic Antigen (CEA)[16]. However, the connection between CXCL and the recurrent nature of GBM is still poorly understood.
Based on the fact of complexity and aggressiveness for the molecular mechanisms underline the GBM, the patients call for a dire need of development of effective biomarker for early diagnosis of recurrent GBM. For this purpose, in this study, a comprehensive differential expression analysis was performed on recurrent patients of GBM. Using an innovative bioinformatics method combined with a clinical experimental verification, we comprehensively explored the functions of CXCL6 as a prospective biomarker for early detection of recurrent GBM, which offered a beneficial guidance for the GBM study in future.
Materials and Methods
Data source:
The Ribonucleic Acid (RNA) sequencing data and clinical information for GBM patients were downloaded from The Cancer Genome Atlas (TCGA, https://tcga-data.nci.nih.gov/tcga/), which included 160 patients with complete survival information.
Survival analysis:
Survival analysis was performed using the R survival package and survminer package based on Kaplan-Meier method to estimate the overall survival rate of different groups. At the same time, the log-rank method was developed to test the differences of survival rate among different groups. The multivariate Cox regression model was generated to analyze whether the factor was independent of others for the GBM patients survival prediction.
Functional enrichment analysis:
For the obtained differentially expressed genes, we used the“clusterProfiler” function package in R language for enrichment analysis of Gene Ontology (GO) (including biological process, molecular function and cellular component) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway. When p<0.05, we considered the corresponding entries to be significantly enriched[17].
Protein-Protein Interaction (PPI) networks:
The Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database is a database analyzing and predicting the functional protein-protein connections. We used the STRING (https://string-db.org/, version 11.0) to generate the functional connections and interactions of candidate proteins[18], of which the interaction pairs with a combined score greater than or equal to 0.4 (confidence score≥0.4) were retained. The Cytoscape (https://cytoscape.org/, version 3.7.2) was developed to visualize the PPI network[19]. We processed the Molecular Complex Detection Method (MCODE) plugin in Cytoscape software to identify significant clustering modules, using MCODE score>2 as a criteria.
Cell lines and culture:
The GBM cell line (U251) was purchased from American Type Culture Collection (ATCC) Company and cultured in Dulbecco’s Modified Eagle Medium (DMEM) supplemented with 10 % Fetal Bovine Serum (FBS, Gibco, United States of America (USA)) as well as 1 % Penicillin+Streptomycin (Gibco, USA). The U251 cells were transfected with CXCL6 plasmid, CXCL6 small interfering RNAs (siRNAs) and Programmed Death Ligand-1 (PD-L1) siRNA to achieve CXCL6 Over-Expression (CXCL6-OE), CXCL Low-Expression (CXCL6-LE) and PD-L1 Low-Expression (PD-L1-LE). CXCL6 siRNAs, PD-L1 siRNAs and corresponding Negative Control (NC siRNA) were constructed by Genewiz Corporation Co. (Tianjin, China). The Lipofectamine™ 3000 was used for plasmid transfection. 1 µM NSC74859 (MedChemExpress (MCE), HY-15146) was utilized to treat U251 cells for Phospho-Signal Transducer and Activator of Transcription 3 (P-STAT3) inhibition.
Western blotting:
The CXCL6, STAT3, P-STAT3, PD-L1, as well as Glyceraldehyde-3-Phosphate Dehydrogenase (GAPDH) antibodies were all purchased and obtained from the Jackson Laboratory. The total proteins of U251 cells were collected. Based on the Bicinchoninic Acid (BCA) measurement, 50 μg proteins each was subjected to Sodium Dodecyl-Sulfate Polyacrylamide Gel Electrophoresis (SDS-PAGE) for analysis. The membranes were transferred on semidry transfer apparatus and blocked with 5 % nonfat dry milk powder at 37° for 1.5 h. The membrane was incubated with corresponding primary antibodies (1:1000 dilution) overnight at 4° and horseradish peroxidase labeled secondary antibodies (1:10 000 dilution) at room temperature for 1 h. Then, the membrane was washed extensively and the following was developed with Enhanced Chemiluminescence (ECL) developer and photographed by ultrasensitive multifunctional imager.
Wound healing assay:
The wound healing assay and transwell assay were followed as per the previously established protocol[20]. U251 cells were inoculated on 6‐well plates with 3´105 cells in each well. A straight line with the same angle and consistent thickness was drawn on the bottom of the 6‐well plate with a 10 μl sterile tip. The wound healing areas were measured and analyzed by Image J software.
Transwell assay:
The transwell assay was established for invasion analysis. The basement Matrigel membrane was obtained from BD Biosciences and established to pre-coat the transwell chamber with a filter of 8 μm pores (Corning, New York). The serum‐free medium of 100 µl containing 1×105 cells per well was added into the upper chamber and the medium with 10 % FBS was added into the lower chamber and the following was placed in a 37° constant temperature cell incubator for 48 h. After that, the cells on the upper chamber were gently wiped off with cotton swabs. The invaded cells in five random areas were counted using Image J software.
The cell proliferation assay was conducted using the Cell Counting Kit-8 (CCK-8) purchased from MCE company. At the same time, the Annexin V-Fluorescein Isothiocyanate (FITC)/Propidium Iodide (PI) double staining of flow cytometry was performed to examine the apoptotic cells.
Statistical analysis:
The Kaplan-Meier method was performed to estimate the overall survival rate of different groups and log-rank was used to test the difference of survival rate between different groups. At the same time, the t-test was used to compare the expression of target genes in different groups, taking p<0.05 as a significant threshold. The statistical analysis in this study was conducted using R software, version 3.5.2.
Results and Discussion
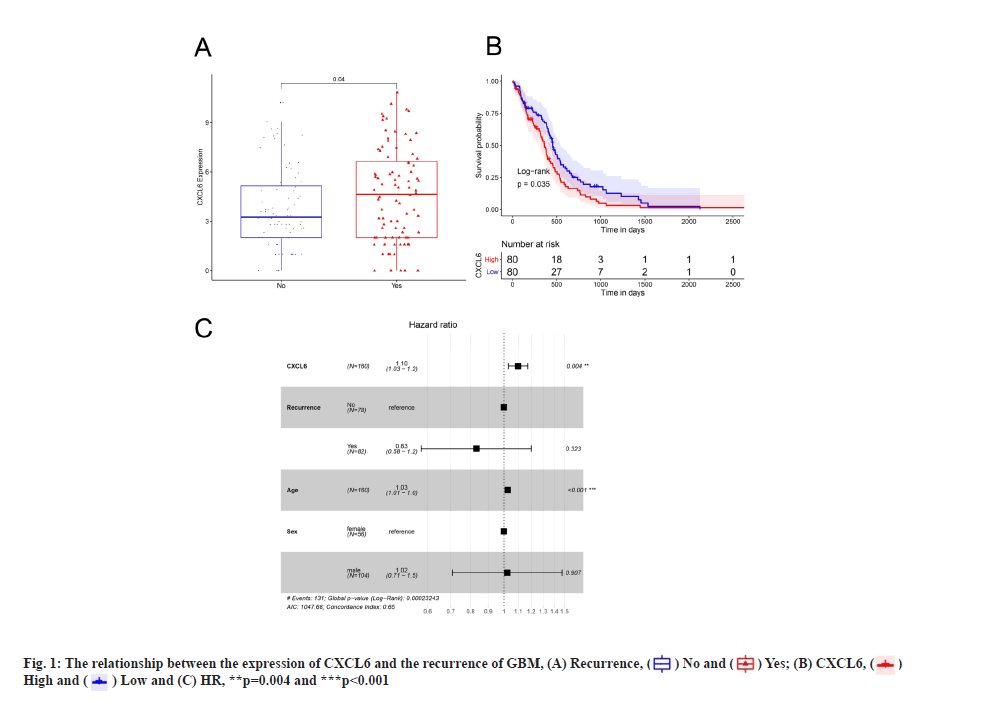
The comparison of CXCL6 expression in recurrent and non-recurrent patients of GBM was shown in fig. 1A. Firstly, we found that the expression of CXCL6 in recurrent patients of GBM was significantly higher than in non-recurrent patients (fig. 1A, p<0.05). Subsequently, the samples were divided into high and low-expression groups, according to the median of CXCL6 expression. Survival analysis demonstrated that the overall survival rate of high-expression group was markedly lower than that of low-expression group of CXCL6. Kaplan-Meier survival curves of GBM patients in CXCL6 high-expression group and low-expression group was shown here. The horizontal axis represents time, the vertical axis represents survival rate and the colors indicate different groups which was shown in fig. 1B. To further determine whether the expression of CXCL6 is an independent prognostic indicator, the factors of age, sex as well as relapse status were recruited to generate a multivariate Cox regression analysis. As shown in fig. 1C, the expression level of CXCL6 was still obviously correlated with the overall survival, whereas the high expression of CXCL6 was associated with a higher risk of mortality and a poor prognostic factor for GBM patients (Hazard Ratio (HR)=1.1, 95 % Confidence Interval (CI): 1.03-1.2, p=0.004). These data suggested that CXCL6 was a key factor for recurrent status of GBM patients.
PD-L1 and STAT signaling were potentially linked to CXCL6 functions as shown in fig. 2. Based on the GO and KEGG enrichment analysis for high and low-expression groups of CXCL6, the related molecular pathways and biological processes were deeply investigated. The top GO term enrichment results with the largest number of genes were shown in fig. 2A. The enrichment results of the KEGG pathways with the largest number of genes were shown in fig. 2B. In the fig. 2A and fig. 2B, the horizontal axis represents the number of enriched genes and the vertical axis represents the name of each GO term respectively. As shown in fig. 2A and fig. 2B, the processes of immunoglobulin complex and STAT3 signaling cascades were significantly enriched in CXCL6 high expression group, which could be the potential molecular mechanisms, underlying in the recurrent patients of GBM. The analysis of PPI network for recurrent GBM was shown in fig. 2C. Different colors represent different clusters of genes for recurrent GBM. At the same time, a STRING database was utilized to construct a PPI network for the genes and the gene interactions with confidence score≥0.4 were selected for visualization with Cytoscape software. The MCODE plugins were performed to identify significant clustering modules, where 6 of them are shown in fig. 2C. The MCODE1 was the most striking one, including Galanin and GMAP Prepropeptide (GAL), C-C Motif Chemokine Ligand 23 (CCL23), Formyl Peptide Receptor 2 (FPR2), Complement Component 5a Receptor 1 (C5AR1), PD-L1 and STAT3 as well as CXCL6. The CXCL6 was significant one with the largest node degree (i.e. 6). Based on these outcomes, PD-L1 and STAT3 signaling were attractive, serving for the primary working mechanism behind CXCL6.
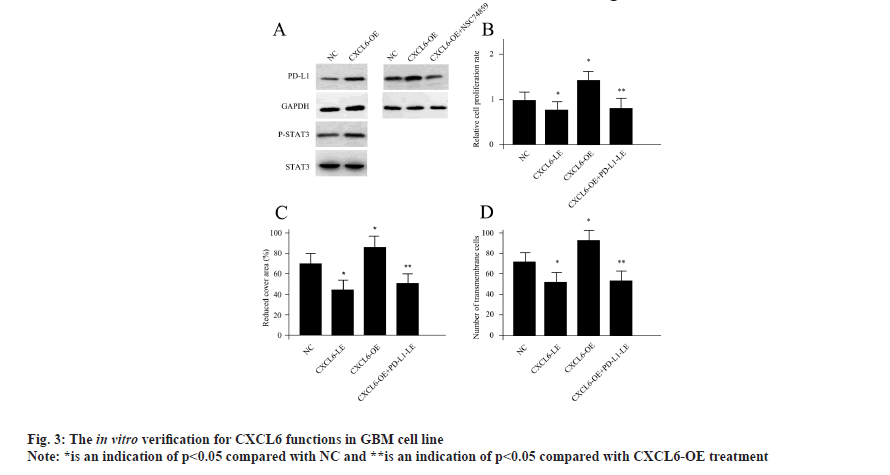
CXCL6 modulated tumor properties of GBM cells via STAT3 dependent PD-L1 signaling was shown in fig. 3. Since the PD-L1 and STAT3 signaling axis were suggested to be closely associated with CXCL6, next, we sought to testify whether there is a direct connection between them. Driven by this question, we obtained CXCL6-OE through CXCL6 plasmid transfection in U251 cells. Compared with control cells, CXCL6-OE cells demonstrated markedly enhanced expression levels for PD-L1, suggesting CXCL6 was a positive regulator for PD-L1. At the same time, the activity of P-STAT3 was also found to be elevated upon CXCL6-OE treatment. Previously, NSC74859 was shown to be a specific inhibitor for P-STAT3[20]. NSC74859 treatment completely abolished the effects of CXCL6-OE for PD-L1 (fig. 3A), indicating CXCL6 modulated PD-L1 via P-STAT3 dependent signaling cascade. As for the cellular functional experiments, CXCL6-OE treatment significantly increased cell proliferation, migration as well as invasion ability, while, CXCL6 siRNAs transfected cells (CXCL6-LE) displayed the opposite pattern. Moreover, the regulation of CXCL6 for U251 cells could be PD-L1 dependent since PD-L1 silencing using siRNA transfection obviously attenuated the functions of CXCL6-OE (fig. 3B-fig. 3D). Based on these results, we hypothesized that CXCL-6 induced tumor properties for GBM cells, which was achieved via PD-L1 dependent of P-STAT3 activity.
The Western blotting of U251 cells upon different treatments was shown in fig. 3A, the cell proliferation assay conducted by CCK-8 assay was shown in fig. 3B, the U251 cells migration ability was measured by wound healing assay as shown in fig. 3C and CXCL6-OE, CXCL6-LE, PD-L1-LE, P-STAT3 and NC was shown in fig. 3D.
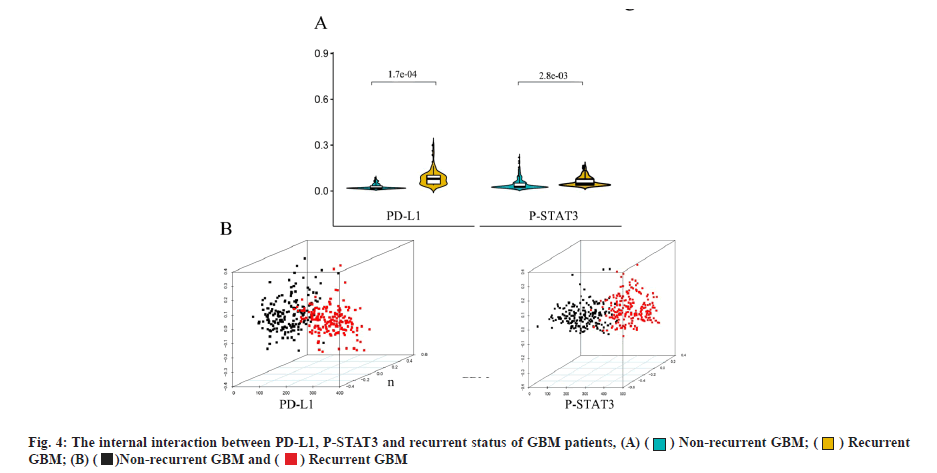
The expressions of PD-L1 and P-STAT3 were closely associated with the recurrent status of GBM patients was shown in fig. 4. Based on the fact that PD-L1 was a direct target of CXCL6, we re-investigated the GBM patients according to PD-L1 and P-STAT3 expression. It could be implied that the expression levels of PD-L1 and P-STAT3 were associated with the recurrent status of GBM patients, as their expression was obviously accelerated for recurrent GBM patients. The violin plot of PD-L1 and P-STAT3 with significant difference between recurrent and non-recurrent GBM patients was shown in fig. 4A and the p-value is calculated by Wilcoxon method. Furthermore, the non-recurrent and recurrent GBM patients could be successfully distinguished by either PD-L1 or P-STAT3 expressions based on Principal Component Analysis (PCA). The PCA analysis for GBM patients based on the expression differences of PD-L1 or P-STAT3 was shown in fig. 4B. These outcomes suggested that CXCL6-P-STAT3-PD-L1 signaling axis was a primary working mechanism underlined GBM recurrence.
The high mortality of GBM has brought many obstacles to clinical treatment all over the world, based on the facts of a high average age of onset, tumor location, as well as high frequency of recurrent. The recurrent tumors are often indistinguishable by Magnetic Resonance Imaging (MRI), when present with similar neurological symptoms. A combination of diffusion and perfusion-weighted MRI might be an alternative method, which could improve diagnostic accuracy by exploiting differences in tissue cellularity and microvasculature respectively. However, this initiates a complex procedure and puts high demands on the clinicians. To address the issues of high rate of misdiagnosis rate and poor understandings of the tumor pathophysiology behind the recurrent GBM, we explored the potential biomarker as well as associated signaling pathways in this study.
The chemokine superfamily consists of a relatively large number of chemokines and chemokine receptors. Currently, more than 50 chemokines and the corresponding receptors have been identified[21,22]. Within these members, the axis of CXCR4/CXCR7-CXCL12 has been deeply studied in multiple cancer types within the last decades. For instance, the metastasis process has been stimulated by irregularity of CXCR4-CXCL12 in prostate cancer, involving Mitogen-Activated Protein Kinase (MAPK)/Extracellular-Regulated Kinase (ERK) signaling pathway[23]. At the same time, the progressive breast tumor growth was suggested to be associated with the increased expression level of CXCR7[24]. Hattermann et al. proposed that the microvascular hyperplasia of the tumor triggered over-expression of CXCR7 in glioblastoma[25]. So that, the GBM patients with poor overall survival rate displayed upregulation of CXCR7[26]. Moreover, a combined administration of X7Ab (a chimeric antibody of CXCR7) and temozolomide (first-line treatment for glioblastoma) could decrease the tumor sizes and prolonged survival in the animal model[27]. In addition to CXCR7, a novel Stromal Derived Factor 1 alpha (SDF-1α/CXCL-12) inhibitor has been shown to reverse the recruitment of macrophages and potentiate the antitumor effect of anti-Vascular Endothelial Growth Factor (VEGF) therapy in the process of GBM[28]. In fact, as a standard chemotherapeutic drug used to treat GBM patients, the temozolomide was demonstrated to function through the expression and secretion of CXCL2, CXCL3 as well as CXCL8 in glioma cells[29]. All the previous investigations provided the evidences for the connection between chemokine superfamily and GBM formation. In this integrated study, we did observed the function of CXCL6 as a biomarker especially for GBM recurrent. However, CXCL6 did not demonstrate a significant difference like other family members in the initiation stage of GBM compared with control health specimen. This may be either due to the fact that CXCL6 played a critical function solely in recurrent stage or there exist a functional switch between different members of this superfamily.
Here, in this study, our data suggested that CXCL6 modulated PD-L1 activity STAT signaling is a causal. Immunotherapies that target Programmed Cell Death 1 (PD-1)/PD-L1 axis have suggested the unprecedented success in a wide variety of human cancers. PD-1 pathway suppresses effector T cells in immune response, thereby causing immune suppression, which has become a hot-spot in cancer research. Previously, in lung cancer cell, STAT3 pathway was shown to regulate expression level of PD-L1 and subsequent Epithelial-to-Mesenchymal Transition (EMT)[30]. Our data also supported the opinion and we also proposed that CXCL6 was a specific upstream regulatory factor for the signaling axis. Based on our search, there were few reports focusing on the connection between CXCL6 and STAT/PD-L1 signaling node for cancer research. In a study of Diabetic Nephropathy (DN), high glucose significantly increased the proliferation of rat renal fibroblasts and Janus Kinase (JAK)-STAT signaling pathway[31]. While, knockdown of CXCL6 ameliorated the pro-proliferation effect of high glucose and decreased the expression of fibrosis-related cytokines, suggesting that CXCL6 promoted fibrosis-related factors to accelerate the development of DN renal interstitial fibrosis by activating JAK/STAT3 signaling pathway. It is worth noting that the primary molecular mechanisms between CXCL6 and STAT/PD-L1 signaling node have not been fully explored here, which call for a great point for the future study. Here, we in-depth re-investigated the GBM patients according to PD-L1 and P-STAT3 expression. It could be demonstrated that the expression levels of PD-L1 and P-STAT3 were closely associated with the recurrent status of GBM patients (fig. 4A). In other words, recurrent status of GBM patients could be distinguished based on CXCL6/STAT/PD-L1 signaling as a casual. The CXCL6 could be a primary biomarker for GBM recurrence, with a high diagnostic value in future clinical study.
Even with years of hard work for the breakthroughs in the recurrent GBM study, the GBM remains at large and the need to discover highly accurate early biomarkers for recurrent GBM. To this end, we systematically compared the recurrent and non-recurrent patients of GBM, which elucidated CXCL6 as a potential biomarker. With enrichment analysis, PPI network construction as well as in vitro verification, the functions of CXCL6 had been deeply explored. All the work here provided novel insights in the administration and evaluating the biomarker investigation for future recurrent GBM research.
Author’s contributions:
Lei Cheng and Xudong Li contributed equally to this work. Lei Cheng collected data, Xudong Li contributed to the conception; Shibo Wang performed the experiments, analyzed and interpreted the data. Yimu Fan analyzed and interpreted the data as well as supervised the work. All authors read and approved the final manuscript.
Funding:
This work was supported by the Science and Technology Development Fund of Tianjin Education Commission for Higher Education (2018KJ079).
Conflict of interests:
The authors declared no conflict of interest.
References
- Batash R, Asna N, Schaffer P, Francis N, Schaffer M. Glioblastoma multiforme, diagnosis and treatment; recent literature review. Curr Med Chem 2017;24(27):3002-9.
[Crossref] [Google scholar] [PubMed]
- Carlsson SK, Brothers SP, Wahlestedt C. Emerging treatment strategies for glioblastoma multiforme. EMBO Mol Med 2014;6(11):1359-70.
[Crossref] [Google scholar] [PubMed]
- Alifieris C, Trafalis DT. Glioblastoma multiforme: Pathogenesis and treatment. Pharmacol Ther 2015;152:63-82.
[Crossref] [Google scholar] [PubMed]
- Hatanpaa KJ, Burma S, Zhao D, Habib AA. Epidermal growth factor receptor in glioma: Signal transduction, neuropathology, imaging, and radioresistance. Neoplasia 2010;12(9):675-84.
[Crossref] [Google scholar] [PubMed]
- England B, Huang T, Karsy M. Current understanding of the role and targeting of tumor suppressor p53 in glioblastoma multiforme. Tumor Biol 2013;34(4):2063-74.
[Crossref] [Google scholar] [PubMed]
- Kim S, Chung JK, Im SH, Jeong JM, Lee DS, Kim DG, et al. 11C-methionine PET as a prognostic marker in patients with glioma: Comparison with 18F-FDG PET. Eur J Nucl Med Mol Imaging 2005;32(1):52-9.
[Crossref] [Google scholar] [PubMed]
- Stoyanov GS, Dzhenkov D, Ghenev P, Iliev B, Enchev Y, Tonchev AB. Cell biology of glioblastoma multiforme: From basic science to diagnosis and treatment. Med Oncol 2018;35(3):1-10.
[Crossref] [Google scholar] [PubMed]
- Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol 2007;114(2):97-109.
[Crossref] [Google scholar] [PubMed]
- Ryskalin L, Busceti CL, Biagioni F, Limanaqi F, Familiari P, Frati A, et al. Prion protein in glioblastoma multiforme. Int J Mol Sci 2019;20(20):5107.
[Crossref] [Google scholar] [PubMed]
- Montemurro N. Glioblastoma multiforme and genetic mutations: The issue is not over yet. An overview of the current literature. J Neurol Surg A Cent Eur Neurosurg 2020;81(1):64-70.
[Crossref] [Google scholar] [PubMed]
- Keeley EC, Mehrad B, Strieter RM. Chemokines as mediators of tumor angiogenesis and neovascularization. Exp Cell Res 2011;317(5):685-90.
[Crossref] [Google scholar] [PubMed]
- Cheng ZH, Shi YX, Yuan M, Xiong D, Zheng JH, Zhang ZY. Chemokines and their receptors in lung cancer progression and metastasis. J Zhejiang Univ Sci B 2016;17(5):342-51.
[Crossref] [Google scholar] [PubMed]
- Zlotnik A, Yoshie O. Chemokines: A new classification system and their role in immunity. Immunity 2000;12(2):121-7.
[Crossref] [Google scholar] [PubMed]
- Saintigny P, Burger JA. Recent advances in non-small cell lung cancer biology and clinical management. Discov Med 2012;13(71):287-97.
[Google scholar] [PubMed]
- Peled A, Wald O, Burger J. Development of novel CXCR4-based therapeutics. Expert Opin Investig Drugs 2012;21(3):341-53.
[Crossref] [Google scholar] [PubMed]
- Pączek S, Łukaszewicz-Zając M, Gryko M, Mroczko P, Kulczyńska-Przybik A, Mroczko B. CXCL-8 in preoperative colorectal cancer patients: Significance for diagnosis and cancer progression. Int J Mol Sci 2020;21(6):2040.
[Crossref] [Google scholar] [PubMed]
- Ritchie ME, Phipson B, Wu DI, Hu Y, Law CW, Shi W, et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 2015;43(7):e47.
[Crossref] [Google scholar] [PubMed]
- Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, et al. STRING v11: Protein-protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 2019;47(1):607-13.
[Crossref] [Google scholar] [PubMed]
- Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, et al. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res 2003;13(11):2498-504.
[Crossref] [Google scholar] [PubMed]
- Zheng S, Shen T, Liu Q, Liu T, Tuerxun A, Zhang Q, et al. CXCL6 fuels the growth and metastases of esophageal squamous cell carcinoma cells both in vitro and in vivo through upregulation of PD‐L1 via activation of STAT3 pathway. J Cell Physiol 2021;236(7):5373-86.
[Crossref] [Google scholar] [PubMed]
- Ogawa E, Sato Y, Minagawa A, Okuyama R. Pathogenesis of psoriasis and development of treatment. J Dermatol 2018;45(3):264-72.
[Crossref] [Google scholar] [PubMed]
- Betakova T, Kostrabova A, Lachova V, Turianova L. Cytokines induced during influenza virus infection. Curr Pharm Des 2017;23(18):2616-22.
[Crossref] [Google scholar] [PubMed]
- Sun YX, Wang J, Shelburne CE, Lopatin DE, Chinnaiyan AM, Rubin MA, et al. Expression of CXCR4 and CXCL12 (SDF‐1) in human prostate cancers (PCa) in vivo. J Cell Biochem 2003;89(3):462-73.
[Crossref] [Google scholar] [PubMed]
- Hernandez L, Magalhaes MA, Coniglio SJ, Condeelis JS, Segall JE. Opposing roles of CXCR4 and CXCR7 in breast cancer metastasis. Breast Cancer Res 2011;13(6):1-7.
[Crossref] [Google scholar] [PubMed]
- Hattermann K, Held-Feindt J, Lucius R, Müerköster SS, Penfold ME, Schall TJ, et al. The chemokine receptor CXCR7 is highly expressed in human glioma cells and mediates antiapoptotic effects. Cancer Res 2010;70(8):3299-308.
[Crossref] [Google scholar] [PubMed]
- Deng L, Zheng W, Dong X, Liu J, Zhu C, Lu D, et al. Chemokine receptor CXCR7 is an independent prognostic biomarker in glioblastoma. Cancer Biomark 2017;20(1):1-6.
[Crossref] [Google scholar] [PubMed]
- Salazar N, Carlson JC, Huang K, Zheng Y, Oderup C, Gross J, et al. A chimeric antibody against ACKR3/CXCR7 in combination with TMZ activates immune responses and extends survival in mouse GBM models. Mol Ther 2018;26(5):1354-65.
[Crossref] [Google scholar] [PubMed]
- Deng L, Stafford JH, Liu SC, Chernikova SB, Merchant M, Recht L, et al. SDF-1 blockade enhances anti-VEGF therapy of glioblastoma and can be monitored by MRI. Neoplasia 2017;19(1):1-7.
[Crossref] [Google scholar] [PubMed]
- Stoyanov GS, Dzhenkov DL. On the concepts and history of glioblastoma multiforme-morphology, genetics and epigenetics. Folia Med 2018;60(1):48-66.
[Crossref] [Google scholar] [PubMed]
- Yang J, Yan J, Liu B. Targeting EGFRvIII for glioblastoma multiforme. Cancer Lett 2017;403:224-30.
[Crossref] [Google scholar] [PubMed]
- Liu F, Hon GC, Villa GR, Turner KM, Ikegami S, Yang H, et al. EGFR mutation promotes glioblastoma through epigenome and transcription factor network remodeling. Mol Cell 2015;60(2):307-18.
[Crossref] [Google scholar] [PubMed]
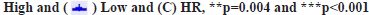 .
.
 .
.